Massively parallel sequencing and rare disease (original) (raw)
Abstract
Massively parallel sequencing has enabled the rapid, systematic identification of variants on a large scale. This has, in turn, accelerated the pace of gene discovery and disease diagnosis on a molecular level and has the potential to revolutionize methods particularly for the analysis of Mendelian disease. Using massively parallel sequencing has enabled investigators to interrogate variants both in the context of linkage intervals and also on a genome-wide scale, in the absence of linkage information entirely. The primary challenge now is to distinguish between background polymorphisms and pathogenic mutations. Recently developed strategies for rare monogenic disorders have met with some early success. These strategies include filtering for potential causal variants based on frequency and function, and also ranking variants based on conservation scores and predicted deleteriousness to protein structure. Here, we review the recent literature in the use of high-throughput sequence data and its analysis in the discovery of causal mutations for rare disorders.
INTRODUCTION
Mendelian disorders have proved to be a rich resource for the study of genes and gene function over the years (1), and to date, close to 3000 Mendelian phenotypes have been solved (2). Conventional methods for disease gene discovery (3) include those based on linkage analysis (4) as well as homozygosity mapping (5), in which markers are used to identify recombination events in pedigrees to narrow the candidate genomic regions segregating with affected status. A typical follow-up is then to re-sequence exons within the candidate region(s) to find protein-altering variants such as missense or nonsense single-base substitutions, or small insertions or deletions (indels).
Many rare disorders are highly amenable to linkage analyses, particularly those that are ‘simple’—monogenic, highly penetrant and inherited in a clear Mendelian fashion. However, some disorders present a challenge for these methods. First, many are extremely rare with only a few affected individuals and families per disorder, which result in underpowered analyses and/or large regions under the linkage peak(s). Second, these disorders are rare because the causal mutations are of large effect and under strong negative selection. As such, these mutations are not often transmitted through many generations and are, in fact, likely to be de novo events, which are not ascertained at all by linkage analyses. To circumvent these challenges, it is often necessary to identify these mutations directly through sequencing. Until recently, however, this has been highly resource-intensive and generally infeasible to do on a large scale or in a genome-wide manner.
Advances in sequencing technology have made it increasingly practical to generate large amounts of sequence data cost-effectively. Known as massively parallel or ‘next-generation' sequencing (6), these technologies have enabled investigators to obtain variant information down to single-base resolution in a rapid, high-throughput fashion on the scale of the whole human genome. Enrichment by either solid-phase or in-solution targeted capture (7) can rapidly isolate candidate regions of interest ranging from hundreds of kilobases in size or capture the entire protein-coding sequence of an individual (the ‘exome', over 30 Mb) for sequencing. However, the major challenge remaining is the interpretation of these sequence data—how can background polymorphisms be distinguished from potentially disease-causing mutations?
A number of recently published studies (Table 1) have successfully employed massively parallel sequencing to Mendelian disease analysis, both within and without the paradigm of linkage. In some, this has simply replaced Sanger re-sequencing of linkage intervals; in other studies, disease genes have been found based on the direct observation of causal mutations, in the absence of linkage information entirely. Here, we review some of the strategies investigators have used to sift through variants to determine the causal mutations for autosomal dominant (8–13), autosomal recessive (14–20) and X-linked recessive disorders (21), as well as to identify the molecular basis (22) or provide evidence for the clinical diagnosis (23) for other diseases.
Table 1.
