Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli (original) (raw)
Abstract
Proper geometric and topological organization of DNA is essential for all chromosomal processes. Two classes of proteins play major roles in organizing chromosomes: condensin complexes and type II topoisomerases. In Escherichia coli, MukB, a structural maintenance of chromosome-like component of the bacterial condensin, and topoisomerase IV (Topo IV), a type II topoisomerase that decatenates the newly replicated daughter chromosomes, are both essential for chromosome segregation in rapidly growing cells. However, little is known about the interplay between MukB and Topo IV. Here we demonstrate a physical and functional interaction between MukB and ParC, a subunit of Topo IV, in vitro. The site of MukB interaction was located on the C-terminal domain of ParC and a loss-of-interaction mutant, ParC R705E R729A, was isolated. This variant retained full activity as a topoisomerase when reconstituted with ParE to form Topo IV. We show that MukB stimulates the superhelical DNA relaxation activity of wild-type Topo IV, but not that of Topo IV reconstituted with ParC R705E R729A.
Coordinating the structural organization of chromosomes is essential for DNA replication, transcription, and chromosome segregation in both prokaryotes and eukaryotes. The assembly of a suitable chromosomal structure is critical during chromosome segregation, when the replicated genome is distributed to the two daughter cells. Failure to achieve proper chromosomal organization during separation can result in DNA breakage, leading to an uneven distribution of the genetic material to the next generation. Whereas DNA replication, condensation, and segregation take place in different phases of the cell cycle in eukaryotes, they occur concomitantly in rapidly growing bacteria where chromosomes can easily get tangled and damaged if not organized properly. Chromosomal organization involves two principal mechanisms: topological maintenance and protein-mediated packaging of the DNA. The former prevents entanglement by regulating the topology of the DNA, resolving unwanted catenanes and knots, and the latter shapes the conformation of chromosomes, increasing the efficiency of any particular macromolecular transaction. A properly organized chromosome is the result of these combined efforts.
The chromosome of Escherichia coli has a single origin of replication from which two forks move bidirectionally to replicate the genome. The circular nature of the chromosome and the θ-type mode of replication demand the decatenation of template strands during and/or at the end of each replication cycle. At the terminal stages of DNA replication, this unlinking process is attained by the strand passage reaction catalyzed by a type II topoisomerase, topoisomerase IV (Topo IV). Topo IV is comprised of two subunits, ParC, the DNA binding, cleavage, and religation subunit, and ParE, the ATPase subunit, which together form a heterotetrameric holoenzyme (1, 2). Topo IV is an essential enzyme in E. coli, and a loss of its function leads to a severe chromosome segregation defect, characterized by extensive filamentation of the cells, unsegregated chromosomes, and anucleate cell formation (1, 3).
The condensin complex is involved in chromosome condensation in both prokaryotes and eukaryotes, but its mechanism of action is not clear. E. coli possesses a condensin complex consisting of three subunits, MukB, MukE, and MukF. MukB belongs to the structural maintenance of chromosomes (SMC) protein family, members of which are composed of a hinge domain, long coiled-coil arms, and head domains that form ATP binding pockets (4, 5). MukE binds to the head domain of MukB via binding to MukF, a kleisin subunit (6), to form the complete complex (5). Deletion of any of the three genes that encode the E. coli condensin complex leads to temperature-sensitive viability and chromosome segregation defects highlighted by increased formation of anucleate cells (7, 8). These observations suggest that the MukBEF complex is important for efficient chromosome segregation.
Since its discovery, there have been many reports on the in vitro activity of the condensin complex that suggest possible mechanisms of action in the cell. Condensin complexes can introduce supercoils into plasmid DNA and knot relaxed circular DNA substrates in the presence of type II topoisomerases, although the chirality of condensin action appears to vary among different organisms (9–12). For instance, the MukB homodimer stabilizes net negative supercoils on plasmid DNA (12), but the yeast Smc2/Smc4 condensin holocomplex has no net effect on superhelicity (11). A recent single-molecule study suggests that MukB acts as a macromolecular clamp and can condense DNA in an ATP-independent manner in vitro (13).
