Generation of VSV Pseudotypes Using Recombinant ΔG-VSV for Studies on Virus Entry, Identification of Entry Inhibitors, and Immune Responses to Vaccines (original) (raw)
. Author manuscript; available in PMC: 2011 Nov 1.
Abstract
Vesicular stomatitis virus (VSV) is a prototypic enveloped animal virus that has been used extensively to study virus entry, replication and assembly due to its broad host range and robust replication properties in a wide variety of mammalian and insect cells. Studies on VSV assembly led to the creation of a recombinant VSV in which the glycoprotein (G) gene was deleted. This recombinant (rVSV-ΔG) has been used to produce VSV pseudotypes containing the envelope glycoproteins of heterologous viruses, including viruses that require high-level biocontainment; however, because the infectivity of rVSV-ΔG pseudotypes is restricted to a single round of replication the analysis can be performed using biosafety level 2 (BSL-2) containment. As such, rVSV-ΔG pseudotypes have facilitated the analysis of virus entry for numerous viral pathogens without the need for specialized containment facilities. The pseudotypes also provide a robust platform to screen libraries for entry inhibitors and to evaluate the neutralizing antibody responses following vaccination. This manuscript describes methods to produce and titer rVSV-ΔG pseudotypes. Procedures to generate rVSV-ΔG stocks and to quantify virus infectivity are also described. These protocols should allow any laboratory knowledgeable in general virological and cell culture techniques to produce successfully replication-restricted rVSV-ΔG pseudotypes for subsequent analysis.
Keywords: Pseudotypes, VSV, transfection, envelope glycoprotein, virus entry, receptor identification, high-throughput screening, vaccines
1. Introduction
The entry of all enveloped viruses requires a membrane fusion event that is mediated by one or more viral glycoprotein found on the surface of the lipid envelope of the virus. These envelope proteins are responsible for binding virus to the cell surface and for inducing fusion of the viral envelope with either the plasma membrane of the host cell, or with an internal membrane following endocytosis of the virion. Identifying host proteins, glycans, or lipids that serve as cellular receptors for viruses has been one of the major goals of virology. However, for viruses that require BSL-3 or BSL-4 containment, the identification of receptors is not trivial and often requires indirect methods that do not utilize live virus. Recent developments that allow pseudotyping of glycoproteins from risk group-3 (RG3) or −4 viruses onto RG-2 viruses that require BSL-2 containment have facilitated the study of virus entry, and have also provided facile methodology to perform high-throughput screening of entry inhibitors for these viruses.
Vesicular stomatitis virus (VSV) is a prototypic nonsegmented, negative-stranded RNA virus that belongs to the family Rhabdoviridae. Laboratory strains of VSV, such as VSV-Indiana, can be handled using BSL-2 containment and have been used as models to study many aspects of negative-strand RNA virus entry and replication. VSV assembly occurs at the plasma membrane and involves budding of virions from the cell surface. During budding, VSV acquires an envelope consisting of a lipid bilayer derived from the plasma membrane and spike proteins consisting of trimers of the VSV glycoprotein (G protein).
One of the remarkable properties of VSV is that VSV virions are not particularly selective in regards to the type of membrane protein that can be incorporated into the viral envelope. Early studies in which cells were coinfected with VSV and other enveloped viruses demonstrated that VSV forms pseudotypes readily (Huang et al., 1974; Weiss et al., 1977; Witte and Baltimore, 1977; Zavada and Rosenbergova, 1972). A pseudotype has the envelope protein of the heterologous virus assembled into the VSV membrane. The ability to form pseudotypes is likely due to the mechanism of VSV budding (Jayakar et al., 2004), which has been shown not to require VSV G protein (Schnell et al., 1997; Takada et al., 1997). Therefore, noninfectious “bald” particles are produced in the absence of G protein, albeit at a much lower efficiency than when G protein is present (Robison and Whitt, 2000). The fact that VSV particles can bud in the absence of G protein, coupled with the promiscuous nature with which heterologous glycoproteins can be incorporated into VSV lead to the development of recombinant viruses in which the VSV glycoprotein gene was deleted and replaced with genes encoding fluorescent reporter proteins (e.g. GFP and RFP), luciferase, or other easily assayable, secreted enzymes (SEAP; secreted human placental alkaline phosphatase). When a glycoprotein or glycoprotein complex from a heterologous virus is expressed transiently in cells infected with these recombinants, pseudotype particles are released at high efficiency. These rVSV-ΔG pseudotypes, when plated on susceptible cells, undergo a single cycle of infection and the entry of the pseudotypes is dictated by the cell tropism and entry properties of the heterologous glycoprotein(s). Since the first description of this system in 1997 (Takada et al., 1997) rVSV-ΔG pseudotypes have been used in numerous studies examining virus entry (Abe et al., 2007; Fukushi et al., 2005; Glende et al., 2008; Hanika et al., 2005; Ito et al., 1999; Kaimori et al., 2004; Matsuura et al., 2001; Okuma et al., 2001; Perez et al., 2001; Saha et al., 2005; Tamura et al., 2005; Tatsuo et al., 2000a), for identification of novel host cell receptors (Tatsuo et al., 2000b), for screening antiviral libraries and development of neutralizing antibody tests (Fukushi et al., 2008; Ge et al., 2006; Ogino et al., 2003; Porotto et al., 2007), and as vaccine vectors (Klas et al., 2002; Lee et al., 2006; Miller et al., 2004; Publicover et al., 2005).
This manuscript provides a detailed description of the methods used to recover, produce, and quantify rVSV-ΔG pseudotypes. This information should allow any laboratory skilled in virological methods and cell culture technology to maintain working stocks of infectious rVSV-ΔG for the purpose of generating rVSV-ΔG pseudotypes for studies of virus entry requirements and high-throughput screening of antiviral compounds, particularly for viruses that require high-level biocontainment or that are inherently difficult to grow.
2. Materials
2.1. Cell Culture, Transfection, Virus Recovery, Titering and Pseudotyping
- –
Dulbecco’ s Modified Eagle’s Medium with L-glutamine (DMEM; Lonza/BioWhittaker cat. #12–604F) supplemented with 5% fetal bovine serum (Atlanta Biologicals; see Note 1). This is called D-5. - –
Serum-free DMEM (SF-DMEM). - –
Solution of trypsin-EDTA in phosphate buffered saline (0. 25% trypsin + 0.1% EDTA in PBS). - –
1.8% Bacto-agar in water. Sterilized and melted by autoclaving. For use, heat in microwave oven until agar is molten. - –
20X NaHCO3: Dissolve 5.92 g NaHCO3 in a total volume of 80 ml Milli-Q-dH2O. Filter sterilize using a 0.2 μ bottle-top filter and store at 4°C. - –
2X DMEM stock solution: Dissolve a 1 -liter packet of powdered DMEM (Invitrogen/Gibco; cat #12800-017) in 450 ml Milli-Q-dH2O. Filter sterilize using 0.2 μ bottle-top filter and store at 4°C. - –
2X DMEM working solution: To prepare a 100 ml 2X DMEM working solution add 5 ml 20X NaHCO3 to 85 ml 2X DMEM stock and then add 10 ml FBS (Final FBS = 10%). - –
Agar Overlay: Melt 1.8% Bacto-agar in a microwave oven or boiling water bath. Add the volume needed for titering to a sterile tube or bottle and cool to 45°C. Mix equal volumes of pre-warmed (to 37°C) 2X DMEM working solution containing 10% FBS with the molten 1.8% agar. Add 2 ml directly to infected cells and let solidify. - –
4% X-gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) in dimethylformamide. Store at −20°C. - –
TransfectACE reagent: Dissolve 0.1g DDAB (dimethyldioctadecyl ammonium bromide; Sigma; cat #D-2779) in 1 ml 100% ethanol by placing in a 37°C water bath with occasional vortex mixing. The DDAB stock must be made fresh each time. Dissolve L-α-phosphatidyl ethanolamine-dioleoyl (Sigma; cat #P1223) in 100% ethanol to 10 mg/ml. Add 40 µl DDAB stock to 1 ml 10mg/ml L-α-phosphatidyl ethanolamine-dioleoyl, mix by vortexing and then inject the ~1 ml lipid/EtOH mix into 9 ml sterile deionized water at room temperature using a pipetteman, while vortexing. Store the liposome suspension at 4°C. The reagent will be good for at least 3 months after preparation. - –
Opti-MEM Reduced Serum Medium (Invitrogen; cat. #11058021). - –
Lipofectamine (Invitrogen; cat. #18324012). - –
Sterile, 15ml or 50ml polystyrene centrifuge tubes. - –
Sterile 3.5 ml snap-cap polystyrene tubes. - –
Baby hamster kidney (BHK-21) cells (clone WI-2; see Note 2) (Kaariainen and Gomatos, 1969). - –
HeLa cells (ATCC #CCL-2).