Published studies utilizing massively parallel sequencing in the analysis of rare disorders
| Disease | Model | Sequencing scope | Reference |
|---|---|---|---|
| Novel disease gene discovery | |||
| Miller syndrome | Autosomal recessive | Whole-genome, one family (two affected siblings and both parents) | 18 |
| Metachondromatosis | Autosomal dominant | Whole-genome, single proband | 12 |
| Miller syndrome | Autosomal recessive | Exome, four cases (two siblings, two other unrelated) | 16 |
| Schinzel–Giedion syndrome | Autosomal dominant | Exome, four unrelated cases | 11 |
| Fowler syndrome | Autosomal recessive | Exome, two unrelated cases | 19 |
| Kabuki syndrome | Autosomal dominant | Exome, 10 unrelated cases | 13 |
| Joubert syndrome 2 | Autosomal recessive | Exomes of 2 individuals (mother and affected daughter) | 15 |
| Non-syndromic hearing loss (DFNB82) | Autosomal recessive | exome, single case | 20 |
| TARP syndrome | X-linked dominant | X chromosome exons, two unrelated carriers | 21 |
| Familial exudative vitreoretinopathy | Autosomal dominant | Linkage interval + 2 candidate genes, single proband | 10 |
| Clericuzio-type poikiloderma with neutropenia | Autosomal recessive | Linkage interval, single case | 14 |
| Sensory/motor neuropathy with ataxia | Autosomal dominant | Linkage interval, proband and both parents | 8 |
| Non-syndromic deafness (DFNB79) | Autosomal recessive | Linkage interval, single case | 17 |
| Clinical diagnosis | |||
| Congenital chloride-losing diarrhea | Autosomal recessive | Exome, single patient with suspected Bartter syndrome | 23 |
| Primary ciliary dyskinesia | Autosomal recessive | Exome, two siblings | |
| Molecular diagnosis | |||
| Charcot–Marie–Tooth disease | Autosomal recessive | Whole-genome, single proband | 22 |
MOLECULAR DIAGNOSIS OF MUTATIONS IN KNOWN GENES
The most straightforward method to identify causal mutations in an individual is by comparison with known mutations and disease-associated genes, like those curated in databases such as Online Mendelian Inheritance in Man (OMIM) (2) and the Human Gene Mutation Database (HGMD) (24). This is of most utility in well-studied diseases and can be used to assist in molecular diagnoses, i.e. finding novel and known mutations in previously identified disease genes. For example, in whole-genome sequencing of an individual with Chacot–Marie–Tooth disease (CMT) (22), analysis of coding variants identified a nonsense mutation in SH3TC2 that was already implicated in CMT, and consistent with the recessive mode of inheritance, a novel missense mutation in the same gene was also found.
In a similar vein, investigators have begun to consider the use of massively parallel sequencing as diagnostic assays for genetic disorders. Re-sequencing of candidate genes for neurofibromatosis type 1 (25), ataxia (26), mitochondrial disorders (27) and ocular birth defects (28), as well as for HLA typing (29), has shown that targeted capture coupled with high-throughput sequencing is increasingly feasible for this task, although further improvements in accuracy and cost may be necessary before its widespread adoption in clinical laboratories.
CLINICAL DIAGNOSIS BASED ON SEQUENCE DATA
Observing known mutations or novel mutations in disease genes may also assist in making a clinical diagnosis for patients. For example, annotation of homozygous variants identified in exome sequencing of a patient from a consanguineous union was used to make a genetic diagnosis of congenital chloride-losing diarrhea (CLD) (23). Initially, the patient was suspected of having Bartter syndrome, but no known disease loci were within homozygous segments. Instead, a homozygous single-base substitution was identified in the gene SLC26A3, in which mutations had previously been found to cause CLD. Clinical follow-up based on this molecular observation confirmed CLD as the primary diagnosis.
In another study, exome sequencing of four individuals affected with Miller syndrome (16) revealed that a subset of cases (siblings) alone had novel variants that were predicted to be damaging in a single gene, DNAH5, which had previously been implicated in primary ciliary dyskinesia. This led to the realization that a cystic fibrosis-like phenotype unique to these siblings was, in fact, not related to Miller syndrome, but instead a superimposition of another disease phenotype caused by mutations in a separate gene.
These studies were the first diagnoses of monogenic disorders based on massively parallel sequencing and suggest its potential to assist in the evaluation and diagnosis of patients, particularly when the diagnosis is uncertain.
