Multiplexed quantitative real-time PCR to detect 22q11.2 deletion in patients with congenital heart disease (original) (raw)
Abstract
22q11.2 Deletion syndrome (22q11.2 DS) [DiGeorge syndrome type 1 (DGS1)] occurs in ∼1:3,000 live births; 75% of children with DGS1 have severe congenital heart disease requiring early intervention. The gold standard for detection of DGS1 is fluorescence in situ hybridization (FISH) with a probe at the TUPLE1 gene. However, FISH is costly and is typically ordered in conjunction with a karyotype analysis that takes several days. Therefore, FISH is underutilized and the diagnosis of 22q11.2 DS is frequently delayed, often resulting in profound clinical consequences. Our goal was to determine whether multiplexed, quantitative real-time PCR (MQPCR) could be used to detect the haploinsufficiency characteristic of 22q11.2 DS. A retrospective blinded study was performed on 382 subjects who had undergone congenital heart surgery. MQPCR was performed with a probe localized to the TBX1 gene on human chromosome 22, a gene typically deleted in 22q11.2 DS. Cycle threshold (Ct) was used to calculate the relative gene copy number (rGCN). Confirmation analysis was performed with the Affymetrix 6.0 Genome-Wide SNP Array. With MQPCR, 361 subjects were identified as nondeleted with an rGCN near 1.0 and 21 subjects were identified as deleted with an rGCN near 0.5, indicative of a hemizygous deletion. The sensitivity (21/21) and specificity (361/361) of MQPCR to detect 22q11.2 deletions was 100% at an rGCN value drawn at 0.7. One of 21 subjects with a prior clinical (not genetically confirmed) DGS1 diagnosis was found not to carry the deletion, while another subject, not previously identified as DGS1, was detected as deleted and subsequently confirmed via microarray. The MQPCR assay is a rapid, inexpensive, sensitive, and specific assay that can be used to screen for 22q11.2 deletion syndrome. The assay is readily adaptable to high throughput.
Keywords: congenital heart defects, DiGeorge syndrome, copy number variant, TBX1
22q11.2 deletion syndrome (22q11.2 DS), also known as DiGeorge syndrome type 1 (DGS1), is estimated to be the most prevalent inheritable genetic deletion syndrome, occurring in ∼1 in 3,000 live births (38). This autosomal dominant disease is characterized by a wide range of clinical phenotypes. While overall phenotypic penetrance for 22q11.2 DS is very high among deleted individuals, there is often marked phenotypic differences between related individuals with identical 22q11.2 microdeletions (2, 9, 11, 12, 23, 27). Clinical abnormalities are diverse and include psychosocial, cognitive, developmental delay, psychiatric illnesses, palatal abnormalities, parathyroid insufficiency, growth retardation, immune defects, congenital heart defects (CHDs), renal anomalies, and abnormal craniofacial findings (23, 24). The early diagnosis of 22q11.2 DS is critically important to effectively treat this disorder. However, despite the obvious phenotypic abnormalities in many patients with 22q11.2 DS, the average age of diagnosis for a person with this syndrome is 6–7 yr (27). Because of its highly variable phenotype, 22q11.2 DS has also been known by a variety of other names (24, 34).
Currently, the definitive diagnosis of a patient with suspected 22q11.2 DS relies on the fluorescent in situ hybridization (FISH) cytogenetic test using a probe localized to the TUPLE1 gene. This probe lies within the ∼3-Mb typical deleted region (TDR) found on chromosome 22q11.2. Unfortunately, as the standard probe used by most clinical cytogenetic reference labs, it often misses 22q11.2 DS subjects who carry smaller microdeletions within the TDR (46). Several laboratories have successfully utilized techniques such as multiplex ligation-dependent probe amplification (MLPA) and quantitative PCR (qPCR) to delineate proximal and distal breakpoints and to detect duplications in this region (17, 37, 46, 47). It is widely believed that one or more of the several dozen genes located within the 22q TDR are responsible for the heterogeneity of 22q11.2 DS (13). One of the earliest candidate genes thought to be responsible for the clinical phenotype of DGS1 was the T-box transcription factor, TBX1; T-box genes encompass a large family of highly conserved transcription factors that are critical in the regulation of a variety of developmental processes (7, 19). A mouse model for the 22q11.2 deletion has shown that haploinsufficiency of the Tbx1 gene is responsible for aortic arch defects in mice. _Tbx1_-null mice develop DGS1 phenotypes such as parathyroid, thymus, and facial abnormalities (22). Taken together, these findings suggest that the human TBX1 gene is a major genetic determinant in the phenotype of 22q11.2 DS (48).
The primary goal of this research was to determine whether multiplexed quantitative real-time PCR (MQPCR) had the potential to be used as a high-throughput screening tool for the specific detection of 22q11.2 DS. To date the majority of phenotypes associated within the TDR are attributed to microdeletions (2, 9, 11, 12, 23, 27). Although a number of microduplications have been reported, associated phenotypes appear to be more variable (28, 30, 36, 37). CHDs are present in 75% of FISH-positive 22q11.2 DS subjects and are the major cause of mortality in >80% of cases; therefore, a secondary focus of this screening assay is to enable timely detection of deleted newborns so that interventions for the heart can be implemented (16, 29, 33, 38). Strict criteria exist for a targeted population-based screen (32). While commercially available MLPA kits have the advantage of looking at multiple probes over the entire 22q11.2 region at higher resolution and whole genome microarrays are capable of delineating copy number variants (CNVs) over a patient's entire genome, because of their higher cost per patient and longer start-to-finish time frame, these technologies are currently more suitable as confirmatory tests in a high-throughput workflow (6, 14, 17, 31, 37, 43).
We developed an MQPCR assay using a single nucleotide polymorphism (SNP)-based target probe near TBX1 to determine relative gene copy number (rGCN). We compared this assay with a commercially available CNV assay located within TBX1. The assays were tested on subjects with congenital heart disease, a population highly enriched for 22q11.2 DS. Confirmation of deletion status on chromosome 22 was performed for all study subjects with the Affymetrix 6.0 SNP array.