2.2. Plasmids and Plasmid Purification
- –
pVSV-ΔG-GFP, a.k.a. pVSV-ΔG* [Fig. 1A; (Takada et al., 1997)] - –
pVSV-ΔG-DsRed, a.k.a. pVSV-ΔG-RFP (Porotto et al., 2007) - –
pVSV-ΔL-GFP (Robison, 2001) - –
pBS-N-Tϕ (Stillman et al., 1995) - –
pBS-P-Tϕ (Stillman et al., 1995) - –
pBS-L-Tϕ (Stillman et al., 1995) - –
pBS-G (Fredericksen and Whitt, 1995) - –
pC-VSVG (Takada et al., 1997), a.k.a. pCAGGS-GInd (Jeetendra et al., 2002). - –
E. coli, strain C600 - –
Luria broth (LB) supplemented with 80 mg/ml ampicillin. - –
GET buffer: glucose (0.9%), EDTA (10mM), Tris (0.25 m_M_ pH 8.0). - –
Alkaline SDS: Sodium dodecyl sulfate (1%), NaOH (0.2 M) - –
Acidic potassium acetate solution: 60 ml 5 M KOAc + 11.5 ml glacial acetic acid + 28.5 ml deionied water. - –
Lysozyme (10 mg/ml) in deionized water. - –
100% Isopropanol - –
70% ethanol - –
TE buffer: Tris (10 m_M_ pH 8.0) + EDTA (1 m_M_). - –
Ethidium bromide (10 mg/ml) in deionized water. - –
Isopropanol equilibrated in cesium chloride-saturated water - –
100% ethanol.
Figure 1.
Recovery, growth and analysis of rVSV-ΔG. (A) Diagram of the cDNA encoding the anti-genome of rVSV-ΔG-GFP. The conserved VSV transcriptional regulatory sequences (octagon = stop sequence; An= polyadenylation sequence; arrow = transcriptional promoter or start sequence) are shown above the diagram. The bacteriophage T7 RNA polymerase promoter (T7) and terminator sequences (Tϕ) and the hepatitis delta virus ribozyme (δ-RBZ) with cleavage site are indicated below the diagram. (B) A flow chart illustrating the key points involved in generating and characterizing recombinant rVSV-ΔG. (I) Initially cells are cotransfected with the plasmid pVSV-ΔG-GFP and the four support plasmids encoding the VSV N, P, G and L proteins. This is followed by virus propagation (II), with subsequent plaque-purification and growth of G-complemented rVSV-ΔG-GFP working stocks (III & IV). The G-complemented virus stocks produced in step IV are titrated, and then used to infect cells that express a heterologous glycoprotein for production of the ΔG-GFP pseudotypes. Figure courtesy of C.S. Robison (Robison, 2001).
2.3. Fluorescence Microscopy
- –
Fix solution: Prepare 3% paraformaldehyde solution by dissolving 4.5 g paraformaldehyde powder in 120 ml deionized water made alkaline by the addition of 75 µl 5 M NaOH. Heat to 50°C to dissolve, and then cool to r.t. Add 15 ml 10X PBS and adjust pH to 7.4 with 5 M HCl. Bring final volume to 150 ml with deionized water, aliquot, and store at −20°C. The fix solution can be frozen and thawed multiple times. At the first thaw a precipitate will form. Heat in a microwave (carefully) with the lid loosened at 10 second intervals and mix by swirling until precipitate dissolves. Cool to r.t. before use. The precipitate does not form during subsequent thaws. - –
Mounting medium: 80% glycerol in phosphate buffered saline and containing 0.1 M n-propyl-gallate (3,4,5-trihydrobenzoic acid n-propyl ester; Sigma, cat #P-3130). - –
10X PBS: 100 m_M_ sodium phosphate monobasic (NaH2PO4 · H2O), 100 m_M_ sodium phosphate dibasic (Na2HPO4 · 7 H2O), 1.5 M sodium chloride (NaCl) dissolved in deionized water. Filter sterilize using a 0.2 μ pore filter and store at r.t. - –
PBS-glycine: Use 10X PBS to prepare a 1X PBS solution that also contains 10 m_M_ glycine. - –
Permeabilization solution: PBS-glycine containing 1% Triton X-100. - –
Primary antibodies: Culture supernatants from mouse hybridomas producing monoclonal antibodies to VSV-M protein (MAb 23H12), or VSV-N protein (MAb 10G4) diluted 1:3 with PBS-glycine (Lefrancois and Lyles, 1982). - –
Secondary antibody: Goat anti-mouse IgG conjugated to rhodamine Red-X (Jackson ImmunoResearch cat #115–295-146), or to FITC (cat #115-095–146).
3. Detailed Procedure
To study the entry mechanism of enveloped viruses that are either difficult to grow, or that require high-level containment (e.g. BSL-3 or BSL-4 agents), the envelope glycoprotein(s) is/are transiently expressed from a plasmid vector and then the cells are infected with the recombinant rVSV-ΔG that has been previously pseudotyped with VSV G protein (e.g. G-complemented rVSV-ΔG). Because the rVSV-ΔG genome does not encode an envelope glycoprotein, virus that is produced will incorporate the heterologous (e.g. non-VSV-G) glycoprotein(s) into the virion after expression of the glycoprotein(s) from the plasmid on the plasma membrane. The consequent infectivity of the pseudotyped progeny virions will be dependent on the host/cell tropism and entry mechanism of the heterologous glycoprotein(s).
The methods described below include those for the initial recovery and production of the G-complemented rVSV-ΔG and are followed by the methods used to generate rVSV-ΔG pseudotyped with heterologous glycoproteins. Important controls used to evaluate the efficiency of virus recovery, and methods used to trouble-shoot the basis for poor recovery efficiencies are included. Finally, methods used to quantify infectivity and to analyze glycoprotein incorporation into rVSV-ΔG particles are described.
3.1. Plasmid Purification
While commercial columns can be used for plasmid purification, more consistent results are obtained using CsCl-EtBr gradient purified plasmids for both recovery of virus and for transfection to produce rVSV-ΔG pseudotypes. Experience indicates that E. coli C600 cells provide the highest yield of plasmids containing VSV genomic cDNAs (e.g. pVSV-ΔG-GFP, or pVSV-ΔL-GFP) when using CsCl-EtBr gradients, although DH5-α cells can also be used.
- Inoculate 250 ml LB-amp broth in a 1-liter Erlenmeyer flask with 250 µl of a fresh overnight culture of E. coli transformed with the desired VSV plasmid and grow for 12–14 hours (see Note 3) at 37°C with vigorous shaking on a platform shaker. Place the flask on ice to prevent overgrowth of the culture.
- Make a glycerol stock by removing 300 µl of the culture and mix with 300 µl 80% glycerol. Place at −80°C for long-term storage of the glycerol stock. An aliquot of the glycerol stock is used to generate the overnight culture for subsequent plasmid preps.
- Transfer the culture to a 250 ml centrifuge bottle and pellet the cells at 8,000 × g for 10 minutes at 4°C in a Sorvall GSA rotor (or equivalent).
- Discard the supernatant, remove residual liquid from the walls of the tube by aspiration and resuspend the cell pellet in 14 ml ice-cold GET buffer. Ensure the pellet is fully resuspended without clumps.
- Freeze the cell suspension by transferring the centrifuge bottle to a −80°C, −20°C, or dry ice-alcohol bath. The suspension should be completely frozen before going to the next step (generally 5–10 minutes in the dry ice bath, 15–20 minutes at −80°C, or >30 minutes at −20°C.
- Thaw rapidly in a 37°C water bath, then place on ice.
- Add 1.6 ml 10 mg/ml lysozyme, mix by swirling and incubate for 5 minutes on ice.
- Add 27 ml freshly prepared alkaline lysis buffer and immediately mix by inverting sharply (use a snapping motion with the wrist to rapidly and completely mix). The cells will lyse upon contact with the lysis buffer and release the genomic DNA making mixing difficult if not done rapidly.
- Incubate on ice for 5–10 minutes, or until turbidity clears.
- Add 21 ml acidic potassium acetate solution and mix completely by rapid inversion as in step 8. Incubate on ice ~5 min.
- Pellet the white potassium dodecyl sulfate precipitate by centrifugation at 10,000 × g for 30 minutes at 4°C in a Sorvall GSA rotor (or equivalent).