Topo IV and MukB both play essential roles in efficient chromosome segregation in E. coli, and it is likely the case that their actions have to be coordinated to achieve faithful and rapid separation of the daughter chromosomes. We detected MukB as an interacting partner of ParC in a yeast two-hybrid screen. In this paper, we validate this finding with purified proteins in vitro and identify the domain on ParC involved in the interaction with MukB. We also identified a loss-of-interaction variant of ParC that retains full activity as a topoisomerase when reconstituted with ParE. Furthermore, we found that MukB is able to stimulate the relaxation activity of Topo IV in vitro. Interestingly, this stimulation activity of MukB was absent when Topo IV was reconstituted with the aforementioned loss-of-interaction ParC variant, suggesting that the physical interaction is important for the stimulation.
Results
MukB and ParC Interact in Vitro and in Vivo.
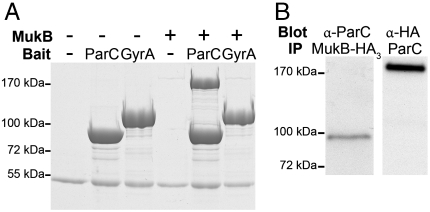
Previously in our laboratory, a yeast two-hybrid screen was executed to identify proteins that interact with the full-length ParC subunit of Topo IV. One protein identified in this screen was MukB. To verify the screening data, the physical interaction between MukB and ParC was tested with purified components in a pull-down assay. Purified MukB served as the prey and HA-tagged ParC and GyrA proteins conjugated to anti-HA antibody agarose beads served as bait. GyrA, the DNA-binding subunit of DNA gyrase, the other type II topoisomerase in E. coli, is homologous to ParC in overall structure, although the two proteins differ distinctly in the organization of their C-terminal domains (14). Hence, GyrA was used as a control to test for the specificity of the MukB-ParC interaction. MukB and the protein beads were mixed in solution and the pull-down fraction was analyzed by SDS-PAGE. HA-ParC physically interacted with MukB, whereas HA-GyrA and the antibody-coated beads alone failed to do so, indicating that the interaction is direct and specific to ParC (Fig. 1A). Although the stiochiometry of the MukB-ParC interaction appears to be 1∶1, we note that in pull-down assays of this type, the array of bait molecules on the surface of the bead can lead to stabilization of weak interactions.
Fig. 1.
MukB and ParC interact in vitro and in vivo. (A) Either HA-ParC protein (86 kDa) or HA-GyrA protein (99 kDa) conjugated to anti-HA antibody agarose beads were incubated with MukB (170 kDa) in binding assays as described. The reaction was analyzed by SDS-PAGE. The 55-kDa band on the gel is the IgG heavy chain. (B Left) Monoclonal anti-HA antibody was used to immunoprecipitate MukB-HA3 (174 kDa) from crude extracts prepared from BW30270_mukB_-HA3 cells, and the gel was Western blotted using polyclonal anti-ParC antibody. (B Right) Polyclonal anti-ParC antibody was used to immunoprecipitate ParC from crude extract prepared from BW30270_mukB_-HA3 cells, and the gel was Western blotted using monoclonal anti-HA antibody.
In order to test their interaction in vivo, a coimmunoprecipitation experiment was conducted with a crude extract from an MG1655 derivative strain where mukB was chromosomally tagged with a triple HA tag. ParC and MukB-HA3 proteins were immunoprecipitated from the crude extract with polyclonal anti-ParC and monoclonal anti-HA antibodies, respectively, and the immunoprecipitated fractions were subjected to Western blot analysis. As in the pull-down experiment, MukB-HA3 was coimmunoprecipitated with ParC and vice versa (Fig. 1B). Therefore, MukB and ParC also interact in vivo.
