Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses (original) (raw)
Abstract
Genetic variation plays a substantial role in variation in strength, but the underlying mechanisms remain poorly understood. The objective of the present study was to examine the mechanisms underlying variation in muscle mass, a predictor of strength, between LG/J and SM/J strains, which are the inbred progeny of mice selected, respectively, for high and low body weight. We measured weight of five hindlimb muscles in LG/J and SM/J males and females, in F1 and F2 intercrosses, and in an advanced intercross (AI), F34, between the two. F2 mice were genotyped using 162 SNPs throughout the genome; F34 mice were genotyped at 3,015 SNPs. A twofold difference in muscle mass between the LG/J and SM/J mouse strains was observed. Integrated genome-wide association analysis in the combined population of F2 and AI identified 22 quantitative trait loci (QTL; genome-wide P < 0.05) affecting muscle weight on Chr 2 (2 QTL), 4, 5, 6 (7 QTL), 7 (4 QTL), 8 (4 QTL), and 11 (3 QTL). The LG/J allele conferred greater muscle weight in all cases. The 1.5-LOD QTL support intervals ranged between 0.3 and 13.4 Mb (median 3.7 Mb) restricting the list of candidates to between 5 and 97 genes. Selection for body weight segregated the alleles affecting skeletal muscle, the most abundant tissue in the body. Combination of analyses in an F2 and AI was an effective strategy to detect and refine the QTL in a genome-wide manner. The achieved resolution facilitates further elucidation of the underlying genetic mechanisms affecting muscle mass.
Keywords: skeletal muscle size, quantitative trait loci, fine mapping
skeletal muscle is the most abundant tissue in the body and is critical for diverse functions, including locomotion, glucose homeostasis, thermoregulation, and protection of bones and viscera. Muscle strength is an important factor of health and fitness as it inversely associated with the risk for coronary heart disease and stroke (33). Because of the world's growing geriatric population, age-related loss of strength and muscle mass (sarcopenia) is an increasingly pressing problem. Understanding the mechanisms underlying skeletal muscle mass and its function is therefore of increasing importance.
Genetic variation accounts for around half of the variation in strength in humans (1, 5, 11, 30, 32, 34, 37), presumably reflecting various aspects of muscle structure and function. Discovery of the relevant gene variants affecting muscle mass may identify the biological pathways affecting skeletal muscle development and atrophy thereby facilitating identification of novel pharmacological targets. Nevertheless, very few specific genes underlying genetic variation in muscle mass have been identified to date (8, 25, 41, 42).
Animal models such as mice (25, 35), sheep (6), and pigs (41) have been used to study the genetics of muscle mass. In these studies, traits that are largely monogenic have been examined, facilitating discovery of the underlying genes. However, variation in skeletal muscle phenotypes is a polygenic trait in all species and attempts to study the polygenic architecture of muscle variation have been initiated in mice (4, 20, 24), pigs (26, 43), and chicken (18, 27, 29, 40). A number of quantitative trait loci (QTL) have been mapped; however, refinement of these QTL and identification of the underlying genes is a challenging problem.
We used a classical F2 intercross and an advanced intercross (AI) population, which allows mapping of QTL to very small intervals (9), in an effort to accelerate progress in this area. By integrating studies of F2 and AI mice, we have combined the power of an F2 population and the resolution of an AI population. The inbred strains used to make this AI were divergently selected for body size (13, 22) and are thus ideally suited for this experiment. Studies in pigs (26, 43), sheep (8), and mice (19, 20) have demonstrated that genetic variation can exert muscle-specific effects. Therefore, we examined five different muscles in the hindlimb of mice in our populations.
METHODS
Animals and phenotype.
The study was carried out on males and females of the LG/J and SM/J inbred strains, LGSMF1, LGSMF2 intercrosses, and a population of the 34th filial generation of AI, LGSMF34, (see Table 1 for details). This set of mice, recently described by Cheng and colleagues (7), was involved in a behavioral study prior to the analyses of muscle phenotypes. Animals were killed at the age of 94 ± 4 days and their carcasses frozen. All procedures through death were conducted at the University of Chicago and approved by the Institutional Animal Care and Use Committee.
Table 1.