- Recover the supernatant by pouring into a 50 ml conical centrifuge tube through 2–3 layers of cheese cloth placed in a funnel. Squeeze the residual supernatant out of the cheese cloth while wearing a latex or nitrile glove.
- Warm to room temperature by placing in a 37°C water bath for ~5 minutes, and then precipitate the plasmid DNA by adding 0.6 volumes of r.t. 100% isopropanol.
- Mix well, incubate at r.t. for ~5 minutes and then pellet at 1,320 × g for 30 min at r.t. in a swinging-bucket table top centrifuge.
- Pour off supernatant and wash pellet by gentle vortexing with ~5 ml of 70% ethanol and place on ice for ~5 min. The pellet should dislodge from the bottom of the tube for complete washing.
- Pellet again at 1,320 × g for 10 min.
- Aspirate supernatant and remove residual ethanol from the walls of the tube.
- Dry the pellet by placing in a vacuum dissector for ~5 min.
- Resuspend the pellet in 3 ml TE buffer. This may take some time depending on the size of the pellet. Can incubate on ice to allow pellet to resuspend if necessary.
- Measure the volume of the fully resuspended DNA solution, transfer to a 15 ml conical centrifuge tube and bring to 4 ml final volume with TE buffer.
- Add 4.24 g CsCl, mix gently by inverting to dissolve, and then add 0.4 ml 10 mg/ml EtBr.
- Mix well by inverting and then centrifuge at 850 × g for 10 min. at r.t. to clarify.
- Transfer supernatant to Beckman vTi65 Quick-seal tubes (or equivalent), balance to within 0.02 g, heat-seal and place in vertical centrifuge rotor (Beckman vTi65 or equivalent) and centrifuge at 372,000 × g for 4 hours, or 178,000 × g for 14–16 hours at 20°C.
- After run is completed, remove tubes, vent by inserting a 16-gauge needle into the top of the tube, and then collect lower (super-coiled) plasmid band by side-puncture using a 16-gauge needle and 3 ml syringe (see Note 4).
- Transfer the collected plasmid band to a 15 ml polystyrene tube and extract EtBr by adding 2–3 volumes CsCl-saturated isopropanol. Mix by inverting 5–10 times and then separate aqueous layer containing the plasmid DNA from the isopropanol layer containing the extracted EtBr by brief (~1 min) centrifugation in a table-top centrifuge at r.t. Aspirate the magenta-colored upper isopropanol layer and repeat 3–4 times until the aqueous layer contains no visible EtBr (pink color).
- Transfer the lower aqueous layer containing the plasmid DNA to a siliconized Corex tube and dilute with 3 volumes of TE buffer. Mix and then add 2 volumes (8 volume equivalents of the original aqueous layer volume) of 100% ethanol and mix well by inverting.
- Incubate on ice for 30 min to precipitate DNA and then pellet at 16,000 × g for 30 min at 4°C in a Sorvall HB-6 swinging-bucket (or equivalent) rotor.
- Discard supernatant and wash pellet with 70% ethanol containing 60 m_M_ sodium acetate. Incubate for 5 min at 4°C and pellet at 10,000 rpm for 10 min.
- Aspirate supernatant, removing all residual liquid from the walls of the tube, and then dry the pellet in a vacuum desiccator for ~10 min.
- Resuspend pellet in 300 µl TE buffer and determine the DNA concentration. Dilute to 1 mg/ml with TE buffer and store at −20°C.
3.2. Preparation of vaccinia virus-T7 stocks
For recovery of VSV from plasmids the vaccinia T7 RNA polymerase expression system developed by Thomas Fuerst and Bernie Moss (Fuerst et al., 1986) is used to express the antigenomic VSV cDNA and for expression of the N, P, G, and L constructs. Several different recombinant vaccinia virus variants that express T7 RNA polymerase have been developed (M. Whitt, unpublished) and the one that is used currently is a modified vaccinia virus derived from the B18R (interferon viroceptor knockout) strain of vaccinia (Symons et al., 1995), into which the bacteriophage T7 polymerase gene was inserted under control of the vaccinia p7.5 early/late promoter (v18R-T7). Either the original vTF7-3 virus or the v18R-T7 virus can be used for rVSV recoveries (Jayakar and Whitt, 2002; Jeetendra et al., 2002; Miller et al., 2004; Mire et al., 2009) and these will be referred to generically as vvT7.
- Plate HeLa cells into 15cm dishes, or T-175 flasks and incubate overnight. Typically, twenty 15 cm dishes are used to produce the vaccinia stocks. The cells should just reach complete confluency before inoculation.
- Rinse the cells two times with serum-free DMEM and infect at an MOI ~ 0.1 – 0.5 in a small volume (generally 3 ml/15 cm dish) of serum-free DMEM. Allow virus to adsorb for 45 min at 37°C. Remove the inoculum and add 20 ml D-5.
- Incubate for 2–3 days at 37°C until CPE (cell rounding) is evident. Generally, ~90–95% of the cells should be rounded and some will have detached from the dish.
- Dislodge the cells by pipetting and transfer with the media to a conical centrifuge bottle. Once the cells are removed, wash the plates twice with a total volume of 15 ml PBS by rinsing the excess cells and media from each plate sequentially. Add the PBS wash to the cell suspension in the conical bottle.
- Pellet the cells by centrifugation at 1,320 × g for 10 min. Discard the supernatant by aspiration into disinfectant (bleach). The cell pellets may be frozen and stored at −80°C, or may be processed (step #6).
- Resuspend the cell pellet in 8 ml ice-cold 10 m_M_ Tris-HCl pH 9.0 by gentle vortexing or pipetting and transfer to a sterile glass dounce homogenizer that has been pre-cooled on ice. Rinse the centrifuge bottles with 4mls ice-cold 10 m_M_ Tris-HCl pH 9.0 and add to the cell suspension in the dounce homogenizer.
- Dounce 60 times on ice in a biosafety cabinet. Use BSL-2 precautions during all phases of the vaccinia purification.
- Transfer the cell extracts to a sterile 50 ml conical centrifuge tube and pellet the nuclei by centrifugation at 140 × g for 5 min at 4°C.
- Collect the post-nuclear supernatant and transfer to a clean sterile 50 ml conical tube.
- Wash the nuclei in the dounce with 10 ml of ice-cold 10mM Tris-HCl pH 9.0 and pellet as above.
- Combine the post-nuclear supernatants and sonicate in a bath sonicator containing ice-cold water for 5 min.
- Centrifuge the sonicates at 500 × g for 5 min to pellet debris. Transfer the supernatants to a clean tube and sonicate again for 5 min. Remove residual debris by centrifuging again at 500 × g for 5 min.
- Transfer the sonicated supernatants to two sterile Beckman SW28 tubes (or equivalent) and under-layer with enough 36% sucrose in 10 m_M_ Tris-HCl pH 9.0 to fill the tube to within 0.5 – 1 cm of the top of the tube.
- Pellet the virus through sucrose cushion by centrifugation at 130,000 × g for 20min at 4°C.
- Resuspend each of the virus pellets in 2 ml of 1 m_M_ Tris-HCl pH 9.0, transfer to a clean sterile tube, and rinse the SW28 tubes with a total of 2 ml of 1 m_M_ Tris-HCl pH 9.0.
- Mix well by vortexing, divide into aliquots and store at −80°C.
- After completely frozen, thaw one of the aliquots and titer on both HeLa cells and on BHK-21 cells.
- To titer, plate cells into 6-well plates and incubate overnight.
- Thaw an aliquot of the frozen vaccinia stock (see Note 5) and make 10-fold serial dilutions in SF-DMEM. Rinse the cells twice with SF-DMEM, add 0.4 ml SF-DMEM and add 0.1 ml of the dilutions to separate wells.
- Adsorb for 1 hr at 37°C with occasional rocking (or on a platform rocker), remove the inoculum and add D-5 (see Note 6).
- Incubate for ~48 hours and then determine the titer by counting plaques. For the v18R-T7 virus, the medium is removed and then the monolayer is overlayered with D-5 containing 0.9% molten Bacto-agar cooled to <50oC and 0.033% X-gal. The v18-T7 expresses β-galactosidase resulting in formation of “blue” plaques upon addition of X-gal. Incubate at 37°C until color formation is detected (~30–60 min).