MATERIALS AND METHODS
Patient samples.
Peripheral blood was drawn from 382 enrolled patients undergoing cardiac surgery at Children's Hospital of Wisconsin (CHW) (Milwaukee, WI) between March 2007 and June 2008 in strict adherence to an Institutional Review Board (IRB)-approved protocol. IRB of Children's Hospital of Wisconsin-approved protocols were the following: the Congenital Heart Disease Tissue Bank (CHW 06/229, GC 300) and Assay for Identification of DiGeorge Syndrome in Children with Congenital Heart Defects (CHW 08/97, GC 699). A chart review of these patients was performed to identify their CHD diagnosis, presence of a DGS1 diagnosis, phenotypes associated with DGS1, and pertinent diagnostic testing such as FISH. At the time of analysis, clinical diagnoses of DGS1 and/or diagnostic testing for DGS1 using FISH were unknown to the analyst as part of the experimental design.
Genomic DNA extraction.
Genomic DNA was obtained from peripheral blood by standard protocols for DNA isolation from Roche Diagnostics, Promega Biotech (Wizard), and Qiagen (Gentra Puregene). Purified genomic DNA was resuspended in 1.0 mM Tris·HCl pH 8.0 and 0.1 mM EDTA. DNA was quality tested by optical density (OD) 260/280 nm ratios, quantified by UV spectrophotometry, and stored at −20°C for this study.
SNP assay target design.
SNPs that localized within and near the TBX1 gene on human chromosome 22, and which did not lie in an area of common copy number variation, were chosen as potential candidates for target assay design (18). SNP rs9618682 (G/A), located 4,660 bp upstream of the TBX1 gene, fit the aforementioned criteria. A commercially available TaqMan SNP assay specific for this region was purchased from Applied Biosystems (ABI; Foster City, CA). It consists of a single tube of forward primer, reverse primer, and two minor groove binding (MGB) probes. Allele probe 1 is conjugated with the reporter dye 6-carboxyfluorescein (6-FAM) and allele probe 2 with the fluorophore VIC. Genomic DNA isolated from ∼300 healthy control subjects was tested by using cycling temperature capillary electrophoresis (CTCE) to determine the homozygosity of the SNP site (heterozygosity = 0, the minor allele did not exist) (4).
TaqMan primers and probe for an internal standard were designed around a homozygous site on human chromosome 19 with Primer Express version 2.0 software (Applied Biosystems). This chromosome was chosen because of its low copy number variation (18). In detail, an 82-bp fragment of the human PTBP1 gene was amplified with forward primer (5′-GTTCCCGTAACTGAAACATCAATG-3′) and reverse primer (5′-AAGGAACCTGAGGATGCTGTGT-3′). Detection was accomplished with a 14-bp NED-labeled DNA probe (5′-CAGGCTCAGCTGGT-3′), which has an emission wavelength of 575 nm, distinct from 6-FAM (518 nm) and VIC (554 nm). This minimizes emission spectrum overlap between the three dyes in a multiplex reaction, thereby reducing signal cross talk. Additionally, amplification efficiency profiles were optimized to have similar log-linear slopes for the experimental and reference targets.
CNV assay target design.
In 2009, commercially available CNV assays became available (ABI). Eighteen FAM-labeled CNV assays targeting TBX1 were tested on a panel of 22q11.2 DS-positive and nondeleted subjects in a blinded fashion. The assay with highest sensitivity and specificity was selected and optimized.
CNV assay ABI Hs_01313390 located within the TBX1 gene on human chromosome 22 was chosen. This assay overlaps intron 11/exon 12 within the TBX1 gene at position 18150843 on chromosome 22 (NCBI Build 36) (20). The TaqMan CNV assays contain a MGB probe labeled with FAM dye and unlabeled primers. A commercially available reference assay was also chosen as the internal standard. This assay detects the Ribonuclease P RNA component H1 (H1RNA) gene (RPPH1) on chromosome 14, cytoband 14q11.2 and utilizes a VIC-labeled TAMRA probe.
Multiplex quantitative real-time PCR assays.
The gene of interest, TBX1, was multiplexed in a single reaction tube with an internal standard for the determination of rGCN. Each sample was amplified in triplicate. In the SNP assay, the 5-μl reaction included 5 ng of genomic DNA template, 1× TaqMan universal PCR master mix, target assay probe and primers at final concentrations of 100 nM and 450 nM, respectively, and internal standard probe and primers at final concentrations of 200 nM and 900 nM, respectively. The reactions were loaded into a 384-well plate (Eppendorf, Westbury, NY), covered with optical adhesive film (ABI), and centrifuged at 2,500 rpm for 2 min at room temperature just before setup. Amplifications were conducted with an ABI HT7900 instrument (ABI). Cycling conditions were 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 92°C for 15 s followed by 60°C for 1 min.
The CNV assay was run according to manufacturer's recommendations with the following modifications: 5 ng of genomic DNA template in a 10-μl reaction containing 1× TaqMan universal PCR master mix, target assay probe and primers, as well as reference assay probe and primers at final concentrations of 250 nM and 900 nM, respectively. Reactions were then handled similarly to the SNP assay.
Data analysis for SNP assay.
Data evaluation was carried out with ABI Prism HT7900 Sequence Detection Software and Microsoft Excel. For each amplification, a computer algorithm calculated the fluorescent signal (ΔRn) generated by the degradation of the hybridized probe and automatically determined the cycle at which each PCR amplification reached a threshold value (Ct) (defaulted to 10 times the standard deviation of the baseline signal). The Ct value is inversely proportional to the log of the number of target copies present in the sample. In addition, the TaqMan universal master mix (ABI) contains the ROX dye as an internal passive reference to which the reporter dye signal can be normalized. Normalization of the reporter dye signal helps increase data precision. Subsequently, the rGCNs of the unknown samples were normalized to an endogenous reference on chromosome 19 and expressed relative to a calibrator sample (normal control TK6 genomic DNA isolated from a human B lymphoblastoid cell line) according to a standard curve (35).