Phenotypes of the LG/J and SM/J strains and their intercross populations
| Sex | Age, days | TA, mg | EDL, mg | Gastroc, mg | Soleus, mg | QF, mg | |
|---|---|---|---|---|---|---|---|
| LG/J | ♂ (n = 18) | 93.9 ± 2.6 | 68.5 ± 4.3 | 12.7 ± 1.0 | 164.8 ± 9.8 | 10.1 ± 0.9 | 321.0 ± 18.6 |
| ♀ (n = 19) | 95.1 ± 1.7 | 56.7 ± 3.0 | 10.0 ± 0.5 | 122.3 ± 7.1 | 8.0 ± 0.8 | 241.4 ± 12.4 | |
| SM/J | ♂ (n = 15) | 90.5 ± 0.9 | 31.2 ± 2.2 | 6.4 ± 0.6 | 80.8 ± 5.7 | 4.8 ± 0.5 | 150.2 ± 9.2 |
| ♀ (n = 10) | 90.7 ± 0.9 | 26.6 ± 2.0 | 5.1 ± 0.5 | 64.1 ± 5.8 | 3.8 ± 0.5 | 117.0 ± 11.0 | |
| LGSMF1 | ♂ (n = 11) | 93.2 ± 1.2 | 58.9 ± 2.9 | 10.2 ± 0.6 | 138.1 ± 7.0 | 8.1 ± 0.5 | 255.3 ± 12.8 |
| ♀ (n = 5) | 91.4 ± 1.3 | 46.5 ± 3.4 | 8.1 ± 0.6 | 100.1 ± 6.0 | 5.9 ± 0.5 | 184.4 ± 13.3 | |
| LGSMF2 | ♂ (n = 252) | 95.3 ± 2.5 | 49.4 ± 6.5 | 9.2 ± 1.2 | 122.8 ± 14.4 | 7.0 ± 1.1 | 227.8 ± 27.9 |
| ♀ (n = 245) | 95.6 ± 2.2 | 38.3 ± 5.9 | 7.0 ± 1.0 | 87.3 ± 11.2 | 5.2 ± 0.8 | 166.6 ± 22.4 | |
| LGSMF34 | ♂ (n = 254) | 93.0 ± 5.4 | 51.8 ± 6.8 | 9.7 ± 1.3 | 128.2 ± 15.8 | 7.7 ± 1.3 | 233.9 ± 29.3 |
| ♀ (n = 238) | 94.1 ± 4.4 | 40.4 ± 5.6 | 7.3 ± 1.0 | 92.4 ± 12.8 | 5.9 ± 1.0 | 172.8 ± 23.1 |
Carcasses were defrosted and tibialis anterior (TA), extensor digitorum longus (EDL), gastrocnemius (Gastroc), soleus, and quadriceps femoris (QF) were dissected under a dissection microscope and weighed on a Mettler AE 50 balance (Mettler Toledo, OH) to the closest 0.1 mg.
Aggregate genetic effects.
Broad sense heritability estimates, H2, were obtained as follows; H2 = (VP − VE)/VP (10), where phenotypic variance, VP, is variance of genetically mixed population (LGSMF2 or LGSMF34) and environmental variance, VE, is variance of LGSMF1 population.
QTL mapping.
Mice were genotyped using SNP markers that were approximately evenly distributed across the genome at 162 (F2) SNPs or 3,015 (AI) single nucleotide polymorphisms (SNPs). We performed genome-wide association analysis in the combined population of the F2 and F34 intercrosses using software developed at the University of Chicago (QTLRel; http://www.palmerlab.org) with some custom modifications. This software allowed us to account for the complex relationships (e.g., sibling, half-sibling, cousins) among the AI mice by using a mixed model as described by Cheng and colleagues (7). Because of sex differences in muscle mass and also the discovery of sex-specific QTL in other studies (19, 20) we explored genetic models where sex was included as either an additive or an interactive covariate.
Significance thresholds.
Muscle-specific thresholds were derived using a gene dropping procedure, which generates genotypic data by simulating meiosis involving individuals in a pedigree, as described in Cheng et al. (7). Briefly, a genotypic data set was simulated, and a genome-wide scan was performed using the simulated genotypic data and the original phenotypic data and using the model for which a significance threshold was being sought. The maximum of the test statistic was recorded. This process was repeated 1,000 times, and the corresponding maxima of the test statistic were pooled to estimate the threshold at a given significance level. This procedure controls the false positive (type I) error rate while preserving correlations among phenotypes and genotypes. We used a similar procedure to calculate significance thresholds on a per-chromosome basis.
There is a substantial difference in muscle size between males and females. Also, a number of loci affecting skeletal growth in a sex-specific manner were reported in this lineage (28). Therefore, to test for the sex-specific effects on muscle weight we compared the model with QTL-by-sex interaction (alternative hypothesis) and the model without the interaction (null hypothesis). The testing procedure was the same as described above, and the testing method was also gene dropping.
In instances of multiple peaks on the chromosome the following criteria were used to discriminate between linked QTL; the peak must be above the genome-wide P < 0.05 threshold and must drop by 3 LOD on each side of that peak (except for the end of the chromosome).
We defined the confidence interval for each QTL as the 1.5-LOD drop off on either side of the peak marker. This interval was expressed in physical map position (Mb) by using the nearest genotyped SNP that flanked the support interval.
Database analyses.
Mouse phenome database (3) was used to identify nonsynonymous SNPs in the QTL regions. The SNP genotypes of the LG/J and SM/J strains were selected from the CGD1 imputed data set (http://phenome.jax.org/db/q?rtn=projects/details&sym=CGD1) (36).
PolyPhen web-based tool (31) was utilized to predict the possible effects of amino acid substitution on the function of a protein (http://genetics.bwh.harvard.edu/pph/). These predictions are based on multiple sequence alignments and functional and structural characterization of the substitution site.
Mouse genome informatics (MGI) database (http://www.informatics.jax.org/) and gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/) were used to examine tissue expression of the genes. The MGI database was also utilized to search for transgenic models influencing skeletal muscle.
Other statistical analyses.