Identification of Variant ParC Proteins That Do Not Interact with MukB.
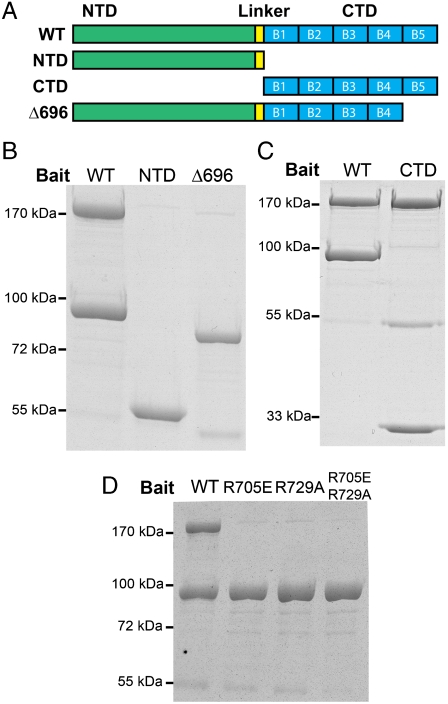
To further examine the MukB-ParC interaction, we created deletion mutants of ParC to identify domains that are involved in the interaction with MukB. ParC consists of two distinct domains, the N-terminal and the C-terminal domains (NTD and CTD, respectively) that are connected by a flexible linker (14). The NTD serves as the main scaffold for the strand passage reaction, containing the DNA gate binding site where cleavage and religation will occur, whereas the CTD has been suggested to be the geometry sensor for specific DNA substrates (14). The CTD consists of five structurally similar units, the blades, which collectively form an open β-pinwheel structure (14). To identify the MukB-interacting domain on ParC, a set of deletion mutants were designed with N-terminal HA tags. These proteins were conjugated to anti-HA antibody beads from extracts of cells and were used in pull-down assays with purified MukB to screen for a loss of interaction. A schematic of the mutant protein design is shown in Fig. 2A.
Fig. 2.
MukB interacts with the CTD of ParC. (A) Schematic of ParC and the designed deletion mutants. ParC protein consists of an NTD (green) and CTD (blue), which are connected by a flexible linker region (yellow). The CTD consists of five structurally similar units of a β-pinwheel (14), the blades, which are denoted as B1, B2, B3, B4, and B5, ordered from the N terminus to the C terminus. The Δ696 mutant lacks the most C-terminal blade, B5. (B) The ParC HA-NTD (59 kDa) and the HA-ParCΔ696 (79 kDa) variant that lacks the fifth blade of the CTD do not interact with MukB. (C) ParC HA-CTD (29 kDa) is sufficient to interact with MukB. (D) Neither HA-ParC R705E, HA-ParC R729A, nor HA-ParC R705E R729A interact with MukB. Binding experiments were as described in the legend to Fig. 1.
ParC NTD and the ParCΔ696 variant lacking the fifth blade of the CTD failed to bring down MukB (Fig. 2B), whereas ParC CTD alone was able to pull down MukB as efficiently as full-length ParC (Fig. 2C). Hence, ParC interacts with MukB via its CTD. This observation augments the result from the pull-down experiment in Fig. 1 because the major structural difference between ParC and GyrA lies in their CTDs. The CTD of GyrA has a closed six-bladed β-pinwheel structure instead of an open five-bladed one as in the ParC CTD (14), and thus it is possible that the open structure of the ParC CTD accommodates an additional surface for protein-protein interactions.