Ct for linear amplification was used to calculate rGCN in the region of the target probe. The following calculations were employed to derive the rGCN:
| ΔΔCt(ddCt)=dCtTK6−dCt Subject Sample | (2) |
|---|
where a nondeleted subject has an rGCN = 1.0 and a subject with a hemizygous deletion has an rGCN = 0.5 (42).
PCR master mixes were prepared for each plate, which introduced a small amount of variation. An average rGCN has been calculated for each assay and was blindly applied to all samples as a normalizing factor.
Data analysis for CNV assay.
Copy Caller software version 1.0 (ABI) was used to analyze the CNV assay. This assay detects the target gene, while the reference detects a gene that is known to be present in two copies in the diploid genome. Relative quantitation was determined with the calibrator sample, TK6 genomic DNA. The Copy Caller software was used to automatically generate the rGCN.
Platform for assessing genomewide copy number variation.
Genetic data was measured via the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA). To reduce the complexity of the genomic DNA (a minimum of 250 ng is required), a digestion with a single restriction enzyme was performed. To achieve complete coverage, two different enzymes (_Sty_I and _Nsp_I) were used. Fragments were ligated with a common set of adaptors. After ligation, the sample was diluted and the complexity of the genome was further reduced via single-primer PCR amplification. This generic primer recognizes the adaptor-ligated fragments and is optimized to select for product sizes ranging between 250 and 2,000 bp. Each sample was set up in quadruplicate, and once amplification was complete products were combined and purified on the Qiagen MinElute 96 UF PCR purification plate. Each fragment was labeled with biotin in which the reaction was catalyzed with terminal deoxynucleotidyl transferase (TdT). Subsequent labeling was performed with an oligonucleotide control reagent, human Cot-1 and herring sperm DNA, tetramethyl ammonium chloride (TMAC), and DMSO. Samples were transferred onto the array and washed for 16–18 h. The Affymetrix Fluidics Station 450 was used to rinse the array, disposing of any unbound DNA products. Staining was performed with streptavidin phycoerythrin (SAPE) stain, followed by an antibody amplification step and a final stain with SAPE. Once completed, each array was scanned with the Scanner 3000. Array-based quality control was assessed by using the 50 control SNPs located on each array, in which 25 control SNPs are typed with a second independent platform (TaqMan, ABI).
After microarray hybridization, samples were tested for standard quality control (QC), including intensity QC and median of absolute values of all pairwise differences (MAPD) QC. Data were acquired with Gene Chip Command Console software, generating sample data intensity files (cel), which were opened with Genotyping Console version 3.0.2 software. Copy number analysis was performed with the HapMap consortium reference model (1), Affymetrix library and annotation files (version NA29), and the following threshold parameters: a minimum of 40 markers and a minimum genomic fragment size of 100 kb.
RESULTS
Chart review.
Review of patients' charts revealed a phenotypically diverse CHD population including many complex lesions as outlined in Table 1 (abbreviations of CHD phenotypes are included).
Table 1.
Breakdown of study subjects by diverse CHD phenotype
| Diagnoses | Subjects | % of Total | Diagnoses | Subjects | % of Total |
|---|---|---|---|---|---|
| Tetralogy of Fallot (TOF) | 37 | 9.7 | Ebstein anomaly | 4 | 1.0 |
| Hypoplastic left heart syndrome (HLHS) | 37 | 9.7 | Aortic insufficiency | 3 | 0.8 |
| Ventricular septal defect (VSD) | Arrhythmia | 3 | 0.8 | ||
| Perimembranous | 28 | 7.3 | Dilated ascending aorta (Marfan syndrome) | 3 | 0.8 |
| Muscular | 4 | 1.0 | Interrupted aortic arch (IAA) | 3 | 0.8 |
| Multiple | 4 | 1.0 | Mitral stenosis | 3 | 0.8 |
| A-V canal (AVC) | 35 | 9.2 | Other thoracic | 3 | 0.8 |
| Single ventricle | 31 | 8.1 | Coronary artery anomaly | 2 | 0.5 |
| Coarctation of the aorta | 29 | 7.6 | Pectus excavatum | 2 | 0.5 |
| Atrial septal defect (ASD) | 28 | 7.3 | Pulmonary stenosis | 2 | 0.5 |
| Aortic stenosis (AS) | 24 | 6.3 | Vascular ring-arch anomaly | 2 | 0.5 |
| Pulmonary atresia (PA) | Cardiac tumor | 1 | 0.3 | ||
| VSD | 14 | 3.7 | Cor triatriatum | 1 | 0.3 |
| IVS | 8 | 2.1 | Double-chamber right ventricle (DCRV) | 1 | 0.3 |
| Transposition of great arteries (TGA) | Pulmonary artery sling | 1 | 0.3 | ||
| D | 20 | 5.2 | Patent foramen ovale (PFO) | 1 | 0.3 |
| L | 1 | 0.3 | Absence of left pulmonary artery (LPA) | 1 | 0.3 |
| Truncus arteriosus (TA) | 15 | 3.9 | Supravalvar mitral ring | 1 | 0.3 |
| Double-outlet right ventricle (DORV) | 14 | 3.7 | Total anomalous pulmonary venous connection | 1 | 0.3 |
| Cardiomyopathy | 8 | 2.1 | Tracheal stenosis | 1 | 0.3 |
| Partial anomalous pulmonary venous return (PAPVR) | 5 | 1.3 | Tricuspid regurgitation (non-Ebstein) | 1 | 0.3 |
In this cohort, 67 patients with phenotypes suggestive of 22q11.2 DS were found to be negative by the FISH assay and were cleared of a DGS1 diagnosis. Twenty-one of the subjects carried the clinical diagnosis of DGS1; however, only 14 had confirmatory FISH (TUPLE1) testing. FISH testing was not performed in the records of the remaining seven subjects.