Muscle weight phenotypes approximated normality in both F2 and F34 populations. Strain effect on muscle weight in progenitors was assessed using two-way ANOVA (strain and sex as independent factors). Pearson's product-moment correlations were calculated between different muscle weights within males and females. First principal component (PC1) was extracted from the five muscles and used as additional phenotype in the QTL analysis.
RESULTS
Phenotypic analyses.
There was a more than twofold difference in muscle weight between the LG/J and SM/J strains (P << 0.0001; Table 1), indicating that selection for body size at 60 days of age (13, 22) resulted in segregation of alleles affecting skeletal muscle weight at 90 days. Phenotypic correlations (within-sex) between weights of various muscles were comparable in the LGSMF2 and LGSMF34 populations and ranged between 0.70 and 0.90 (all P < 0.01) in males and 0.69 and 0.91 (P < 0.01) in females.
Broad sense heritability ranged between 0.60 and 0.82, was similar among the five muscles and tended to be somewhat higher in males than females (Table 2). Heritability estimates derived from the LGSMF34 variances were comparable with those based on the LGSMF2.
Table 2.
Broad sense heritability of muscle weight in males and females
| TA | EDL | Gastroc | Soleus | QF | |
|---|---|---|---|---|---|
| LGSMF2 | |||||
| males | 0.80 | 0.76 | 0.76 | 0.82 | 0.79 |
| females | 0.67 | 0.62 | 0.79 | 0.60 | 0.65 |
| LGSMF34 | |||||
| males | 0.82 | 0.81 | 0.80 | 0.88 | 0.81 |
| females | 0.63 | 0.62 | 0.78 | 0.73 | 0.67 |
QTL analysis.
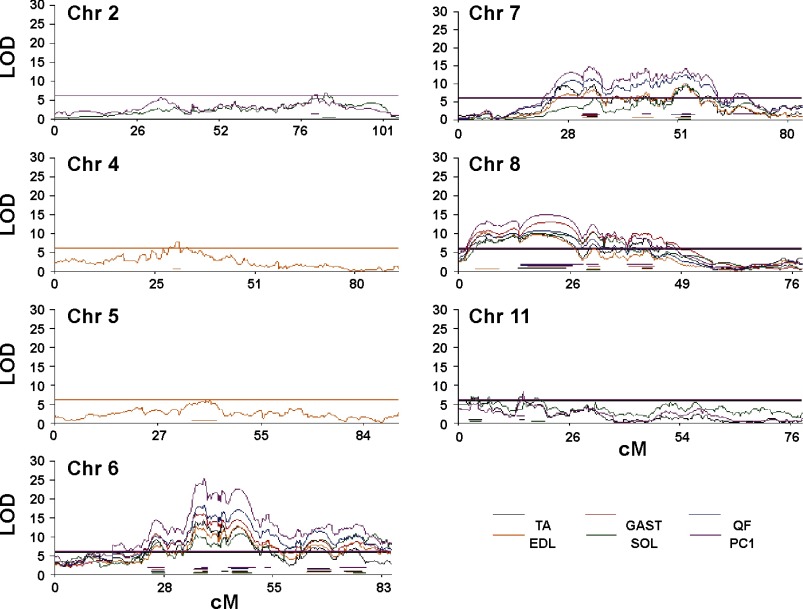
Multiple linked QTL emerged as a characteristic feature of the genetic architecture of muscle weight in the cross of LG/J and SM/J strains. Association analysis of individual muscles and of PC1 in the integrated LGSMF2 and LGSMF34 population mapped 22 QTL (genome-wide P < 0.05) to Chromosomes 2 (2 loci), 4, 5, 6 (7 loci), 7 (4 loci), 8 (4 loci), and 11 (3 loci) (Fig. 1). Characteristics of the genetic architecture of muscle weight are summarized in Table 3. Size of the QTL effect did not differ between males and females statistically significantly. Among the QTL, two (_Skmw28_, _Skmw37_) explained >10% of phenotypic variance, and 10 loci explained 5% or more of phenotypic variation in one or more muscles and/or in PC1. The LG/J allele always conferred greater muscle weight. The QTL support interval (1.5-LOD drop-off) of individual traits ranged between 0.3 and 13.4 Mb (median 3.7 Mb).
Fig. 1.
LOD plots of chromosomes harboring muscle weight quantitative trait loci (QTL) identified by genome-wide association analyses of 5 hindlimb muscles and the first principal component (PC1) in integrated population of LGSMF2 and LGSMF34 mice. Horizontal lines represent genome-wide muscle-specific thresholds (P < 0.05).
Table 3.