NOVEL DISEASE GENE DISCOVERY
Disorders without known mutations or disease-associated genes require different filtering strategies to isolate pathogenic mutations. In general, the following assumptions are made about causal mutations underlying ‘simple', monogenic, Mendelian disease: (i) a single mutation is sufficient to cause the disease, which would (ii) be rare, and probably private to affected individuals, and (iii) because they are of large effect, they are most likely coding and (iv) highly penetrant. As such, investigators look first for variants that change protein sequence, i.e. missense and nonsense substitutions, coding indels as well as splice acceptor and donor site changes, and second for variants that are very rare or novel. Where necessary, a further assumption is often made that the disease is genetically homogenous, i.e. unrelated affected individuals have mutations in the same gene, at least for the individuals chosen for the study.
The first report of the potential of using massively parallel sequencing to identify mutations by this strategy was for Freeman–Sheldon syndrome (FSS) (9). In this study, the exomes of four unrelated affected individuals were sequenced, and for each individual, genes that had at least one private protein-altering variant were shortlisted, consistent with a dominant disease model. After intersecting the genes from all four individuals, it was found that only one gene was common among all—MYH3, which had previously been shown to be causal for FSS. Although this was not a novel identification of the disease-associated gene, it was nonetheless a proof-of-concept experiment that showed how massively parallel sequencing could be applied on a genome-wide scale to find causal mutations for monogenic diseases even without linkage or pedigree information, nor any information related to disease mechanism.
The same strategy was employed successfully in at least two other studies for autosomal dominant disorders—one for Schinzel–Giedion syndrome (11) and another for Kabuki syndrome (13). Notably, consistent with the sporadic nature of these syndromes, the majority of the causal mutations identified in these studies were found to be de novo, which highlights the advantage of using exome sequencing over linkage studies for these cases.
To extend the strategy to recessive disease, shortlisted genes were required to have at least two private protein-altering variants instead of just one. This accounts for two situations: under a simple recessive model, the disease mutation should be homozygous, and under a compound heterozygous model, two different mutations on different haplotypes are expected instead. This approach was applied to a presumed recessive disease, Miller syndrome (16). Four affected individuals, of whom two were siblings, were exome sequenced, and a manual review of the intersecting two genes found compound heterozygous mutations in DHODH to be causal. A similar analysis in Fowler syndrome (19) also was successful in identifying compound heterozygous causal mutations in FLVCR2, using only two affected individuals.
It is not always necessary to do a genome-wide analysis, especially in the cases where linkage intervals have already been determined or other familial information is available. Sequencing only one or two individuals will often suffice, particularly because the potential causal variant lists are not long. Using linkage information in whole-genome sequencing of a patient with metachondromatosis (12), exome sequencing of a proband and her mother in Joubert syndrome 2 (15) and exome sequencing of a proband with non-syndromic hearing loss DFNB82 (20) helped to narrow identify PTPN11, TMEM216 and GPSM2, respectively, as disease-associated genes.
Familial information was also used in whole-genome sequencing of Miller syndrome (18). Two affected siblings and their parents were sequenced, which allowed the resolution of haplotype inheritance for each sibling. The investigators then focused their disease analysis on the 22% of the genome for which both maternal and paternal haplotypes were inherited identically in both siblings and could shortlist potential causal variants to within these intervals, even though no linkage analysis was available.
In other studies that utilized linkage information, only the linkage intervals were captured and sequenced, likely because it is presently more cost-effective. In familial exudative vitreoretinopathy (10), clericuzio-type poikiloderma with neutropenia (14) and non-syndromic hearing loss DFNB79 (17), a single proband was sequenced to find the causal mutation within linkage intervals; in sensory/motor neuropathy with ataxia (8), a proband with one or two parents were sequenced; and in X-linked TARP syndrome (21), only exons on the X chromosome were captured and sequenced from two carriers. As sequencing costs drop, however, it is possible that the cost of targeted capture becomes disproportionately high when compared with sequencing costs, which may favor more whole-genome sequencing for single samples instead.