3.3. Recovery of rVSV-ΔG from Plasmids
The recovery of rVSV-ΔG from plasmids using reverse genetics involves the transfection of cells that have been infected with recombinant vaccinia expressing bacteriophage T7 RNA polymerase (vvT7). Cells are transfected with 5 different plasmids which express the VSV antigenomic-sense (or positive-sense) RNA and cDNAs for the VSV N, P, G, and L proteins, all of which are under control of T7 promoters (Fig. 1). A fraction of the T7-expressed anti-genomic RNA will be encapsidated by the VSV N protein and this encapsidated RNA will serve as the template for synthesis of genomic RNA by the VSV polymerase complex consisting of the VSV L and P proteins. Once genomic RNA has been produced, which can only occur through the activity of the VSV polymerase complex acting on the encapsidated anti-genome, viral mRNAs are synthesized and the replication cycle is completed. In addition to more N, P, and L proteins, the VSV matrix (M) protein is produced from the virally encoded M gene. The M protein is required for RNP condensation and virus budding. Because rVSV-ΔG does not encode the viral glycoprotein (G), this must be provided in trans to produce infectious G-pseudotyped rVSV-ΔG virions. The protocol to recover and titer G-pseudotyped rVSV-ΔG (G*ΔG-VSV) is divided into two steps as outlined below.
3.3.1 Step #1-Primary recovery
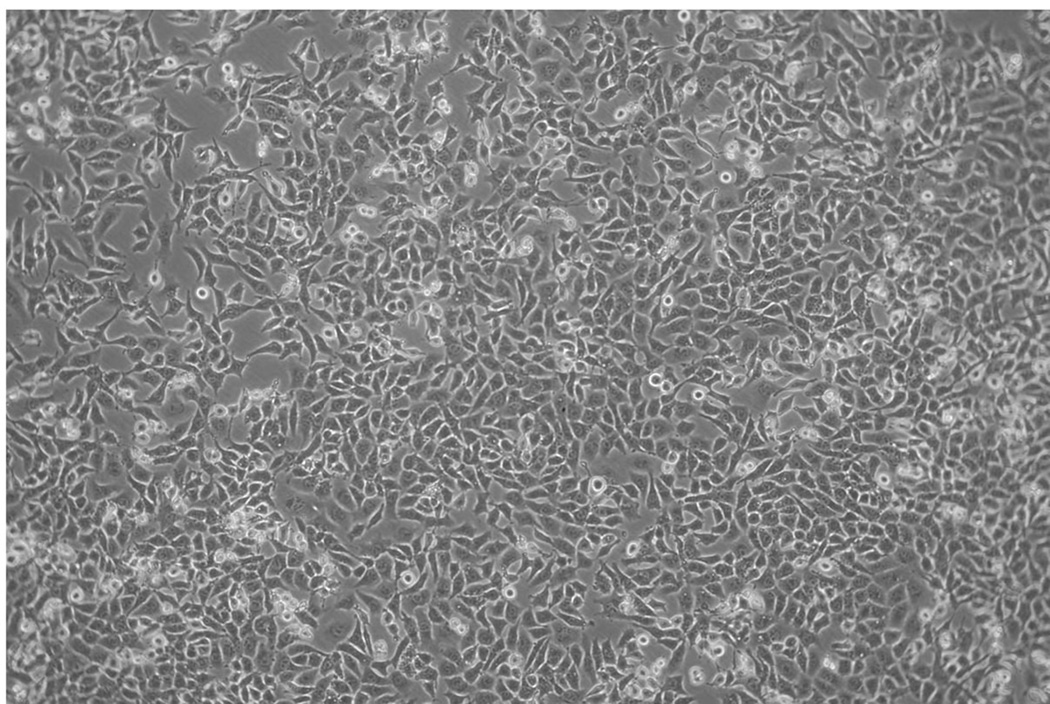
- Cell plating and vvT7 infection: Approximately 16–24 hours before starting the primary recovery, seed BHK cells into 6-well plates. Add between 5 × 105 to 9 × 105 cells/well. When plating, ensure that the cells are evenly disbursed in the well (see Note 7). Rock the plate back-and-forth in both directions to achieve an evenly distributed monolayer. Do not swirl the plate since this causes the cells to accumulate in the middle of the well. Several different seeding densities can be used depending on the time of day the cells will be plated and the time interval between seeding the 6-well plates and the start of the recovery. Incubate the cells at 37°C in a humidified incubator containing 6%–8% CO2 for 14–16 hours (see Note 8). The cells should be at ~90–95% confluency the next day for virus recovery (Fig. 2).
- First, the cells are infected with a recombinant vaccinia virus expressing T7 RNA polymerase (vvT7). Begin by removing the vvT7 stock from the −80°C freezer. Thaw rapidly in a 37°C water bath and place on ice.
- In a 15ml sterile conical tube, prepare the vvT7 inoculum in SF-DMEM pre-warmed to 37°C. Use sufficient vvT7 to give an MOI = 5. Each well should receive 500 µl of the inoculum. Always include an extra “wells-worth” of inoculum to ensure there is a sufficient amount of inoculum for the recovery. Mix by vortexing.
- Remove the growth medium from the 6-well plates and rinse each well with 1 −3 ml pre-warmed SF-DMEM. Aspirate and add 500 µl of the vvT7 inoculum to each well. Adsorb by occasional rocking, or by placing the plate on the platform rocker in a 37°C, 6%–8% CO2 incubator for ~45 minutes.
- Next, prepare the transfection mix. The transfection mix should be prepared in a biological safety cabinet to maintain sterility. While the vaccinia virus is adsorbing, prepare a master mix containing the recovery support plasmids (N, P, G, and L) in a 15ml or 50ml sterile polystyrene conical. The amount of transfection mix to be prepared will depend on the number of recoveries being done. Each well will receive an N, P, G, and L plasmid mixture that has a ratio of 3:5:8:1 µg of each plasmid, respectively. The final amount of support plasmid DNAs added will be 17µg per well.
- For each well add 1 ml of SF-DMEM to a polystyrene tube. This will be for the support plasmid master mix. Always include one extra recovery well-equivalent in the calculations to ensure a sufficient amount of the master mix is available.
- Add the appropriate volume of the 3:5:8:1 (N:P:G:L) support plasmid solution to the tube. Vortex the DNA master mix and add 1 ml of the mix into the appropriate number of labeled 3.5 ml snap-cap polystyrene tubes. Label the tubes according to recovery number and/or name.
- Next add 5 µg of the pVSV-ΔG plasmid to be recovered to the snap-cap tubes. Each virus to be recovered should be done in duplicate. In addition, 2 wells will be used for positive control transfections using the pVSV-ΔL-GFP plasmid. Replace the cap on the tubes and vortex. Be sure that no medium is trapped in cap.
- Next prepare the transfection reagent solution. In a 15ml or 50ml polystyrene tube, prepare a master mixture of TransfectACE. For each well, add 1ml SF-DMEM to the conical and add 3.5 µl of TransfectACE for each µg of DNA added to the conical (see Note 9). Therefore, in a typical transfection containing 17 µg of support plasmids and 5 µg of pVSV plasmid, add 77 µl of TransfectACE. Include one extra well-equivalent in the calculations as described above. Vortex gently to mix.
- Add 1ml of the TransfectACE master mix solution to a single snap-cap tube containing 1 ml of recovery plasmid DNA and immediately mix by gently inverting the tube several times to ensure homogeneous formation of lipid-DNA complexes. Do not add the transfection reagent to all tubes first, and then mix as this seems to increase variability in the efficiency of recovery. Remove any trapped media from the snap cap by “flicking” the tube. Prepare the DNA and TransfectACE suspensions approximately 20 minutes prior to the end of the vvT7 adsorption.
- Incubate the DNA:lipid mixture in the biological safety cabinet for 20–30 minutes.
- After the incubation, remove the vvT7 inoculum from each recovery well. When finished rinse the pipette or pipette tip with 20% bleach to inactivate the vaccinia virus.
- Add 2 ml of the DNA:lipid mix from each tube to the appropriately labeled well.
- Put the plate in a 37°C, 6%–8% CO2 incubator and incubate for 3–5 hours.
- After the incubation, remove the transfection mix and then add 1.5 to 2.5 ml/well D-5 and incubate at 37°C for 40–48 hours. Typically, virus recovery supernatants will be harvested between 42 and 46 hours post-transfection.
Figure 2.
Phase contrast micrograph of BHK-21 cells taken immediately before starting a recovery experiment for rVSV-ΔG viruses. The cells should be evenly distributed and should ~90–95% confluent. The image was collected using a digital camera connected to a phototube with a 2.5X lens on an Olympus CK2 microscope equipped with a 10X phase objective. The image brightness was adjusted in Canvas 11 using the “levels” function to optimize visualization of individual cells.