Summary of the genetic architecture of muscle weight in the LG/J and SM/J strains
| %Var | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | Chr | Start | End | Size | Low | High | Candidate Genes | QTL | Locus |
| P | 2 | 153.445 | 156.369 | 2.9 | 2.9 | _Ncoa6_TM, _Zfp341_NS | Lbn2.2 | Skmw21 | |
| S | 157.958 | 162.160 | 4.2 | 1.6 | _Slc32a1_TM, _Lbp_NS, _Plcg1_NS | Bdlnq7, Lbn2.2 | Skmw22 | ||
| E | 4 | 53.650 | 55.833 | 2.2 | 3.5 | _Fktn_TM, _Tmem38b_TM | N/A | Skmw23 | |
| E | 5 | 69.173 | 82.557 | 13.4 | 2.3 | _Corin_TM, _Sgcb_TM, _Pdgfra_TM, _Kdr_TM, _Hopx_TM | N/A | Skmw24 | |
| T,G,S,P | 6 | 49.942 | 53.809 | 3.9 | 1.9 | 5.9 | _Hoxa1_TM, _Hoxa2_TM, _Hoxa3_TM | Lbn6.1b | Skmw25 |
| T,E,G,S,Q | 79.491 | 83.038 | 3.5 | 2.8 | 9.7 | _Htra2_SP | N/A | Skmw26 | |
| T | 92.586 | 93.829 | 1.2 | 5.0 | N/A | N/A | Skmw27 | ||
| T,E,S,Q,P | 94.796 | 100.209 | 5.4 | 3.6 | 12.3 | _Foxp1_TM | N/A | Skmw28 | |
| P | 110.596 | 113.993 | 3.4 | 6.9 | Cav3TM, Mtmr14TM, VhlTM , Ttll3_NS, Il17rc_NS_, Fancd2_NS_, Irak2_NS | N/A | Skmw29 | ||
| T,E,G,Q | 127.916 | 128.186 | 0.3 | 3.6 | 4.9 | N/A | N/A | Skmw30 | |
| T,E,S,P | 136.083 | 138.966 | 2.9 | 2.5 | 5.5 | N/A | Lbn6.2 | Skmw31 | |
| T,E,G,Q,P | 7 | 73.789 | 75.508 | 1.7 | 2.5 | 6.1 | _Igf1r_TM | N/A | Skmw32 |
| E,P | 90.115 | 96.666 | 6.6 | 2.9 | 5.6 | _Fes_TM, _Fzd4_TM | Lbn7.1 | Skmw33 | |
| T,E,S,Q,P | 118.012 | 122.071 | 4.1 | 2.5 | 5.7 | _Mrvi1_TM, _Tead1_TM | N/A | Skmw34 | |
| P | 135.755 | 141.073 | 5.3 | 3.8 | Ate1TM, Bub3TM, Fgfr2TM, Hmx3/Hmx2TM, Plekha1GT | N/A | Skmw35 | ||
| E | 8 | 10.522 | 15.505 | 5.0 | 5.1 | N/A | Lbn8.2 | Skmw36 | |
| T,G,Q,P | 31.960 | 38.083 | 6.1 | 7.6 | 10.7 | _Nrg1_TM | Bdywt, Lbn7 | Skmw37 | |
| E,G,S,P | 48.755 | 55.272 | 6.5 | 2.6 | 5.8 | _Hand2_TM, _Wdr17_NS | N/A | Skmw38 | |
| T,E,G,P | 85.897 | 87.972 | 2.1 | 1.6 | 4.7 | _Gipc1_GT, _Cacna1_SP, TM, _Calr_TM, _Dnase2a_TM | N/A | Skmw39 | |
| T,S | 11 | 6.827 | 9.697 | 2.9 | 1.6 | 2.8 | _Igfbp3_TM | Bglq8 | Skmw40 |
| T,G,Q,P | 28.239 | 29.465 | 1.2 | 2.0 | 3.3 | _Efemp1_TM | W6q3 | Skmw41 | |
| S | 31.225 | 35.337 | 4.1 | 1.7 | Hba-a1TM , Kcnmb1TM | W6q3 | Skmw42 |
Consistent with the positive phenotypic correlation between different muscles, the majority of the QTL exerted pleiotropic effects on more than one muscle. Weight of the TA muscle was affected by 12 (together accounting for 54% of phenotypic variance, based on single QTL model estimates), EDL by 12 (45%), Gastroc by 7 (42%), soleus by 9 (19%), and QF by 6 (39%) QTL. The genetic architecture of the PC1 largely overlapped with that of the individual muscles. In addition, PC1 analysis permitted refinement of three additional loci (Table 3). A total of 14 QTL were identified for PC1.
Candidate genes.
The mapping resolution provided by our analyses guided us to a manageable number of genes in many loci; for instance, Skmw27 and Skmw30 harbored fewer than 10 genes and three loci; Skmw23, Skmw32, and Skmw40 harbored fewer than 20. The SNP analysis of all QTL identified ∼30 genes with nonsynonymous polymorphisms between the two strains that were clustered within 10 QTL regions. We examined these genes using the PolyPhen tool (http://genetics.bwh.harvard.edu/pph/), which relies on the sequence and phylogenetic and structural information characterizing the substitution (31). Most of these polymorphisms were predicted to have no effect of the protein function. However, polymorphisms in the zinc finger protein 341, Zfp341, (Skmw21); lipopolysaccharide binding protein, Lbp, phospholipase γ 1, Plcg1 (Skmw22); tubulin tyrosine ligase-like family member 3, Ttll3; interleukin 17 receptor C, Il17rc; Fanconi anemia complementation group D2, Fancd2; interleukin-1 receptor-associated kinase 2, Irak2 (Skmw29); and WD repeat containing protein 17, Wdr17 (Skmw38) genes are predicted to alter protein function. Based on the available microarray data, transcripts of all eight genes are detected in mouse skeletal muscle (GDS2840, GDS2698) providing support for further scrutiny of the candidacy of these genes.