FILTERING BASED ON FUNCTION
The primary filter most investigators use to identify potentially causal mutations is based on variant function—namely, if the variant affects coding regions (missense, nonsense, coding indels and splice acceptor and donor sites) or other non-coding RNA transcripts. The main rationale given for this is that these variants tend to be of larger effect than non-coding variants, and also because it is difficult to predict the effects of non-coding and synonymous variants with any certainty. As such, in order to reduce noise when analyzing possible disease-causing variants, non-coding and synonymous variants are often ignored or greatly down-weighted.
For some disorders, it is possible to filter variants even further, by focusing only on those that are loss-of-function (i.e. nonsense and frameshift mutations). Since there are only a limited number of such mutations in any genome (<50), the candidate list is shortened very quickly. This filter was particularly useful to identify mutations in RBM10 as causal for TARP syndrome (21), and also in MLL2 for Kabuki syndrome (13).
For all the studies reviewed here, at least, restricting the analyses to coding variants has been justified—all have identified variants that affect protein function, mainly missense and nonsense mutants, and also frameshifting indels, and in one case a mutation at a splice acceptor site. This will most certainly not always be the case, as intronic, regulatory and synonymous variants are certainly known to affect disease (reviewed in 30), and as more disorders are studied, there will be a growing need for functional annotation of non-coding regions and tools to analyze the same.
RANKING VARIANTS BY EFFECT AND CONSERVATION
Variants can also be ranked by potential effect on protein structure and function, and also by conservation scores, as estimated by tools like SIFT (31), PolyPhen (32,33), CDPred (21), PhyloP (34) and GERP (35,36), with the rationale that mutations which are disruptive to proteins and/or at more conserved sites are more likely to be pathogenic. These tools have limited specificity and sensitivity (37), however, and mutations ultimately determined to be causal will rank highly, but potentially not first. As such, these rankings are normally used in conjunction with other strategies and not as a stand-alone filter.
FILTERING FOR RARE VARIANTS
Rare diseases, by definition, have an individual incidence of less than 1/1150–1/2000 in the population (38,39), and it is expected that mutations underlying these rare diseases will be at correspondingly rare frequencies, and most likely private to affected individuals. This is especially so for mutations that are highly penetrant—these variants of major effect and distinctive phenotype are not expected to be found in the population at large, and hence will not be seen in genome-wide scans for variants [e.g. the 1000 Genomes Project (40)], nor in polymorphism repositories [e.g. dbSNP (41)]. Exclusion from these data sets is typically an important criterion in defining a rare, novel or private variant.
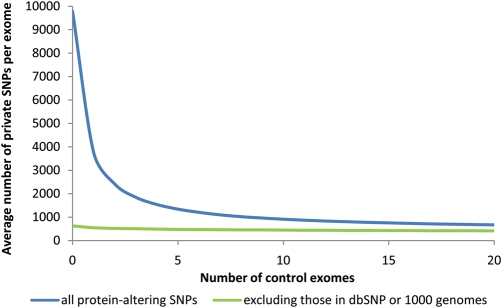
From empirical analysis of published exomes (9,13,16), we estimate that there are ∼20 000 single-nucleotide variants in a given exome [as defined by the Consensus Coding Sequence database (CCDS)], with about half affecting protein sequence. Using dbSNP and 1000 genomes data together as a filter suggests the number of novel SNPs is at most 1/10th of that (Fig. 1), which is a reflection of the breadth of ascertainment available in these data sets. However, a caveat to note is that phenotypic information is not always available for the samples used in these data sets, and it is possible that pathogenic mutations are present in them. In the case of recessive mutations, particularly, there is a chance that a normal carrier could have been genotyped and the recessive disease-causing mutation deposited in the database.
Figure 1.
Effect of increasing number of control exomes on private variants observed in a single exome, with and without the use of dbSNP and 1000 genomes data. The number of private mutations observed in an individual from sequential addition of control exomes was averaged from 10 000 permutations of 21 published exomes of non-African ancestry (9,13,16).