MQPCR assays.
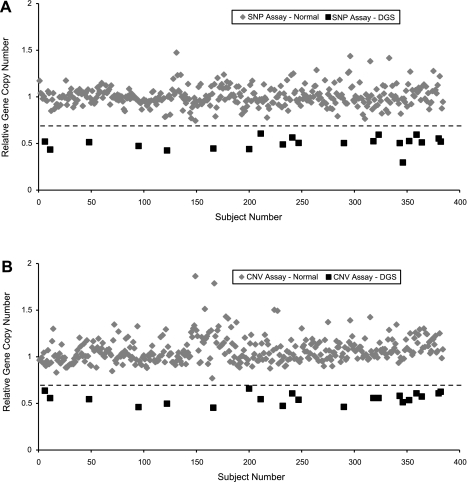
Figure 1 illustrates scatter plots of each subject's rGCN as determined by the SNP assay (Fig. 1_A_) and the CNV assay (Fig. 1_B_). The distribution of subjects by rGCN appeared to naturally separate into two distinct populations. Those that grouped near an rGCN of 0.5 were tentatively labeled as 22q11.2 deletions. Those that grouped near an rGCN of 1.0 were labeled as nondeleted. Both assays identified the same 21 subjects as deletions. Additionally, two duplications were found to occur outside the 95% confidence interval of the nondeleted subjects but were intermixed with outliers from subjects with normal 22q11.2 copies.
Fig. 1.
Scatter plots showing multiplexed quantitative real-time PCR (MQPCR) analyses over the TBX1 gene locus on human chromosome 22 for n = 382 subjects with congenital heart disease. A: single nucleotide polymorphism (SNP) assay. B: copy number variant (CNV) assay. A calculated relative gene copy number (rGCN) of 1.0 indicates a nondeleted subject (2 copies), whereas a rGCN of 0.5 indicates a hemizygous deletion (1 copy). Twenty-one subjects, denoted by squares, were labeled as deleted. The remaining 361 subjects, denoted by diamonds, were nondeleted. The dashed line represents a threshold value of 0.7, where the sensitivity and specificity of both assays is 100%. DGS, DiGeorge syndrome.
Affymetrix confirmation.
Three hundred eighty-two of the subjects met Affymetrix DNA purification standards, allowing microarray confirmation of the assay results.
All 21 subjects recognized as having a deletion from the MQPCR analysis were confirmed to have a 22q11.2 deletion by Affymetrix 6.0 hybridization. Table 2 lists the CHD diagnosis of these 21 subjects and the frequency of deletion for each phenotype. This frequency is in agreement with previously published data (29).
Table 2.
Prevalence of 22q11.2 deletion among specific CHD diagnoses
| Diagnoses | Total Subjects | 22q11.2 Del Subjects | Deletion Frequency, % |
|---|---|---|---|
| TOF | 37 | 7 | 19 |
| TA | 15 | 5 | 33 |
| PA | |||
| VSD | 14 | 3 | 21 |
| IVS | 8 | ||
| VSD | |||
| Perimembranous | 28 | 2 | 7 |
| Muscular | 4 | ||
| Multiple | 4 | ||
| AS | 24 | 1 | 4 |
| DORV | 14 | 1 | 7 |
| IAA | 3 | 1 | 33 |
| Absence of LPA | 1 | 1 | 100 |
| Diagnoses without deletion | 230 | ||
| Total subjects | 382 | 21 | 5 |
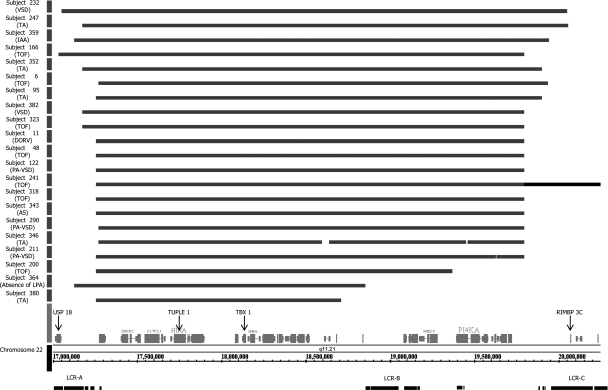
As seen in Fig. 2, the TDR for these subjects ranged in length from 1.5 to 3.0 Mb and localized near low copy number repeats LCR-A and LCR-C (46). Affected genes for all 21 subjects were defined on the 5′-end by the ubiquitin-specific protease 18 gene (USP18) and on the 3′-end by the RIMS-binding protein 3C gene (RIMBP 3C). Interestingly, subject 241 was found to carry a 1.8-Mb distal 22q.11.2 duplication overlapping the same locus as the previously published 22q11.2 distal deletion (3). This duplication was contiguous with the proximal DGS1 deletion found in this subject.
Fig. 2.
Diagram of deletions that mapped to the 22q11.2 region with Affymetrix Genome-Wide SNP 6.0 microarray analysis. Twenty-one subjects were confirmed by gene chip analysis, as illustrated by bars, to carry a deletion in the 22q11.2 region ranging from 1.5 to 3 Mb in length. All subjects diagnosed with DiGeorge syndrome type 1 (DGS1) were confirmed in this manner. The proximal deletion was bounded on the 5′-side by the USP18 gene and on the 3′-end by the RIMPB 3C gene. Both the gene targeted for 22q11.2 FISH analysis (TUPLE1) and the target of the MQPCR probes in the present study (TBX1) are noted in black. A 1.8-Mb distal duplication, located contiguous to the proximal 22q deletion, was noted in subject 241 (extended bar). The low copy number repeat regions (LCR-A, -B, and -C) are shown for reference (46). Congenital heart defect (CHD) phenotypes are indicated in parentheses (see Table 1 for CHD abbreviations).