We examined available transgenic models for possible effects on muscle tissue using the MGI database and found that allelic variations (due to spontaneous or induced mutations) in a number of genes from the QTL regions can affect the skeletal muscle (summarized in Table 3). There were no nonsynonymous polymorphisms between the two strains in these genes. However, polymorphisms causing expression variation could be the underlying mechanism of the QTL effects.
DISCUSSION
We carried out the genome-wide mapping of QTL affecting skeletal muscle mass in an F2 and AI population of mice derived from strains selected for high and low body weight at 60 days of age. We hypothesized that such selection would have segregated alleles affecting skeletal muscle weight, the most abundant tissue in the body. Indeed, the LG/J and SM/J strains exhibited a twofold difference in weight of five hindlimb muscles and heritability estimates ranged between 0.60 and 0.88 for the weight of different muscles. We mapped 22 QTL affecting weight of one or more muscles. The LG/J allele conferred an increasing effect in all QTL. The analyses in the integrated F2 and AI population permitted us to achieve a genome-wide fine mapping of the loci to median resolution of 3.7 Mb. In four muscles, the aggregate effect of the identified QTL accounted for between 38 and 59% of phenotypic variation and somewhat less, 19%, in soleus. The underlying mechanism of this contrast between soleus and other muscles is not clear but could be partially due to the fiber type composition differences. Mouse soleus consists of a mixture of type I and IIA fibers, whereas four other tested muscles predominantly consist of type IIA and IIB fibers (47, 49).
Analyses of body and organ weights as well as skeleton and body growth have proven the LG/J × SM/J lineage a suitable model for studying the mechanisms underlying variation in body size components (17, 44). Because of the contribution of muscle tissue to body size and growth some overlap between QTL affecting muscle weight and integrative phenotypes was expected. In accordance with this assumption muscle weight QTL mapped to the similar genomic regions on Chr 2, 4, 6, 7, 8, and 11 to those implicated in body weight and growth traits (17, 44). The comparison with the genetic architecture of the long bone (28) indicated possible muscle/bone pleiotropy of seven loci scattered through chromosomes 2 (Skmw21, Smw22), 6 (Skmw25, Skmw31), 7 (Skmw33), and 8 (Skmw36, Skmw37). Several scenarios can be offered to explain the pleiotropy. Considering a single causative gene per QTL, variation in muscle mass could be secondary to that in bone length. Alternatively, it can be a result of a concomitant influence on both tissues. However, it is also possible that pleiotropy is an outcome of the effects of different genes influencing muscle and bone phenotypes. Identification of the causative genes will be required for understanding of the underlying mechanisms of pleiotropy.
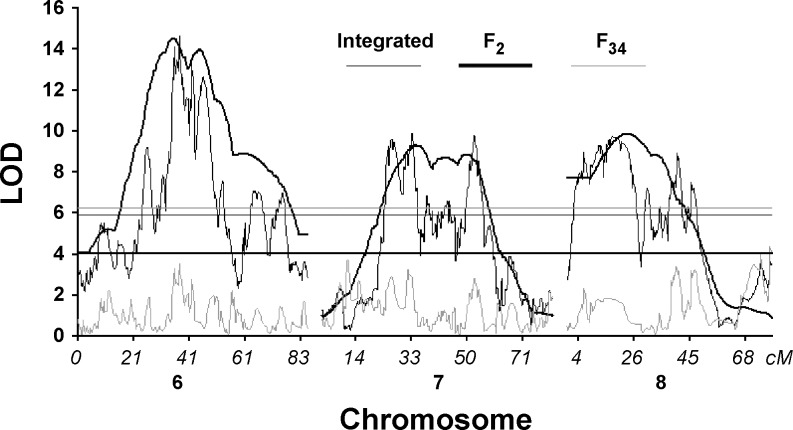
We were able to refine nine loci to <3 Mb size. The strength of the classical QTL mapping in an F2 intercross lies in its ability to identify the entire genetic architecture contributing to phenotypic variation. The refinement of the QTL, however, poses a significant hurdle. Typically, the fine mapping is carried out via congenic strain approach on a locus by locus basis aiming to narrow down the region of interest to a manageable list of candidate genes (38). An advanced intercross is an appealing alternative, particularly for the phenotype with rich genetic architecture, because it allows screening of the entire genome in one study (9). However, the power to detect QTL in AI appeared more limited than in F2s. For instance, none of the QTL affecting TA weight on Chr 6, 7, or 8 in LGSMF2 intercross was replicated in a separate analysis within the LGSMF34 population (Fig. 2). We hypothesize that this is an outcome of the partitioning of QTL into several linked loci. The effect size of fractioned QTL might have been beyond detection power of the available sample size. Partitioning of the QTL has been reported previously (45) and suggested as a possible cause of replication failure in addition to the false positives and the Beavis effect (2). A QTL scan in the integrated F2 and AI population of the same lineage offers a superior strategy of data analysis by increasing the sample size, combining the detection power of the former and accuracy of the latter. To illustrate the aspect of accuracy, we continue an example presented in Fig. 2. A 1.5-LOD drop off interval of the TA weight QTL on Chr 6 spanned between 54.1 and 104.8 Mb (50.7 Mb long) in the LGSMF2 population. The QTL broke down into six smaller loci in the combined analysis of the LGSMF2 and LGSMF34 populations (Supplemental Table S1).1 The size of the refined loci ranged between 0.3 and 5.4 Mb, a >10-fold reduction. Importantly, three out of six loci were outside of the LGSMF2 1.5-LOD drop off interval, and thus, candidacy of the genes within those regions would not even be considered in the analysis of solely F2 population.