As a complementary approach then, control individuals with known phenotype and family history are often sequenced along with the affected cases, also to be used as a filter for common variants. This has the advantage of allowing for population-matched controls, especially for populations that may be under-represented in the current databases, and also to control for technical artifacts that may arise during the sequencing or alignment of sequence reads.
The effect of adding more control exomes is illustrated in Figure 1, which shows that the average number of novel or private SNPs in a given individual drops exponentially as more control exomes are used and starts to plateau by the time 15–20 controls are added (Fig. 1, blue line). This suggests that a limited number of control exomes is sufficient to filter for private variants by this method even without external datasets—adding beyond 20 controls to these data clearly has a diminishing rate of return.
SEQUENCING SCOPE
Since most current analyses are restricted to protein-coding regions, a prior decision is made in some studies to restrict the sequencing scope to genic regions, whether within a linkage region or as a whole exome, primarily based on cost. It is notable that there are many potential definitions for the exome. Earlier studies used the genes from the CCDS (42), a set of well-annotated, highly conserved proteins, with the rationale being that although this is a relatively conservative set of genes, it is also less likely to contain pseudogenes and potentially unverified protein transcripts, which would result in spurious variant calls and add noise to the analysis of potential mutations.
However, the CCDS is by no means complete, and failure to find a disease gene could be the result of missing sequence or annotation. In one example, mutations in the gene associated with Kabuki syndrome (13) were identified only because the exome definition was expanded to the RefSeq database (43). With the CCDS definition, this gene would not have been captured nor sequenced, and the mutations would have been missed entirely. Hence, to reduce the likelihood of this happening, more recent studies have started to use larger, more inclusive gene sets instead, like the aforementioned RefSeq database (43), the Ensembl database (44), genes from the UCSC browser (45) and the GENCODE set (46), particularly as capture technology limitations are overcome. It is worth noting that even when whole-genome sequencing becomes affordable enough such that targeted methods are not necessary, these definitions will still be important in the annotation of coding variants.
SPURIOUS GENE IDENTIFICATIONS
Using the aforementioned filters may not always be completely specific to the disease-associated gene. In a number of studies, spurious genes were also identified, but later dismissed upon manual review for various reasons.
In the Schinzel–Giedion syndrome study, all affected individuals had the exact same variant in 10 genes, suggesting that these could be polymorphisms that were missed in the controls (11), or possibly that there was a systematic artifact in variant calling. In another gene in the same study, CTBP2, the investigators note that it had a high variation rate, even in controls, suggesting that this could be due to the presence of other paralogous loci. A similar observation was made in the Miller syndrome study, where CDC27 was identified as a potential candidate, but noted to have an unannotated processed pseudogene that contributed to misalignments and an inflated variant call rate.
Another factor that could affect the number of variant calls is the length of the gene since many of these only have limited variant calls in the existing databases. For example, MUC16 was shortlisted in the Kabuki syndrome study (13) as the only gene to be common to all 10 cases, but was determined to be a false-positive due to its long length (>14 500 amino acids), which could have contributed to a high number of variant calls across all individuals.
MISSING MUTATIONS
It is also possible that not all affected individuals will have sequenced mutations in the same gene. This could be due to the disease model (e.g. genetic heterogeneity or non-coding variants) or technical issues (e.g. missing variant calls due to low coverage). As an example, of the 10 affected individuals in the Kabuki syndrome study (13), only 7 had causal mutations identified by exome sequencing. Misalignments and low coverage led to missed frameshift indel calls in two of the remaining three (later identified by Sanger sequencing), and the causal mutation in the last is still unknown. To deal with these issues, it is possible to sequence more affected individuals or to change the filtering approach to require that only a subset of individuals have a shared gene, rather than all, but the trade-off is that the candidate gene lists are greatly inflated.
CONCLUSIONS
Massively parallel sequencing has been applied successfully to find causal mutations for a number of monogenic disorders, including several that have been intractable to linkage analysis. In some studies, massively parallel sequencing was used to re-sequence genes within linkage intervals; in others, whole-genome or exome sequencing was used to find variants on a genome-wide scale. However, we note that the overall rate of success for these methods is uncertain, as only successful studies have been reported to date.