3.3.2 Step #2- Amplification of the primary recovery virus
- The same day that the transfection for the primary recovery is performed, plate a fresh set of BHK cells in 6 well plates and grow overnight. These cells will be transfected with a plasmid (pCAGGS-G) that expresses VSV G protein from a strong pol-II promoter to amplify the recovered viruses obtained in Step #1. The cells should be evenly distributed as described in Step 1 and should be at ~85% confluency the next morning.
- Replace the D-5 medium on the cells with ~1.5 ml Opti-MEM pre-warmed to 37°C and put the cells back into the 37°C incubator.
- Prepare a transfection mix consisting of 2 µg pCAGGS-G and 10 µl Lipofectamine in 2 ml Opti-MEM per well as follows. This can be prepared as a Master Mix and aliquoted into the 6-well plates. To one sterile 15 ml or 50 ml polystyrene tube add the appropriate volume of Opti-MEM to give 1ml per well. To a separate tube add the same volume of Opti-MEM.
- To one of the tubes add the appropriate amount of pCAGGS-G plasmid to give 2 µg per well. Vortex gently to mix.
- To the other tube add the appropriate volume of Lipofectamine to give 10 µl per well. Vortex to mix.
- Immediately add the lipid mix to the DNA-OptiMEM mix and gently invert 5–6 times.
- Incubate 20 min at r.t.
- Remove the Opti-MEM from the cell monolayers and add 2 ml of the transfection mix directly to cells.
- Rock the plate 2–3 times and then incubate for ~4 hrs at 37°C.
- After the incubation remove the transfection mix and replace with D-5.
- Incubate the cells in a 37°C, 6%–8% CO2 incubator for 24–30 hrs. Evaluate the efficiency of the transfection by examining cells for syncytia starting at 24 hrs post-transfection. You should see small (3–4 cell syncytia) distributed throughout the monolayer by 20–24 hours (Fig. 3). This indicates that sufficient G protein has been expressed on the cell surface to induce cell-cell fusion (see Note 10).
- After confirming that the pCAGGS-G transfection was successful as indicated by syncytia formation, the next step is to filter the primary recovery supernatants to remove the vaccinia virus and to transfer the filtrates onto the pCAGGS-G transfected cells (Fig. 1B; see Note 11).
- First, label each well of the pCAGGS-G transfected 6-well plate with the name and/or number of the virus that is being recovered.
- Second, remove the plunger from a sterile 3ml syringe and attach a 0.22 µm Millex-GS syringe filter onto the syringe. You can pre-assemble all the syringe-filters that will be needed, or do this one at a time. Use a single syringe-filter for each supernatant.
- Third, remove the medium from one of the pCAGGS-G transfected plates at a time to prevent the cells from drying out, then collect the supernatant from one of the primary recovery wells from Step #1 above with a pipette and add to the syringe.
- Filter the supernatant (see Note 12) directly onto the appropriate well of the new 6-well plate. Note that the ΔL-GFP supernatant does not need to be filtered and transferred since this is used only to determine the efficiency of the recovery transfection by the expression of GFP.
- Place the plate(s) with the recovery supernatants in a 37°C 6%–8% CO2 incubator and incubate for 24 to 72 hours.
- To determine the efficiency of the primary transfection, aspirate the supernatant from the ΔL-GFP control wells and rinse the wells with 2 ml PBS. Remove the PBS, add ~2ml Fix solution and incubate for ~20 min to inactive the vaccinia virus. Remove the Fix solution and rinse with PBS-glycine, then examine the cells for GFP expression using a fluorescence microscope. If between 10–20% of the cells fluoresce green (Fig. 4) due to the expression of GFP, then the chances that the primary recovery was successful is good. If there are fewer than 5% GFP-positive cells in the ΔL-GFP recovery wells, then the support recovery plasmids may need to be reoptimized. The primary recovery plate can then be discarded in the biological hazardous waste.
- The new plate(s) containing the filtered recovery supernatants should be monitored daily for the development of VSV-induced cytopathic effects (CPE), which are seen as areas (or foci) of rounded, refractile cells (Fig. 5). Generally cell rounding will be obvious by 48 hr, but may be observed by 24 hr if the recovery is very efficient.
- When 40% – 100% of the cells show signs of VSV-induced CPE, the supernatants can be harvested. Ideally it is best to wait until most of the cells are showing signs of VSV CPE, however, if the recovery is inefficient (for example only one infectious particle is generated in the recovery) then the percentage of cells showing signs of CPE after 72 h may be low (e.g. ~40%). Do not incubate more that 72 h as virus will lose infectivity. Generally, CPE is evident after 24 – 36 hours.
- Collect the supernatants and transfer to a sterile conical centrifuge tube. Remove cells and cell debris by centrifugation for 10 min at ~450 × g.
- Aliquot the cleared supernatants into appropriately labeled sterile freezer vials/tubes and store frozen at −80°C. The supernatants can be stored at −80°C indefinitely.
- Determine the titer, plaque-purify, and generate a large working stock as described below.
Figure 3.
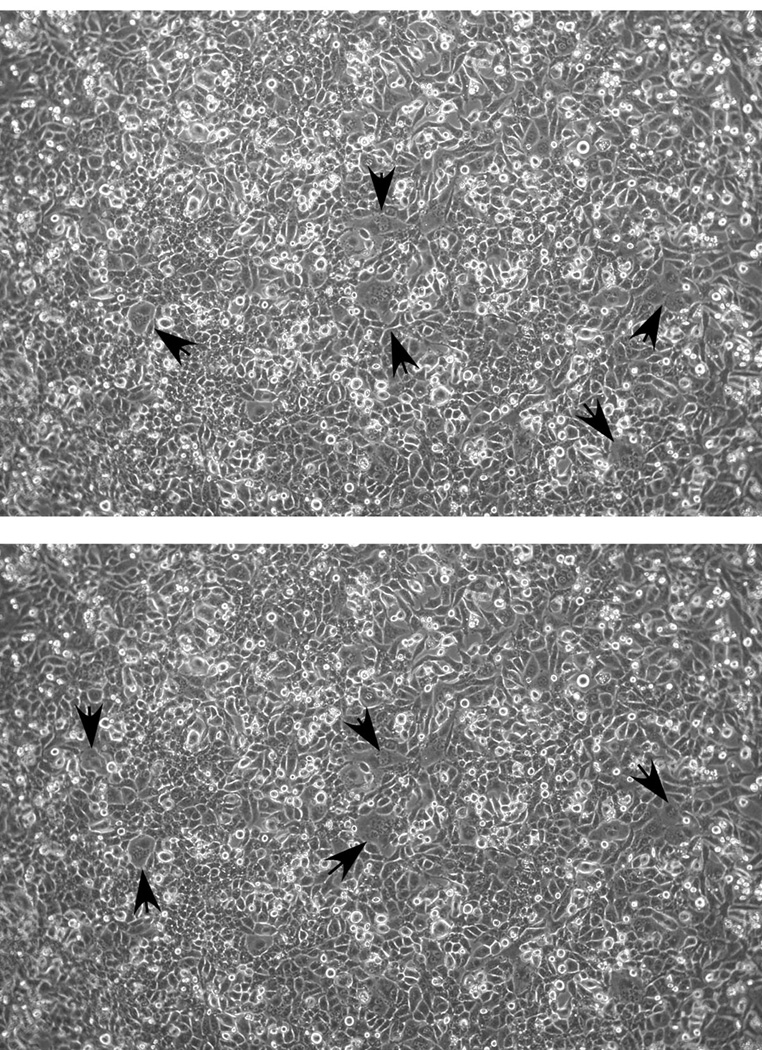
Phase contrast images of two fields of cells at 22 hours post-transfection with pCAGGS-G. Arrowheads indicate regions of small syncytia beginning to form. Images were captured and adjusted to optimize cell visualization as described for Fig. 2.
Figure 4.
Immunofluorescence/phase contrast micrograph of ΔL-GFP transfected cells 24 hours post-transfection. Cells were imaged using low-level brightfield illumination and fluorescence was achieved by excitation using a FITC-filter set on a Zeiss Axiophot using Axiovision software. Quantification of GFP-positive cells revealed that 21.5% of the cells were replicating ΔL-GFP.
Figure 5.