Two subjects, subjects 170 and 315, were discovered to harbor a 2.6-Mb duplication encompassing the TDR (8). The duplication was defined by the DiGeorge syndrome critical region 6 (DGCR6) gene on the 5′-end and spanned to the proximal side of the RIMBP 3C gene on the 3′-end, within the TDR. It is important to note that neither of these subjects carried a clinical diagnosis of DGS1. However, subject 315 carried a syndromic phenotype, including flat facies, developmental delay, and chronic infections. These characteristics could also be attributed to a large 14-Mb duplication found by microarray analysis on chromosome 11q23.3-q25. Subject 209, with a diagnosis of truncus arteriosus (TA) and no deletion in the TDR, was found to have a 1.2-Mb distal deletion. This subject did not have other phenotypes associated with the diagnosis of DGS1. Also, multiple non-DGS1 subjects were found to have smaller 100- to 500-kb microduplications and deletions within the recombination hot spots of LCR-A and -B (46).
The microarray results demonstrated that the MQPCR assays correctly identified all 14 subjects with a previous positive FISH test. Of the seven remaining patients clinically diagnosed with DGS1 (FISH not performed), MQPCR detected deletions in six (all subsequently confirmed by microarray). The seventh patient (subject 179), phenotyped as “partial DiGeorge,” was found to be negative for deletion of 22q11.2 by MQPCR. This finding was confirmed by microarray. Importantly, one subject previously without a clinical diagnosis of DGS1 (subject 382) was found by MQPCR to be deleted in 22q11.2. The diagnosis of 22q11.2 DS was confirmed by both microarray and subsequent FISH, and then reported to the caregivers and family. The stair-step illustration of the Affymetrix data in Fig. 3 demonstrates the copy number state of subjects 179 and 382. Chromosome 10p13 deletions have also been implicated in phenotypes associated with DGS (10). In the study population, microarray analysis did not detect deletions at this locus.
Fig. 3.
Schematic of Affymetrix Genome-Wide SNP 6.0 microarray data depicting copy number state on human chromosome 22q11.2. A represents a nondeleted control subject, whereas the stair step view shown by B indicates the typical 3-Mb deletion over the 22q11.2 locus of a control subject diagnosed with DGS1. Gene chip analysis of subject 179 (C) confirms the absence of a deletion over the 22q region, despite a clinical phenotype of “partial DiGeorge.” Likewise, a previously unknown deletion in subject 382 (D) was confirmed.
Statistical analysis.
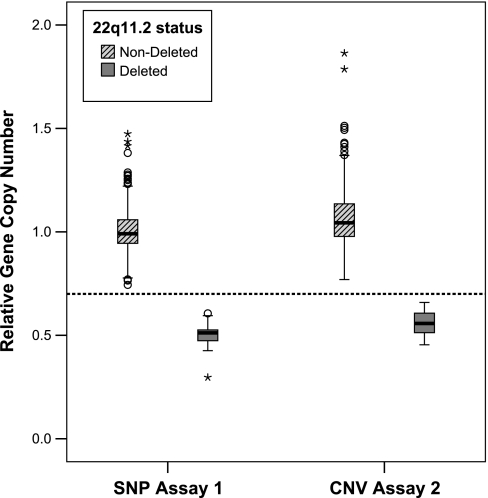
A box plot was used to explore the statistical significance of the MQPCR assay data. Figure 4 illustrates that both assays reliably distinguished between subjects with 22q11.2 DS and those subjects that were not deleted. For both assays, a cut point value of 0.7 can be drawn, with a dashed line, to indicate a threshold where there is no crossover between the deleted and nondeleted subjects.
Fig. 4.
Box plot assessing the statistical significance of the 2 MQPCR assays: SNP vs. CNV assay. The dark line in the middle of the box represents the median. The box represents the middle 50%, called the interquartile range (IQR). The whiskers or lines extending from the boxes represent 1.5× IQR. The open circles represent outliers that are >1.5× IQR from the median, and the asterisks represent extremes that are >2× IQR from the median. The median values for rGCN for both assays were in good agreement with theoretical values of 1.0 for a nondeleted subject representing 2 genomic copies and 0.5 representing a deleted subject with 1 genomic copy. For both assays, a cut point value of 0.7 can be drawn, with a dashed line, to indicate a threshold where there is no crossover between deleted and nondeleted.
As seen in Table 3, a median value of 0.99 (SNP) vs. 1.04 (CNV) for rGCN was found for nondeleted subjects and 0.51 (SNP) vs. 0.56 (CNV) for deleted subjects. These values closely reflect the theoretical rGCNs of 1.0 for homozygous individuals and 0.5 for those with hemizygous deletions.
Table 3.
Statistical summary of MQPCR assays
| Assay | Deletion Status | n | Mean | SD | Median | Maximum | Minimum |
|---|---|---|---|---|---|---|---|
| SNP assay | Nondeleted | 361 | 1.00 | 0.11 | 0.99 | 0.74 | 1.47 |
| Deleted | 21 | 0.50 | 0.07 | 0.51 | 0.30 | 0.61 | |
| CNV assay | Nondeleted | 361 | 1.07 | 0.14 | 1.04 | 0.77 | 1.86 |
| Deleted | 21 | 0.55 | 0.06 | 0.56 | 0.45 | 0.66 |
To assess the best cut point to screen for 22q11.2 deletion syndrome, we applied receiver operating curve (ROC) analysis to the assay data (Table 4). In this analysis, the sensitivity is plotted against the specificity and the aim is to choose a value that best optimizes desired characteristics. For example, if the aim of a test is to screen a population, it is desirable to identify 100% of the cases. Therefore, a high sensitivity is desired even at the expense of specificity. For the SNP assay, 100% of the cases of 22q11.2 DS would be identified with a threshold value between 0.6 and 0.75, while for the CNV assay 100% of the cases would be identified with a threshold value between 0.65 and 0.8. It is important to note that there was no crossover of values for deleted and nondeleted subjects in either assay. In fact, a value of 0.7 would work as a cut point for screening for 22q11.2 deletions using either assay.