Fig. 2.
LOD plots of chromosomes 6, 7, and 8 of QTL analyses carried out on TA muscle weight in the integrated population of the F2 and F34 intercrosses and separately. Horizontal line represents empirically determined genome-wide 5% thresholds for integrated, F2, and F34 populations.
Mapping resolution led us to a manageable list of the candidate genes in most of the loci. Among them we identified eight genes, Zfp341, Lbp, Plcg1 Ttll3, Il17rc, Fancd2, Irak2, and Wdr17, with nonsynonymous polymorphisms that are likely to influence function of the proteins between the two strains. These genes are expressed in muscle tissue and thus are good candidates for follow-up analysis. Several other genes in the QTL regions are likely functional candidates as well. For instance, mutation in mitochondrial protease Omi coding gene, Htra2 (Skmw26), leads to muscle wasting (16), overexpression of caveolin-3, Cav3, gene results in fiber phenotypes resembling Duchenne muscle dystrophy (12), and disruption of type 1 IGF receptor gene, Igf1r (Skmw32), causes severe suppression of muscle growth (21), whereas Tead1 (Skmw34) (39), Tead4 (Skmw30) (48), and Mef2a (Skmw32) (48) genes are transcription factors abundantly expressed in skeletal muscle. The genomic sequence of the LG/J and SM/J strains and the transcriptome analysis will provide additional leverage for further nomination of candidate genes underlying those QTL.
The overall genetic architecture was richer than that of any individual muscle indicating that not all muscles were affected (or at least were not affected equally) by the same locus. Six out of 22 loci were found influencing only EDL (three loci), soleus (two loci), or TA (one locus) muscle. Specificity of the Skmw22 and Skmw42 loci to soleus might be related to the distinct fiber type composition of this muscle (a mixture of type 1 and type 2A fibers) compared with the other examined muscles (predominantly a mixture of type 2A and type 2B fibers) as well as its function (soleus is a frequently used postural muscle). The effects specific to EDL (Skmw23 and Skmw24 loci) but not, for instance, to its synergist TA were also reported in the intercross between the C57BL/6J and DBA/2J strains (19, 20), although the underlying mechanisms of the specificity between muscles of similar type remain unclear. Specificity of the loci affecting TA (Skmw27) and EDL (Skmw36) muscles most likely is an artifact arising from inability to discriminate between the linked loci according to the selected criteria, i.e., 3 LOD drop between the peaks, in other muscles (Fig. 1). Muscle-specific effects were observed earlier in various species (15, 19, 20, 23, 26, 43), and a gene underlying variation of such nature was recently identified in sheep (46). Identification of the genes and pathways affecting different muscles in a variable manner will help in understanding myopathies that often affect some but not all muscles (14).
In conclusion, we identified and refined the genetic architecture consisting of 22 QTL affecting variation in muscle weight in the LG/J × SM/J lineage. The resolution and effect size of the QTL are favorable for a fruitful pursuit of the underlying genes.
GRANTS
National Institute of Health Grants AR-052879 to A. Lionikas, AR-056280 to D. A. Blizard, DA-024845 to A. A. Palmer, MH-020065 to A. A. Palmer. Schweppe Foundation Career Development Award to A. A. Palmer.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
Table S1
ACKNOWLEDGMENTS
We acknowledge the valuable technical and scientific assistance of Greta Sokoloff, Kaitlin Samocha, Tanya Cebollero, Michaelanne Munoz, Claudia Wing, Sima Lionikaitė, and Viktė Lionikaitė. We also thank the laboratory of Dr. James Cheverud for making the F33 mice available to us.
Footnotes
1
The online version of this article contains supplemental material.