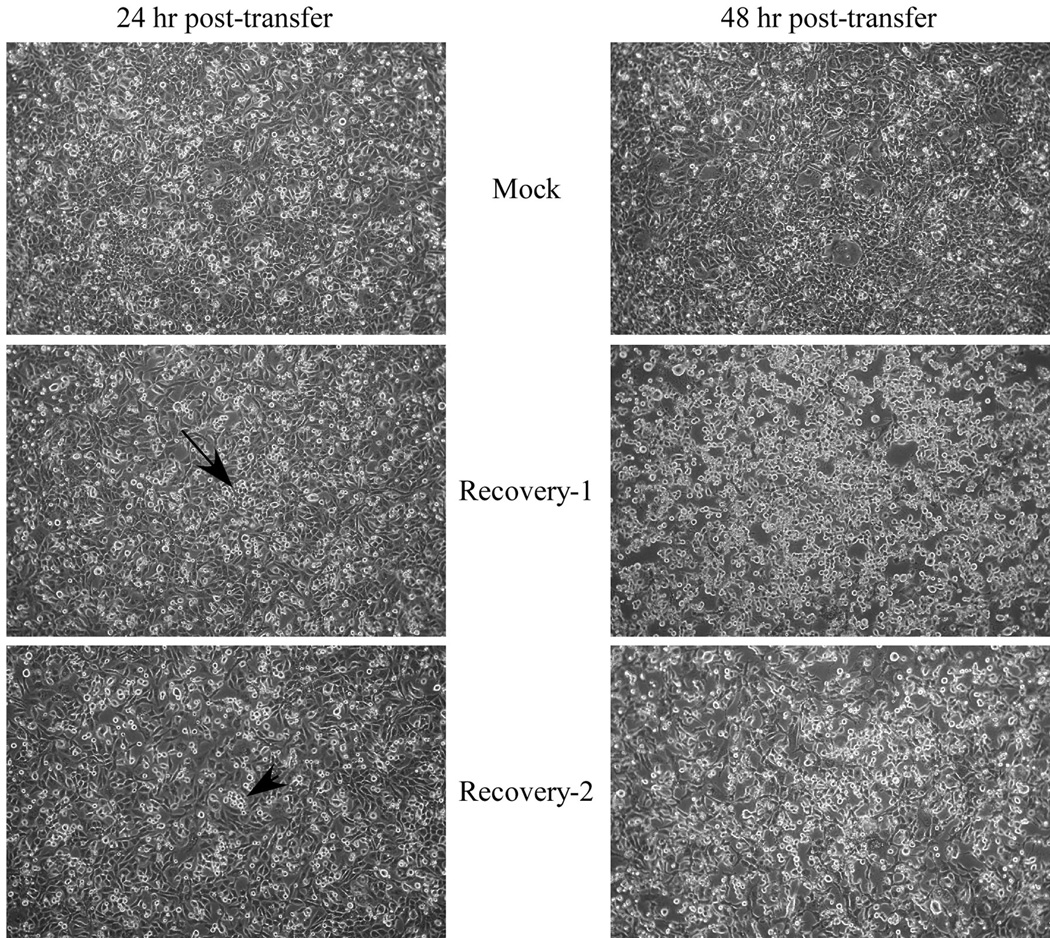
Phase contrast images of cells 24 or 48 hours post-transfer of two independent supernatants from a recovery experiment. The cells had been transfected with pCAGGS-G 24 hours prior to transfer of 0.2 µ filtered recovery supernatants. The top panels show cells after transfer of the supernatant from a culture that was infected with vvT7 and transfected with only the support plasmids. The middle and lower panels are from two independent recovery experiments. The middle panel has a small area of infected cells (arrow) near the middle of the image which are showing VSV-induced CPE. The lower panel also has a small, but much less obvious area of CPE (arrowhead). By 48 hours cells that were infected with the two recovery trial supernatants show extensive cell rounding, characteristic of VSV-induced CPE while the mock culture (top, right panel) does not.
3.4. Plaque Purification and Titering of G*ΔG-VSV
To ensure homogeneity the recovered viruses should be plaque-purified. The following procedure describes a general method for titering G*ΔG-VSV by plaque assay. Dilutions that generate well-isolated plaques can be used to obtain plaque isolates. Plaque purification also ensures that there is no residual vaccinia virus that may have carried over during the initial ΔG-VSV amplification after filtering.
- Transfect 1 × 10 cm dish of BHK cells with pCAGGS-G using Lipofectamine as follows.
- Replace growth medium on cells with ~8 ml Opti-MEM and put cells back into incubator.
- Add 4 ml Opti-MEM to a 15 ml polystyrene tube, then add 16 µg pCAGGS-G and vortex gently.
- In another tube add 4 ml Opti-MEM and then 80 µl Lipofectamine, vortex briefly and gently.
- Immediately add the lipid mix to the DNA-OptiMEM mix and gently invert 5–6 times.
- Incubate 20 min at r.t.
- Remove the Opti-MEM from the cells and add the transfection mix directly to cells.
- Rock the plate 2–3 times and then incubate for 4 hrs at 37°C in a 6%–8% CO2 incubator.
- After 4 hrs replace the transfection medium with D-5 and put back in the incubator.
- Approximately 30 hrs post-transfection, trypsinize the cells and divide into two 6-well plates. Let the cells attach for at least 6 hrs, but not more that 18 hrs since the cell density will get too high.
- Make 10-fold serial dilutions of the virus stock up to 10−7 using SF-DMEM as diluent.
- Remove the medium from the 6-well plates, add 0.3ml of SF-DMEM to each well and then add 100 µl of the serial dilutions to the appropriately labeled well.
- Rock the plates manually every 10 minutes, or place on a platform rocker in a CO2 incubator and adsorb the virus for 60 min at 37°C.
- Leave the inoculum on the cells and overlay directly with agar-overlay (D-5 containing 0.9% agar).
- Let the overlay solidify and then move the plates to the 37°C incubator.
- Incubate the plates until plaques are visible (typically 24 hrs, but may be longer depending on cell density at time of infection).
- For plaque isolates choose plaques that are well separated from other plaques.
- Use a sterile Pasteur pipette and insert into the agar all the way to the bottom of the plate to remove the plaque and agar plug.
- Put the agar plug into 0.5 ml D-5 and let virus elute 1–2 hours at 4°C.
- Store the eluted plaque isolate at −80°C, or use an aliquot directly to amplify the virus to make a P1 (first passage) stock.
3.5. Amplification G*ΔG-VSV Plaque Isolates and Generation of G*ΔG-VSV Working Stocks
The following procedure is essentially the same as that used to pseudotype heterologous envelope proteins onto ΔG-VSV, except that to generate working stocks; a) the virus is pseudotyped with VSV-G, b) it is performed at a larger scale, and c) the infection is done at a lower multiplicity to prevent accumulation of defective-interfering (DI) particles. Investigators may request an aliquot of the G*ΔG-VSV working stock from my laboratory rather than recovering the virus from plasmids. Approximately 0.5 ml of a working stock will be provided to laboratories that make this request. After receiving an aliquot of the working stock it will be necessary to produce additional working stocks by amplifying an aliquot of the G*ΔG-VSV P1 stock on cells transfected with pCAGGS-G.
- Begin with a 10 cm dish of BHK cells. The cells must not be overgrown and must be evenly distributed over the plate (see Note 7) at ~85% confluency.
- Aspirate the growth medium and replace with 10 ml Opti-MEM. Incubate the cells for approximately 20 min at 37°C in a 6–8% CO2 incubator while prepare the transfection mix is prepared. The cells can be incubated for as long as 60 min, but 20 min is recommended.
- Immediately prepare the DNA:liposome transfection cocktail in the biosafety cabinet in the following manner:
- Add 4 ml Opti-MEM to a 15 ml polystyrene tube, then add 16 µg pCAGGS-G and vortex gently.
- In another tube add 4 ml Opti-MEM and then 80 µl Lipofectamine, vortex briefly and gently.
- Immediately add the lipid mix to the DNA-OptiMEM mix and gently invert 5–6 times.
- Incubate 20 min at r.t.
- Remove the Opti-MEM and add the transfection mix directly to cells.
- Rock the plate 2–3 times and then incubate for 4 hrs at 37°C in a 6%–8% CO2 incubator.
- After 4 hrs replace the transfection medium with D-5 and put back in the incubator.
- Incubate for 24–30 hours. Evaluate the transfection efficiency by examining cells for syncytia (multi-nucleated fused cells) starting at 24 hours post-transfection.
- By 24–30 hours, the cells should have become confluent and there should be between 3 and 10 syncytia per microscope field using a 10X objective. At this point, the plates are ready to be infected.
- To amplify virus from a plaque isolate, remove the medium from the transfected cells, add 4 ml SF-DMEM and then add 0.1 ml of the eluted plaque. This will correspond to ~1–5 × 105 infectious units (IU). For amplification of a virus stock, infect using a multiplicity of no more than 0.1 (~1 × 106 IU). Adsorb for 1–2 hours and then add 8 ml D-5 directly to the plate (e.g. do not remove the inoculum).