Table 4.
Receiver operating curve analysis of MQPCR assays
| Assay | Positive if Greater or Equal to | Sensitivity | Specificity |
|---|---|---|---|
| SNP assay | 0.600 | 1.000 | 0.952 |
| 0.675 | 1.000 | 1.000 | |
| 0.754 | 0.997 | 1.000 | |
| CNV assay | 0.648 | 1.000 | 0.952 |
| 0.714 | 1.000 | 1.000 | |
| 0.800 | 0.997 | 1.000 |
Figure 5 graphically represents a comparison of the SNP assay and the CNV assay. In the scatter plot, if the two assays agree they will lie on the line Y = X. Both MQPCR assays were able to accurately distinguish the population of 22q11.2 DS deleted subjects from those that were nondeleted.
Fig. 5.
Statistical comparison of the two MQPCR assays by scatter plot. If the two assays agree with one another, they will lie on the line Y = X.
DISCUSSION
Although 22q11.2 DS is the most common inheritable genetic deletion syndrome, the diagnosis is frequently delayed. Subspecialists evaluating patients for developmental delay, speech abnormalities, growth delay, and immunodeficiencies are often the first clinicians to make the diagnosis of DGS1. Although >75% of infants diagnosed with 22q11.2 DS have a CHD, >50% of DGS1 patients diagnosed later in life were also found to have a CHD (16, 27, 45). Given the phenotypic variability, a multidisciplinary approach is necessary to effectively treat these patients. With early diagnosis, there are targeted interventions for 22q11.2 DS individuals that can significantly improve/prevent many of the associated comorbidities and possible mortality. These interventions include surgery for congenital heart and palate defects, intravenous immunoglobulin for immunodeficiency/autoimmune defects, thymic transplantation for complete absence of normal T cells, growth hormone replacement for pituitary/thyroid defects, calcium replacement for parathyroid insufficiency, gastroenterology/feeding team interventions for palatal and gastrointestinal structural abnormalities, and individual educational plans for subjects with speech, motor, cognitive, or language delays (38).
Early clinical intervention in the treatment of DGS1 patients is both clinically important and effective; therefore a sensitive, specific, and cost-effective screening test would be extremely beneficial. Quantitative real-time PCR is a method that has enabled researchers to measure changes in nucleic acid levels in DGS1 (46). MQPCR has the advantage of having an internal standard within an assay, eliminating interassay variability, and has been previously applied in a small number of patients to confirm deletions and duplications of the dystrophin gene with TaqMan chemistry (44). The purpose of this study was to see whether an MQPCR assay could be applied as a rapid, high-throughput, primary screen for 22q11.2 DS and ultimately identify patients who may require echocardiogram screening for CHD. Two MQPCR assays were tested, one developed by this group and one commercially available. Both assays demonstrated robust and similar results.
In this population of phenotypically diverse subjects with congenital heart disease, the 22q11.2 deletion was detected in 5% of cases. One patient was not previously known to carry the deletion and was identified by MQPCR and confirmed by FISH and microarray analysis. Another patient who was previously characterized as partial DiGeorge was found not to have the deletion. Previous studies have shown that many forms of CHD are observed in DGS1 patients including tetralogy of Fallot (TOF), interrupted aortic arch (IAA) (type B), TA, and ventricular septal defect (VSD) (5, 9, 15, 23–25, 29, 39, 40). 22q11.2 DS was detected in our CHD population with phenotypes of TOF, TA, pulmonary atresia (PA), VSD, double-outlet right ventricle (DORV), IAA, and aortic stenosis (AS), and although the numbers were small, deletion was not detected with other common CHD diagnoses such as transposition of great arteries (TGA), hypoplastic left heart syndrome (HLHS), and atrial septal defect (ASD). These results confirm previous observations that these CHDs may arise through different molecular pathways (15, 21, 26, 29, 41). This underscores the need for physicians to have a high level of suspicion for 22q11.2 DS, in particular CHD phenotypes, and emphasizes the need for all 22q11.2 DS individuals to be thoroughly screened for heart defects (29).
There are strict guidelines for population-based screens, both for the primary screening tool used as well as the secondary confirmation test employed (32). The MQPCR assay is an attractive primary screening tool for high-throughput workflow because of its low cost and short time frame. Currently, results are reported in less than 4 h at an expense of less than $2.00 per patient. Subsequently, those identified as positive would be rescreened, tested for DNA quality, and verified with FISH or higher-resolution methods such as MLPA or microarray, which offer the advantage of delineating boundaries of deletions/duplications and are not limited in their ability to detect atypical CNVs (6, 14, 31, 43). Commercially available MLPA kits offer the advantage of detecting both deletions and duplications over the entire 22q11.2 region using multiple probe sets; unfortunately, the cost and time of MLPA would not meet the criteria for a population-based, primary screening tool specifically targeting patients with 22q11.2 DS and cardiac defects (17, 32, 37). While whole genome microarrays would be a practical choice for confirming copy number anomalies across a patient's entire genome, they are not suitable as a primary screening tool because of the expense and time required per patient on a large population basis (6, 14, 31, 43).
The majority of CNVs reported in the literature for 22q11.2 are due to microdeletions in the TDR; therefore the MQPCR assay target probe was designed very specifically over TBX1 to capture as many deleted individuals from the population as possible. The probe employed identified all of the large deletions (>100 kb) in the 22q11.2 region for our cohort. Notably, as a screening tool, use of a single target gene may miss a small percentage of patients with atypical deletions in the 22q11.2 region and may not reliably detect duplications with high specificity (3). Although duplications may have a milder phenotypic effect, the ability to detect them in addition to deletions to assess their associations to congenital cardiac disease would be an attractive feature (28, 30, 36, 37). Additional gene targets within the same assay would likely help identify those patients by offering higher resolution and potentially increase the specificity of detecting duplications, but will require further investigation.