REFERENCES
- 1.Arden NK, Spector TD. Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J Bone Miner Res 12: 2076– 2081, 1997. [DOI] [PubMed] [Google Scholar]
- 2.Beavis W. The power and deceit of QTL experiments: lessons from comparative QTL studies. Proceedings of the Forty-Ninth Annual Corn & Sorghum Industry Research Conference, Washington, DC: American Seed Trade Association, 1994, p. 250–266. [Google Scholar]
- 3.Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica 122: 71– 74, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F(2) variance of growth and obesity in DU6i × DBA/2 mice. Genome Res 10: 1941– 1957, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmelli D, Reed T. Stability and change in genetic and environmental influences on hand-grip strength in older male twins. J Appl Physiol 89: 1879– 1883, 2000. [DOI] [PubMed] [Google Scholar]
- 6.Charlier C, Segers K, Karim L, Shay T, Gyapay G, Cockett N, Georges M. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet 27: 367– 369, 2001. [DOI] [PubMed] [Google Scholar]
- 7.Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 2010. May 3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockett NE, Jackson SP, Shay TL, Nielsen D, Moore SS, Steele MR, Barendse W, Green RD, Georges M. Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proc Natl Acad Sci USA 91: 3019– 3023, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199– 1207, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falconer DS, Mackay TFC. Components of variance. In: Introduction to Quantitative Genetics (4 ed.). Harlow: Pearson Education, 1996, p. 122–125. [Google Scholar]
- 11.Frederiksen H, Gaist D, Petersen HC, Hjelmborg J, McGue M, Vaupel JW, Christensen K. Hand grip strength: a phenotype suitable for identifying genetic variants affecting mid- and late-life physical functioning. Genet Epidemiol 23: 110– 122, 2002. [DOI] [PubMed] [Google Scholar]
- 12.Galbiati F, Volonte D, Chu JB, Li M, Fine SW, Fu M, Bermudez J, Pedemonte M, Weidenheim KM, Pestell RG, Minetti C, Lisanti MP. Transgenic overexpression of caveolin-3 in skeletal muscle fibers induces a Duchenne-like muscular dystrophy phenotype. Proc Natl Acad Sci USA 97: 9689– 9694, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodale H. A study of the inheritance of body weight in the albino mouse by selection. J Hered 29: 101– 112, 1938. [Google Scholar]
- 14.Griggs RC, Markesbery WR. Distal myopathies. In: Myology (2 ed.), edited by Engel AG, Franzini-Armstrong C: McGraw-Hill, 1994, p. 1246–1257. [Google Scholar]
- 15.Jackson SP, Miller MF, Green RD. Phenotypic characterization of Rambouillet sheep expressing the callipyge gene: II. Carcass characteristics and retail yield. J Anim Sci 75: 125– 132, 1997. [DOI] [PubMed] [Google Scholar]
- 16.Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 425: 721– 727, 2003. [DOI] [PubMed] [Google Scholar]
- 17.Kenney-Hunt JP, Vaughn TT, Pletscher LS, Peripato A, Routman E, Cothran K, Durand D, Norgard E, Perel C, Cheverud JM. Quantitative trait loci for body size components in mice. Mamm Genome 17: 526– 537, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Lagarrigue S, Pitel F, Carre W, Abasht B, Le Roy P, Neau A, Amigues Y, Sourdioux M, Simon J, Cogburn L, Aggrey S, Leclercq B, Vignal A, Douaire M. Mapping quantitative trait loci affecting fatness and breast muscle weight in meat-type chicken lines divergently selected on abdominal fatness. Genet Sel Evol 38: 85– 97, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lionikas A, Blizard DA, Gerhard GS, Vandenbergh DJ, Stout JT, Vogler GP, McClearn GE, Larsson L. Genetic determinants of weight of fast- and slow-twitch skeletal muscle in 500-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol Genomics 21: 184– 192, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Lionikas A, Blizard DA, Vandenbergh DJ, Glover MG, Stout JT, Vogler GP, McClearn GE, Larsson L. Genetic architecture of fast- and slow-twitch skeletal muscle weight in 200-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol Genomics 16: 141– 152, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75: 59– 72, 1993. [PubMed] [Google Scholar]
- 22.MacArthur J. Genetics of body size and related characters. I. Selection of small and large races of the laboratory mouse. Am Nat 78: 142– 157, 1944. [Google Scholar]
- 23.Malek M, Dekkers JC, Lee HK, Baas TJ, Prusa K, Huff-Lonergan E, Rothschild MF. A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. II. Meat and muscle composition. Mamm Genome 12: 637– 645, 2001. [DOI] [PubMed] [Google Scholar]
- 24.Masinde GL, Li X, Gu W, Davidson H, Hamilton-Ulland M, Wergedal J, Mohan S, Baylink DJ. Quantitative trait loci (QTL) for lean body mass and body length in MRL/MPJ and SJL/J F(2) mice. Funct Integr Genomics 2: 98– 104, 2002. [DOI] [PubMed] [Google Scholar]
- 25.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387: 83– 90, 1997. [DOI] [PubMed] [Google Scholar]
- 26.Milan D, Bidanel JP, Iannuccelli N, Riquet J, Amigues Y, Gruand J, Roy PL, Renard C, Chevalet C. Detection of quantitative trait loci for carcass composition traits in pigs. Genet Sel Evol 34: 705– 728, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadaf J, Pitel F, Gilbert H, Duclos MJ, Vignoles F, Beaumont C, Vignal A, Porter TE, Cogburn LA, Aggrey SE, Simon J, Le Bihan-Duval E. QTL for several metabolic traits map to loci controlling growth and body composition in an F2 intercross between high- and low-growth chicken lines. Physiol Genomics 38: 241– 249, 2009. [DOI] [PubMed] [Google Scholar]