Using simple filters based on variant function and frequency, as well as careful choices of cases and controls, has been highly useful in isolating pathogenic mutations from background polymorphisms. In particular, priority is given to variants that are private to affected individuals, under the assumption that the mutations underlying these monogenic disorders are highly penetrant and rare. A second assumption is that the underlying mutations have large effect and are likely to be coding, and third that the disorder is genetically homogeneous in the samples being studied.
As less simple syndromes and disorders are studied, these assumptions are less likely to hold. In cases where genetic heterogeneity is present, mutations are less penetrant and non-coding variation is causal, the current strategies outlined here will be too stringent. Allowing for these more complex models will inflate candidate lists, and more sophisticated approaches will need to be developed to conduct candidate prioritization.
Massively parallel sequencing has the potential to accelerate the pace of disease gene discovery. Early successes have particularly shown its potential to revolutionize the way that the genetic bases of Mendelian disorders are studied. Recently initiated projects also aim to apply exome and whole-genome sequencing to common, genetically complex diseases, and this will bring its own set of challenges with respect to study design and statistical analysis.
Conflict of Interest statement. None declared.
FUNDING
Our work was supported in part by grants from the US National Institutes of Health (NIH)–National Heart, Lung and Blood Institute (5R01HL094976 to D.A.N. and J.S.), the NIH–National Human Genome Research Institute (5R21HG004749 to J.S., 1RC2HG005608 to M.J.B., D.A.N. and J.S.), NIH–National Institute of Environmental Health Sciences (HHSN273200800010C to D.A.N.), the Life Sciences Discovery Fund (2065508 and 0905001), the Washington Research Foundation and the NIH–National Institute of Child Health and Human Development (1R01HD048895 to M.J.B.). S.B.N. is supported by the Agency for Science, Technology and Research, Singapore.
References
- 1.Antonarakis S.E., Beckmann J.S. Mendelian disorders deserve more attention. Nat. Rev. Genet. 2006;7:277–282. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- 2.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein D., Risch N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat. Genet. 2003;33(suppl.):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 4.Lander E.S., Botstein D. Mapping complex genetic traits in humans: new methods using a complete RFLP linkage map. Cold Spring Harb. Symp. Quant. Biol. 1986;51:49–62. doi: 10.1101/sqb.1986.051.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Lander E.S., Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 6.Metzker M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 7.Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 8.Brkanac Z., Spencer D., Shendure J., Robertson P.D., Matsushita M., Vu T., Bird T.D., Olson M.V., Raskind W.H. IFRD1 is a candidate gene for SMNA on chromosome 7q22-q23. Am. J. Hum. Genet. 2009;84:692–697. doi: 10.1016/j.ajhg.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikopoulos K., Gilissen C., Hoischen A., van Nouhuys C.E., Boonstra F.N., Blokland E.A., Arts P., Wieskamp N., Strom T.M., Ayuso C., et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am. J. Hum. Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoischen A., van Bon B.W., Gilissen C., Arts P., van Lier B., Steehouwer M., de Vries P., de Reuver R., Wieskamp N., Mortier G., et al. De novo mutations of SETBP1 cause Schinzel–Giedion syndrome. Nat. Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 12.Sobreira N.L., Cirulli E.T., Avramopoulos D., Wohler E., Oswald G.L., Stevens E.L., Ge D., Shianna K.V., Smith J.P., Maia J.M., et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volpi L., Roversi G., Colombo E.A., Leijsten N., Concolino D., Calabria A., Mencarelli M.A., Fimiani M., Macciardi F., Pfundt R., et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am. J. Hum. Genet. 2010;86:72–76. doi: 10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edvardson S., Shaag A., Zenvirt S., Erlich Y., Hannon G.J., Shanske A.L., Gomori J.M., Ekstein J., Elpeleg O. Joubert syndrome 2 (JBTS2) in Ashkenazi Jews is associated with a TMEM216 mutation. Am. J. Hum. Genet. 2010;86:93–97. doi: 10.1016/j.ajhg.