In summary, MQPCR can reliably detect haploinsufficiency of TBX1 at 22q11.2 with high sensitivity and specificity. The approach is rapid—after DNA extraction turnaround time to results for 90 subjects is 4 h. Finally, the approach is cost-effective at less than $2.00 per patient. The approach is very amenable to high-throughput analysis and shows tremendous promise for population-based screening.
GRANTS
This work was supported by funding from the National Institute of Child Health and Human Development (1R21-HD-060309-01, 3R2-HD-060309-02S1), Biotechnology and Bioengineering Center, Children's Research Institute, Division of Cardiothoracic Surgery, Herma Heart Center, and the American Association for Thoracic Surgery.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
The authors gratefully acknowledge the hard work and enthusiastic support of M. Krolikowski for IRB support, the Physician Assistants M. Barnes, J. Dunham-Ingle, T. Fehrenbacher, M. Madrzak, E. May, and R. Smith (CHW, Pediatric Cardiothoracic Surgery), as well as M. Krolikowski for IRB support at CHW, D. Jones and OR staff for their tireless contributions to the Surgical Tissue Bank, R. Lorier for her contributions to the Affymetrix hybridizations, M. Goetsch for processing the DNA, M. Olivier for his contribution to assay design, and B. Bilicki for his support with the CN Assays from ABI.
REFERENCES
- 1.The International HapMap Consortium The International HapMap Project. Nature 426: 789–796, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Baldini A. DiGeorge syndrome: an update. Curr Opin Cardiol 19: 201–204, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, Lalani SR, Chinault AC, Cheung SW, Lupski JR, Patel A. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet 82: 214–221, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorheim J, Abrahamsen TW, Kristensen AT, Gaudernack G, Ekstrom PO. Approach to analysis of single nucleotide polymorphisms by automated constant denaturant capillary electrophoresis. Mutat Res 526: 75–83, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O'Leary LA, Wong LY, Elixson EM, Mahle WT, Campbell RM. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics 112: 101–107, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Brunet A, Armengol L, Heine D, Rosell J, Garcia-Aragones M, Gabau E, Estivill X, Guitart M. BAC array CGH in patients with velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21.1. BMC Med Genet 10: 144, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML. Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics 43: 267–277, 1997 [DOI] [PubMed] [Google Scholar]
- 8.Courtens W, Schramme I, Laridon A. Microduplication 22q11.2: a benign polymorphism or a syndrome with a very large clinical variability and reduced penetrance?—Report of two families. Am J Med Genet A 146A: 758–763, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Cuneo BF. 22q11.2 Deletion syndrome: DiGeorge, velocardiofacial, and conotruncal anomaly face syndromes. Curr Opin Pediatr 13: 465–472, 2001. Erratum Curr Opin Pediatr 14 (Apr): 286, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Daw SC, Taylor C, Kraman M, Call K, Mao J, Schuffenhauer S, Meitinger T, Lipson T, Goodship J, Scambler P. A common region of 10p deleted in DiGeorge and velocardiofacial syndromes. Nat Genet 13: 458–460, 1996 [DOI] [PubMed] [Google Scholar]
- 11.de La Rochebrochard C, Joly-Helas G, Goldenberg A, Durand I, Laquerriere A, Ickowicz V, Saugier-Veber P, Eurin D, Moirot H, Diguet A, de Kergal F, Tiercin C, Mace B, Marpeau L, Frebourg T. The intrafamilial variability of the 22q11.2 microduplication encompasses a spectrum from minor cognitive deficits to severe congenital anomalies. Am J Med Genet A 140: 1608–1613, 2006 [DOI] [PubMed] [Google Scholar]
- 12.De Silva D, Duffty P, Booth P, Auchterlonie I, Morrison N, Dean JC. Family studies in chromosome 22q11 deletion: further demonstration of phenotypic heterogeneity. Clin Dysmorphol 4: 294–303, 1995 [DOI] [PubMed] [Google Scholar]
- 13.Emanuel BS, McDonald-McGinn D, Saitta SC, Zackai EH. The 22q11.2 deletion syndrome. Adv Pediatr 48: 39–73, 2001 [PubMed] [Google Scholar]
- 14.Erdogan F, Larsen LA, Zhang L, Tumer Z, Tommerup N, Chen W, Jacobsen JR, Schubert M, Jurkatis J, Tzschach A, Ropers HH, Ullmann R. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J Med Genet 45: 704–709, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Goldmuntz E, Clark BJ, Mitchell LE, Jawad AF, Cuneo BF, Reed L, McDonald-McGinn D, Chien P, Feuer J, Zackai EH, Emanuel BS, Driscoll DA. Frequency of 22q11 deletions in patients with conotruncal defects. J Am Coll Cardiol 32: 492–498, 1998 [DOI] [PubMed] [Google Scholar]
- 16.Goldmuntz E, Driscoll DA, Emanuel BS, McDonald-McGinn D, Mei M, Zackai E, Mitchell LE. Evaluation of potential modifiers of the cardiac phenotype in the 22q11.2 deletion syndrome. Birth Defects Res A Clin Mol Teratol 85: 125–129, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Zhu X, Yang Y, Mo X, Sheng M, Yao J, Wang D. Incidences of micro-deletion/duplication 22q11.2 detected by multiplex ligation-dependent probe amplification in patients with congenital cardiac disease who are scheduled for cardiac surgery. Cardiol Young 19: 179–184, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 27: 286–291, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 12: 996–1006, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewin MB, Lindsay EA, Jurecic V, Goytia V, Towbin JA, Baldini A. A genetic etiology for interruption of the aortic arch type B. Am J Cardiol 80: 493–497, 1997 [DOI] [PubMed] [Google Scholar]
- 22.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410: 97–101, 2001 [DOI] [PubMed] [Google Scholar]