- Incubate overnight and then examine the cells for the development of VSV-induced CPE.
- Harvest the supernatant when most of the cells show signs of VSV-induced CPE.
- Transfer the supernatant to a sterile conical tube and centrifuge for 10 min at ~450 × g.
- The cleared supernatants are then aliquoted into appropriately labeled sterile eppendorf tubes or screw-top freezing vials and stored frozen at −60°C to −80°C.
3.6. Pseudotyping ΔG-VSV with Heterologous Envelope Proteins
To successfully generate ΔG-VSV pseudotypes using heterologous (e.g. non-VSV) envelope proteins, the cDNA for the desired envelope glycoprotein should be cloned into a high-level expression vector such as pCAGGS (Niwa et al., 1991). Other factors that may affect the efficiency of pseudotype production include: a) the rate of transport of the envelope glycoprotein to the cell surface, b) accumulation of the envelope protein on the plasma membrane [e.g. envelope proteins that are rapidly endocytosed often produce low amounts of pseudotyped particles], c) the type of plasma membrane sub-domain to which the envelope protein localizes [e.g. proteins that accumulate in cholesterol-rich rafts do not pseudotype well with VSV (Johnson et al., 1998)].
Most studies that utilize ΔG-VSV pseudotypes require only small amounts of the pseudotypes to be produced. For example, many studies will examine virus infectivity on a panel of cells to identify susceptible vs. non-susceptible cell types. The titers of ΔG-VSV pseudotypes are often high (e.g. >1 × 107 IU/ml), therefore 2 ml of pseudotypes produced in a 35 mm dish or single well of a 6-well plate is sufficient for most studies.
The pseudotyping procedure can be adapted also for high-throughput screening of antiviral entry inhibitors as described by Porotto et al (Porotto et al., 2007). In this assay cells expressing the heterologous envelope protein are infected at a low multiplicity and the virus is allowed to grow in a multicycle fashion such that replication and spread is dependent on multiple rounds of pseudotyping within the culture. This is essentially what occurs when titering rVSV-ΔG viruses as described in Section 3.4, except the heterologous glycoprotein, rather than VSV G protein, is expressed.
The following procedure outlines the method used for small-scale pseudotype production. If larger volumes are needed, then the procedure can be modified similar to that used to amplify ΔG-VSV stocks (Section 3.5), except that the heterologous envelope glycoprotein would be expressed. Pseudotypes are stable if stored in DMEM containing at least 5% FBS. Alternatively, if the protein profile of the pseudotyped virus needs to be examined by staining of an SDS-polyacrylamide gel with Coomassie blue or silver stain the virus can be grown in serum-free medium, pelleted through a 20% sucrose cushion by centrifugation at 100,000 × g for 35 min and then resuspended in 10 m_M_ Tris-150 m_M_ NaCl containing 10% sucrose (TN-sucrose). Most pseudotypes are stable for weeks to months without significant loss of infectivity in TN-sucrose if stored at −80°C.
- Plate BHK-21 cells so they will be ~80%–90% confluent the next day (see Note 7).
- The next morning the cells should look flat and be evenly distributed without clumps. Replace the growth medium with Opti-MEM and incubate for ~20 minutes at 37°C while transfection mix is being prepared.
- To prepare the transfection mix, add the appropriate volume of Opti-MEM to a sterile polystyrene tube and then add the appropriate amount of plasmid encoding the heterologous envelope protein and vortex gently.
- In a separate tube add the appropriate volume of Opti-MEM and the appropriate amount of Lipofectamine and vortex gently to mix.
- Immediately add the lipid mix to the DNA-Opti-MEM mix and gently invert 5–6 times.
- Incubate for 20 min at r.t.
- Remove the Opti-MEM from the cell monolayer and add the transfection mix directly to cells.
- Rock the plate 2–3 times and then incubate for 4 hrs at 37°C.
- After transfecting for ~4 hrs, replace the transfection medium with D-5 and incubate for ~24–30 hrs.
- The following day the cells should have become confluent. Examine the cells beginning ~24 hrs post-transfection and infect when the monolayer reaches confluency, or when the monolayer begins to show CPE due to expression of the envelope protein.
- Infect the transfected cells at a multiplicity of ~3–5 to ensure every cell is infected. Incubate for approximately 24 – 30 hrs, or until the majority of cells show VSV-induced CPE and then collect the supernatant. Clarify the supernatant by low-speed centrifugation (1,320 × g for 10 min) and use immediately or store aliquots at −80°C.
Acknowledgements
The methods described in this manuscript were developed with support from NIH grant GM-53726 and the efforts of numerous graduate students and post-doctoral fellows including Brenda Fredericksen, Elizabeth Stillman, Clint Robison, Himangi Jayakar, Jeetendra Eswaraka, Hideki Tani, Makiko Watanabe and Chad Mire.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1
The FBS used for cell culture does not need to be heat-inactivated.
2
The BHK-21 cells that are used are not those available from the ATCC. The cells from ATCC are very fibroblast-like. Instead the BHK-21 clone WI-2 cells are more epithelial-like. These cells have much higher transfection efficiencies which appear to be one of the critical factors that influence the efficiency of virus recovery from plasmids.
3
When growing bacteria transformed with pVSV plasmids containing the full-length VSV cDNA, do not grow for more than 14 hours since this will dramatically reduce the yield.
4
The bands should be visible under ordinary room lighting as a dark red band. If there is a large amount of sheared bacterial genomic DNA, this may appear as a band above the lower supercoiled plasmid band. The sheared genomic DNA should be removed first by side-puncture if present.
5
The vaccinia-T7 stocks can be frozen and thawed approximately 4 times without significant loss of infectivity.
6
No solidifying medium such as agarose or methylcellulose is necessary since vaccinia virus is very cell-associated. Distinct plaques will form on cells grown in liquid medium.
7
When plating, ensure that the cells are evenly distributed in the well or on the dish. Use cells that have not been allowed to overgrow at any time, not even once. Generally, cells used for pseudotyping should be passaged every day to ensure that they do not overgrow and will produce a homogeneous cell suspension after trypsinizing. It is important that the cells are not clumpy after plating. They must sit down homogeneously. To evenly distribute the cells move the plate back and forth and side to side multiple times to achieve a very even monolayer. Do not swirl the plate since this causes the cells to accumulate in the middle of the well or dish and this will adversely affect cell transfection and pseudotype production.
8
The BHK-21 clone WI-2 cells grow best in a 6%–8% CO2 environment. All cell culture work used for virus recover, pseudotyping, and virus entry characterization is performed in 6%–8% CO2 incubator.
9
It may be necessary to determine the optimal TransfectACE:DNA ratio for “in-house” prepared TransfectACE. Typically, between 3 to 5µl of TransfectACE per µg of plasmid DNA results in high-efficiency transfections.
10
VSV G protein is a pH-dependent fusion protein, meaning that low pH (∼ pH 6.3–6.1) induces a conformational change in G protein which results in activation of its membrane fusion activity. However, when G protein is expressed at sufficiently high levels on the cell surface of certain cell types (such as BHK-21), a fraction of the G protein trimers “flip” to the low-pH conformation and can induce cell-cell fusion in the absence of a low pH trigger.
11
VSV is sufficiently small to pass through a 0.22µm filter; however, the vaccinia virus will be retained by the filter. This step should remove all of the vaccinia virus, but caution should be used because occasionally some vaccinia will pass through the membrane, possibly due to membrane rupture.
12
Pass the supernatant through the filter very slowly since the high hydrostatic pressure obtained using a 3ml syringe can rupture the filter.