- 28.Norgard EA, Jarvis JP, Roseman CC, Maxwell TJ, Kenney-Hunt JP, Samocha KE, Pletscher LS, Wang B, Fawcett GL, Leatherwood CJ, Wolf JB, Cheverud JM. Replication of long-bone length QTL in the F9–F10 LG,SM advanced intercross. Mamm Genome 20: 224– 235, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park HB, Jacobsson L, Wahlberg P, Siegel PB, Andersson L. QTL analysis of body composition and metabolic traits in an intercross between chicken lines divergently selected for growth. Physiol Genomics 25: 216– 223, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Perusse L, Lortie G, Leblanc C, Tremblay A, Theriault G, Bouchard C. Genetic and environmental sources of variation in physical fitness. Ann Hum Biol 14: 425– 434, 1987. [DOI] [PubMed] [Google Scholar]
- 31.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30: 3894– 3900, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed T, Fabsitz RR, Selby JV, Carmelli D. Genetic influences and grip strength norms in the NHLBI twin study males aged 59–69. Ann Hum Biol 18: 425– 432, 1991. [DOI] [PubMed] [Google Scholar]
- 33.Silventoinen K, Magnusson PK, Tynelius P, Batty GD, Rasmussen F. Association of body size and muscle strength with incidence of coronary heart disease and cerebrovascular diseases: a population-based cohort study of one million Swedish men. Int J Epidemiol 38: 110– 118, 2009. [DOI] [PubMed] [Google Scholar]
- 34.Silventoinen K, Magnusson PK, Tynelius P, Kaprio J, Rasmussen F. Heritability of body size and muscle strength in young adulthood: a study of one million Swedish men. Genet Epidemiol 32: 341– 349, 2008. [DOI] [PubMed] [Google Scholar]
- 35.Szabo G, Dallmann G, Muller G, Patthy L, Soller M, Varga L. A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome 9: 671– 672, 1998. [DOI] [PubMed] [Google Scholar]
- 36.Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome 19: 199– 208, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomis MA, Van Leemputte M, Maes HH, Blimkie CJ, Claessens AL, Marchal G, Willems E, Vlietinck RF, Beunen GP. Multivariate genetic analysis of maximal isometric muscle force at different elbow angles. J Appl Physiol 82: 959– 967, 1997. [DOI] [PubMed] [Google Scholar]
- 38.Tomida S, Mamiya T, Sakamaki H, Miura M, Aosaki T, Masuda M, Niwa M, Kameyama T, Kobayashi J, Iwaki Y, Imai S, Ishikawa A, Abe K, Yoshimura T, Nabeshima T, Ebihara S. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat Genet 2009. May 24 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39.Tsika RW, Schramm C, Simmer G, Fitzsimons DP, Moss RL, Ji J. Overexpression of TEAD-1 in transgenic mouse striated muscles produces a slower skeletal muscle contractile phenotype. J Biol Chem 283: 36154– 36167, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uemoto Y, Sato S, Odawara S, Nokata H, Oyamada Y, Taguchi Y, Yanai S, Sasaki O, Takahashi H, Nirasawa K, Kobayashi E. Genetic mapping of quantitative trait loci affecting growth and carcass traits in F2 intercross chickens. Poult Sci 88: 477– 482, 2009. [DOI] [PubMed] [Google Scholar]
- 41.Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425: 832– 836, 2003. [DOI] [PubMed] [Google Scholar]
- 42.Varga L, Szabo G, Darvasi A, Muller G, Sass M, Soller M. Inheritance and mapping of Compact (Cmpt), a new mutation causing hypermuscularity in mice. Genetics 147: 755– 764, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varona L, Ovilo C, Clop A, Noguera JL, Perez-Enciso M, Coll A, Folch JM, Barragan C, Toro MA, Babot D, Sanchez A. QTL mapping for growth and carcass traits in an Iberian by Landrace pig intercross: additive, dominant and epistatic effects. Genet Res 80: 145– 154, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Vaughn TT, Pletscher LS, Peripato A, King-Ellison K, Adams E, Erikson C, Cheverud JM. Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genet Res 74: 313– 322, 1999. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Le Roy I, Nicodeme E, Li R, Wagner R, Petros C, Churchill GA, Harris S, Darvasi A, Kirilovsky J, Roubertoux PL, Paigen B. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res 13: 1654– 1664, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White JD, Vuocolo T, McDonagh M, Grounds MD, Harper GS, Cockett NE, Tellam R. Analysis of the callipyge phenotype through skeletal muscle development; association of Dlk1 with muscle precursor cells. Differentiation 76: 283– 298, 2008. [DOI] [PubMed] [Google Scholar]
- 47.Wirtz P, Loermans HM, Peer PG, Reintjes AG. Postnatal growth and differentiation of muscle fibres in the mouse. I. A histochemical and morphometrical investigation of normal muscle. J Anat 137: 109– 126, 1983. [PMC free article] [PubMed] [Google Scholar]
- 48.Yasunami M, Suzuki K, Ohkubo H. A novel family of TEA domain-containing transcription factors with distinct spatiotemporal expression patterns. Biochem Biophys Res Commun 228: 365– 370, 1996. [DOI] [PubMed] [Google Scholar]
- 49.Yu F, Gothe S, Wikstrom L, Forrest D, Vennstrom B, Larsson L. Effects of thyroid hormone receptor gene disruption on myosin isoform expression in mouse skeletal muscles. Am J Physiol Regul Integr Comp Physiol 278: R1545– R1554, 2000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1