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehman A.U., Morell R.J., Belyantseva I.A., Khan S.Y., Boger E.T., Shahzad M., Ahmed Z.M., Riazuddin S., Khan S.N., Friedman T.B. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am. J. Hum. Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roach J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M., et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalonde E., Albrecht S., Ha K.C., Jacob K., Bolduc N., Polychronakos C., Dechelotte P., Majewski J., Jabado N. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum. Mutat. 2010;31:918–923. doi: 10.1002/humu.21293. [DOI] [PubMed] [Google Scholar]
- 20.Walsh T., Shahin H., Elkan-Miller T., Lee M.K., Thornton A.M., Roeb W., Abu Rayyan A., Loulus S., Avraham K.B., King M.C., et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston J.J., Teer J.K., Cherukuri P.F., Hansen N.F., Loftus S.K., Chong K., Mullikin J.C., Biesecker L.G. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am. J. Hum. Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupski J.R., Reid J.G., Gonzaga-Jauregui C., Rio Deiros D., Chen D.C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D.A., et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloglu A., Ozen S., Sanjad S., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenson P.D., Mort M., Ball E.V., Howells K., Phillips A.D., Thomas N.S., Cooper D.N. The human gene mutation database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou L.S., Liu C.S., Boese B., Zhang X., Mao R. DNA sequence capture and enrichment by microarray followed by next-generation sequencing for targeted resequencing: neurofibromatosis type 1 gene as a model. Clin. Chem. 2010;56:62–72. doi: 10.1373/clinchem.2009.132639. [DOI] [PubMed] [Google Scholar]
- 26.Hoischen A., Gilissen C., Arts P., Wieskamp N., van der Vliet W., Vermeer S., Steehouwer M., de Vries P., Meijer R., Seiqueros J., et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum. Mutat. 2010;31:494–499. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- 27.Vasta V., Ng S.B., Turner E.H., Shendure J., Hahn S.H. Next generation sequence analysis for mitochondrial disorders. Genome Med. 2009;1:100. doi: 10.1186/gm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raca G., Jackson C., Warman B., Bair T., Schimmenti L.A. Next generation sequencing in research and diagnostics of ocular birth defects. Mol. Genet. Metab. 2010;100:184–192. doi: 10.1016/j.ymgme.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley G., Higuchi R., Hoglund B., Goodridge D., Sayer D., Trachtenberg E.A., Erlich H.A. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74:393–403. doi: 10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper D.N., Chen J.M., Ball E.V., Howells K., Mort M., Phillips A.D., Chuzhanova N., Krawczak M., Kehrer-Sawatzki H., Stenson P.D. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Hum. Mutat. 2010;31:631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- 31.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepel A, Pollard K., Haussler D. New methods for detecting lineage-specific selection. Proceedings of the 10th International Conference on Research in Computational Molecular Biology (RECOMB 2006) 2006:190–205. [Google Scholar]
- 35.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper G.M., Goode D.L., Ng S.B., Sidow A., Bamshad M.J., Shendure J., Nickerson D.A. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods. 2010;7:250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rare Diseases Act. 2002. http://frwebgate.access.gpo.gov/cgi-bin/getdoc.cgi?dbname=107_cong_public_laws&docid=f:publ280.107 .
- 39.EURORDIS: Rare Diseases Europe. http://www.eurordis.org/about-rare-diseases .
- 40.1000 Genomes Project. http://www.1000genomes.org. (last accessed June 23, 2010)
- 41.Database of Single Nucleotide Polymorphisms (dbSNP) Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine; http://www.ncbi.nlm.nih.gov/SNP/ (last accessed June 23, 2010). [Google Scholar]
- 42.Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R., Searle S., Farrell C.M., Loveland J.E., Ruef B.J., et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubbard T.J., Aken B.L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhn R.M., Karolchik D., Zweig A.S., Wang T., Smith K.E., Rosenbloom K.R., Rhead B., Raney B.J., Pohl A., Pheasant M., et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]