References
- Abe K, Nozaki A, Tamura K, Ikeda M, Naka K, Dansako H, Hoshino HO, Tanaka K, Kato N. Tandem repeats of lactoferrin-derived anti-hepatitis C virus peptide enhance antiviral activity in cultured human hepatocytes. Microbiol. Immunol. 2007;51:117–125. doi: 10.1111/j.1348-0421.2007.tb03882.x. [DOI] [PubMed] [Google Scholar]
- Fredericksen BL, Whitt MA. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J. Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier RW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc.Natl.Acad.Sci.USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S, Mizutani T, Saijo M, Matsuyama S, Miyajima N, Taguchi F, Itamura S, Kurane I, Morikawa S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005;86:2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- Fukushi S, Watanabe R, Taguchi F. Pseudotyped vesicular stomatitis virus for analysis of virus entry mediated by SARS coronavirus spike proteins. Methods Mol. Biol. 2008;454:331–338. doi: 10.1007/978-1-59745-181-9_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Wen Z, Wang X, Hu S, Liu Y, Kong X, Chen H, Bu Z. Generating vesicular stomatitis virus pseudotype bearing the severe acute respiratory syndrome coronavirus spike envelope glycoprotein for rapid and safe neutralization test or cell-entry assay. Ann. NY Acad. Sci. 2006;1081:246–248. doi: 10.1196/annals.1373.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glende J, Schwegmann-Wessels C, Al-Falah M, Pfefferle S, Qu X, Deng H, Drosten C, Naim HY, Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381:215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanika A, Larisch B, Steinmann E, Schwegmann-Wessels C, Herrler G, Zimmer G. Use of influenza C virus glycoprotein HEF for generation of vesicular stomatitis virus pseudotypes. J. Gen. Virol. 2005;86:1455–1465. doi: 10.1099/vir.0.80788-0. [DOI] [PubMed] [Google Scholar]
- Huang AS, Palma EL, Hewlett N, Roizman R. Pseudotype formation between enveloped RNA and DNA viruses. Nature. 1974;252:743–745. doi: 10.1038/252743a0. [DOI] [PubMed] [Google Scholar]
- Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar HR, Jeetendra E, Whitt MA. Rhabdovirus assembly and budding. Virus Research. 2004;106:117–132. doi: 10.1016/j.virusres.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Jayakar HR, Whitt MA. Identification of two additional translation products from the matrix (M) gene that contribute to vesicular stomatitis virus cytopathology. J Virol. 2002;76:8011–8018. doi: 10.1128/JVI.76.16.8011-8018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeetendra E, Robison CS, Albritton LM, Whitt MA. The membrane-proximal domain of vesicular stomatitis virus G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 2002;76:12300–12311. doi: 10.1128/JVI.76.23.12300-12311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Rodgers W, Rose JK. A plasma membrane localization signal in the HIV-1 envelope cytoplasmic domain prevents localization at sites of vesicular stomatitis virus budding and incorporation into VSV virions. Virology. 1998;251:244–252. doi: 10.1006/viro.1998.9429. [DOI] [PubMed] [Google Scholar]
- Kaariainen L, Gomatos PJ. A kinetic analysis of the synthesis in HK21 cells of RNAs specific for Semliki Forest virus. J.Gen.Virol. 1969;5:251–265. doi: 10.1099/0022-1317-5-2-251. [DOI] [PubMed] [Google Scholar]
- Kaimori A, Kanto T, Kwang Limn C, Komoda Y, Oki C, Inoue M, Miyatake H, Itose I, Sakakibara M, Yakushijin T, Takehara T, Matsuura Y, Hayashi N. Pseudotype hepatitis C virus enters immature myeloid dendritic cells through the interaction with lectin. Virology. 2004;324:74–83. doi: 10.1016/j.virol.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Klas SD, Robison CS, Whitt MA, Miller MA. Adjuvanticity of an IL-12 fusion protein expressed by recombinant deltaG-vesicular stomatitis virus. Cellular Immunology. 2002;218:59–73. doi: 10.1016/s0008-8749(02)00575-0. [DOI] [PubMed] [Google Scholar]
- Lee BH, Yoshimatsu K, Araki K, Okumura M, Nakamura I, Arikawa J. A pseudotype vesicular stomatitis virus containing Hantaan virus envelope glycoproteins G1 and G2 as an alternative to hantavirus vaccine in mice. Vaccine. 2006;24:2928–2934. doi: 10.1016/j.vaccine.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Lefrancois L, Lyles DS. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology. 1982;121:157–167. [PubMed] [Google Scholar]
- Matsuura Y, Tani H, Suzuki K, Kimura-Someya T, Suzuki R, Aizaki H, Ishii K, Moriishi K, Robison CS, Whitt MA, Miyamura T. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology. 2001;286:263–275. doi: 10.1006/viro.2001.0971. [DOI] [PubMed] [Google Scholar]
- Miller MA, Lavine CL, Klas SD, Pfeffer LM, Whitt MA. Recombinant replication-restricted VSV as an expression vector for murine cytokines. Protein Expression & Purification. 2004;33:92–103. doi: 10.1016/j.pep.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Mire CE, Dube D, Delos SE, White JM, Whitt MA. Glycoprotein-dependent acidification of vesicular stomatitis virus enhances release of matrix protein. J. Virol. 2009;83:12139–12150. doi: 10.1128/JVI.00955-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Ogino M, Ebihara H, Lee BH, Araki K, Lundkvist A, Kawaoka Y, Yoshimatsu K, Arikawa J. Use of vesicular stomatitis virus pseudotypes bearing hantaan or seoul virus envelope proteins in a rapid and safe neutralization test. Clin. Diagn. Lab. Immunol. 2003;10:154–160. doi: 10.1128/CDLI.10.1.154-160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma K, Matsuura Y, Tatsuo H, Inagaki Y, Nakamura M, Yamamoto N, Yanagi Y. Analysis of the molecules involved in human T-cell leukaemia virus type 1 entry by a vesicular stomatitis virus pseudotype bearing its envelope glycoproteins. J. Gen. Virol. 2001;82:821–830. doi: 10.1099/0022-1317-82-4-821. [DOI] [PubMed] [Google Scholar]
- Perez M, Watanabe M, Whitt MA, de la Torre JC. N-terminal domain of Borna disease virus G (p56) protein is sufficient for virus receptor recognition and cell entry. J Virol. 2001;75:7078–7085. doi: 10.1128/JVI.75.15.7078-7085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porotto M, Carta P, Deng Y, Kellogg GE, Whitt M, Lu M, Mungall BA, Moscona A. Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. J. Virol. 2007;81:10567–10574. doi: 10.1128/JVI.01181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Publicover J, Ramsburg E, Rose JK. A single-cycle vaccine vector based on vesicular stomatitis virus can induce immune responses comparable to those generated by a replication-competent vector. J Virol. 2005;79:13231–13238. doi: 10.1128/JVI.79.21.13231-13238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison CS. Microbiology & Immunology, Vol. Ph.D. Memphis: University of Tennessee Health Science Center; 2001. Assembly and fusion-potentiation domains of the vesicular stomatitis virus glycoprotein and their utility in the development of cell-type-targeted rhabdoviral vectors; p. 157. [Google Scholar]
- Robison CS, Whitt MA. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 2000;74:2239–2246. doi: 10.1128/jvi.74.5.2239-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha MN, Tanaka A, Jinno-Oue A, Shimizu N, Tamura K, Shinagawa M, Chiba J, Hoshino H. Formation of vesicular stomatitis virus pseudotypes bearing surface proteins of hepatitis B virus. J. Virol. 2005;79:12566–12574. doi: 10.1128/JVI.79.19.12566-12574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, Johnson JE, Buonocore L, Rose JK. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection [see comments] Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- Stillman EA, Rose JK, Whitt MA. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J. Virol. 1995;69:2946–2953. doi: 10.1128/jvi.69.5.2946-2953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. A novel system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Oue A, Tanaka A, Shimizu N, Takagi H, Kato N, Morikawa A, Hoshino H. Efficient formation of vesicular stomatitis virus pseudotypes bearing the native forms of hepatitis C virus envelope proteins detected after sonication. Microbes Infect. 2005;7:29–40. doi: 10.1016/j.micinf.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Tatsuo H, Okuma K, Tanaka K, Ono N, Minagawa H, Takade A, Matsuura Y, Yanagi Y. Virus entry is a major determinant of cell tropism of Edmonston and wild-type strains of measles virus as revealed by vesicular stomatitis virus pseudotypes bearing their envelope proteins. J.Virol. 2000a;74:4139–4145. doi: 10.1128/jvi.74.9.4139-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000b;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- Weiss RA, Boettiger D, Murphy HM. Pseudotypes of avian sarcoma viruses with the envelope properties of vesicular stomatitis virus. Virology. 1977;76:808–825. doi: 10.1016/0042-6822(77)90261-6. [DOI] [PubMed] [Google Scholar]
- Witte ON, Baltimore D. Mechanism of formation of pseudotypes between vesicular stomatitis virus and murine leukemia virus. Cell. 1977;11:505–511. doi: 10.1016/0092-8674(77)90068-x. [DOI] [PubMed] [Google Scholar]
- Zavada J, Rosenbergova M. Phenotypic mixing of vesicular stomatitis virus with fowl plaque virus. Acta Virol. 1972;16:103–114. [PubMed] [Google Scholar]