- 23.McDonald-McGinn DM, Emanuel BS, Zackai EH. 22q11.2 Deletion syndrome. Gene Reviews. www.genetests.org, 2005 [PubMed]
- 24.McDonald-McGinn DM, Kirschner R, Goldmuntz E, Sullivan K, Eicher P, Gerdes M, Moss E, Solot C, Wang P, Jacobs I, Handler S, Knightly C, Heher K, Wilson M, Ming JE, Grace K, Driscoll D, Pasquariello P, Randall P, Larossa D, Emanuel BS, Zackai EH. The Philadelphia story: the 22q11.2 deletion: report on 250 patients. Genet Couns 10: 11–24, 1999 [PubMed] [Google Scholar]
- 25.McElhinney DB, Clark BJ, 3rd, Weinberg PM, Kenton ML, McDonald-McGinn D, Driscoll DA, Zackai EH, Goldmuntz E. Association of chromosome 22q11 deletion with isolated anomalies of aortic arch laterality and branching. J Am Coll Cardiol 37: 2114–2119, 2001 [DOI] [PubMed] [Google Scholar]
- 26.McElhinney DB, Driscoll DA, Levin ER, Jawad AF, Emanuel BS, Goldmuntz E. Chromosome 22q11 deletion in patients with ventricular septal defect: frequency and associated cardiovascular anomalies. Pediatrics 112: e472, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Oskarsdottir S, Persson C, Eriksson BO, Fasth A. Presenting phenotype in 100 children with the 22q11 deletion syndrome. Eur J Pediatr 164: 146–153, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, Lenzi T, Keegan CE, Sutton VR, Belmont J, Chinault AC, Lupski JR, Cheung SW, Roeder E, Patel A. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med 10: 267–277, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Pierpont ME, Basson CT, Benson DW, Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 3015–3038, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Portnoi MF. Microduplication 22q11.2: a new chromosomal syndrome. Eur J Med Genet 52: 88–93, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Richards AA, Santos LJ, Nichols HA, Crider BP, Elder FF, Hauser NS, Zinn AR, Garg V. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatr Res 64: 358–363, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Routes JM, Grossman WJ, Verbsky J, Laessig RH, Hoffman GL, Brokopp CD, Baker MW. Statewide newborn screening for severe T-cell lymphopenia. JAMA 302: 2465–2470, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, Aurias A, Raymond FL, Clayton-Smith J, Hatchwell E, McKeown C, Beemer FA, Dallapiccola B, Novelli G, Hurst JA, Ignatius J, Green AJ, Winter RM, Brueton L, Brondum-Nielsen K, Scambler PJ. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet 34: 798–804, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shprintzen RJ. Velo-cardio-facial syndrome: 30 years of study. Dev Disabil Res Rev 14: 3–10, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skopek TR, Liber HL, Penman BW, Thilly WG. Isolation of a human lymphoblastoid line heterozygous at the thymidine kinase locus: possibility for a rapid human cell mutation assay. Biochem Biophys Res Commun 84: 411–416, 1978 [DOI] [PubMed] [Google Scholar]
- 36.Sparkes R, Chernos J, Dicke F. Duplication of the 22q11.2 region associated with congenital cardiac disease. Cardiol Young 15: 229–231, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Stachon AC, Baskin B, Smith AC, Shugar A, Cytrynbaum C, Fishman L, Mendoza-Londono R, Klatt R, Teebi A, Ray PN, Weksberg R. Molecular diagnosis of 22q11.2 deletion and duplication by multiplex ligation dependent probe amplification. Am J Med Genet A 143A: 2924–2930, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Sullivan KE. Chromosome 22q11.2 deletion syndrome: DiGeorge syndrome/velocardiofacial syndrome. Immunol Allergy Clin North Am 28: 353–366, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Sullivan KE. The clinical, immunological, and molecular spectrum of chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Curr Opin Allergy Clin Immunol 4: 505–512, 2004 [DOI] [PubMed] [Google Scholar]
- 40.Sullivan KE, McDonald-McGinn D, Driscoll DA, Emanuel BS, Zackai EH, Jawad AF. Longitudinal analysis of lymphocyte function and numbers in the first year of life in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). Clin Diagn Lab Immunol 6: 906–911, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi K, Kido S, Hoshino K, Ogawa K, Ohashi H, Fukushima Y. Frequency of a 22q11 deletion in patients with conotruncal cardiac malformations: a prospective study. Eur J Pediatr 154: 878–881, 1995 [DOI] [PubMed] [Google Scholar]
- 42.Thiel CT, Kraus C, Rauch A, Ekici AB, Rautenstrauss B, Reis A. A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2–12 duplications and deletions leading to HMSN/HNPP. Eur J Hum Genet 11: 170–178, 2003 [DOI] [PubMed] [Google Scholar]
- 43.Thienpont B, Mertens L, de Ravel T, Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR, Devriendt K. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J 28: 2778–2784, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Traverso M, Malnati M, Minetti C, Regis S, Tedeschi S, Pedemonte M, Bruno C, Biassoni R, Zara F. Multiplex real-time PCR for detection of deletions and duplications in dystrophin gene. Biochem Biophys Res Commun 339: 145–150, 2006 [DOI] [PubMed] [Google Scholar]
- 45.van Engelen K, Topf A, Keavney BD, Goodship JA, van der Velde ET, Baars MJ, Snijder S, Moorman AF, Postma AV, Mulder BJ. 22q11.2 Deletion syndrome is under-recognised in adult patients with tetralogy of Fallot and pulmonary atresia. Heart 96: 621–624 [DOI] [PubMed] [Google Scholar]
- 46.Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EW, Squire JA. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics 6: 180, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weksberg R, Stachon AC, Squire JA, Moldovan L, Bayani J, Meyn S, Chow E, Bassett AS. Molecular characterization of deletion breakpoints in adults with 22q11 deletion syndrome. Hum Genet 120: 837–845, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet 362: 1366–1373, 2003 [DOI] [PubMed] [Google Scholar]