ParCΔ696 possessed all of the blades of the CTD except the last one but still failed to interact with MukB, suggesting the crucial involvement of the fifth blade in the interaction. To identify the residues that are important for the interaction, charge reversal and charge ablation point mutations were introduced to the fifth blade of the ParC CTD of the full-length HA-tagged ParC, and those mutants were screened for loss of interaction with MukB in the pull-down assay. Two such mutants were identified, ParC R705E and ParC R729A, both of which failed to interact with MukB (Fig. 2D). The double mutant protein that combines these two mutations also failed to interact with MukB (Fig. 2D). Interestingly, the two mutated amino acid residues, Arg705 and Arg729, are adjacent in the ParC structure (14), suggesting a candidate surface for the interaction with MukB.
ParC R705E R729A Is Active when Reconstituted as Topo IV in Vitro.
To study the significance of the MukB-ParC interaction, we hoped to obtain a loss-of-interaction variant of ParC that retained full activity as a topoisomerase when combined with ParE to form the Topo IV holoenzyme. The ParC R705E R729A mutant satisfied the first criterion, and thus we tested whether it can reconstitute a functional topoisomerase.
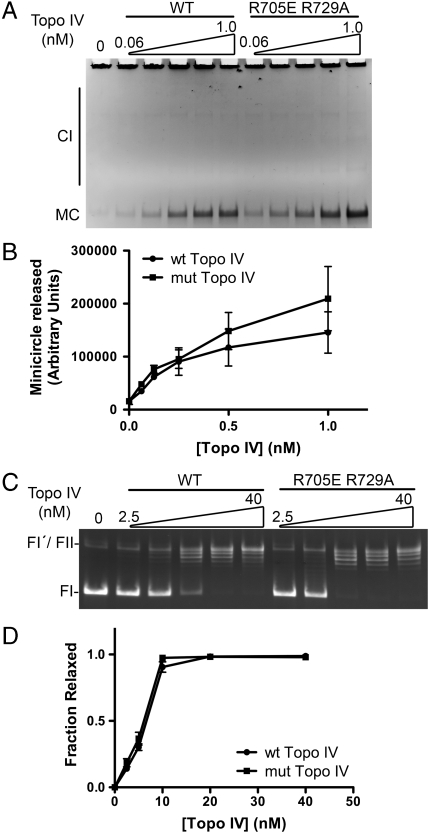
Purified wild-type and the ParC R705E R729A proteins were incubated with ParE to reconstitute Topo IV and tested for their activities in vitro. We measured the activity of the wild-type and mutant enzymes in decatenation of kinetoplast DNA (kDNA). kDNA is a large DNA complex found in the mitochondria of trypanosomes that consists of thousands of circular DNA molecules catenated together (15). The high molecular weight of kDNA prevents its entry to an agarose gel; however, the individual constituent minicircles can be easily observed under routine electrophoresis conditions. The catenated minicircles can be unlinked by treatment with a type II topoisomerase such as Topo IV. The kDNA decatenation activity of the wild-type and mutant Topo IV was measured, and no significant difference in activity was observed between the two enzymes (Fig. 3 A and B). Production of minicircles was already apparent at 0.06 nM of Topo IV in both wild-type and mutant cases, and the amount of minicircles released was comparable between the wild-type and mutant Topo IVs within each experiment, indicating that they have the same specific activity for decatenation of kDNA.
Fig. 3.
Topo IV reconstituted with ParE and ParC R705E R729A is active. (A) ParC R705E R729A is as active as wild-type ParC in the unlinking of DNA rings. Assays for the decatenation of kinetoplast DNA by the indicated concentrations of Topo IV reconstituted with ParE and either wild-type or ParC R705E R729A were as described. MC, minicircle; CI, catenated intermediates. (B) The amount of minicircles released as shown in A was determined, and the means from three independent experiments were plotted. SEMs are indicated. (C) ParC R705E R729A is as active as wild-type ParC in the removal of negative supercoils. Assays for the relaxation of negatively supercoiled plasmid DNA by the indicated concentrations of Topo IV reconstituted with ParE and either wild-type ParC or ParC R705E R729A were as described. (D) The fraction of relaxed plasmid DNA as shown in C was determined, and the means from three independent experiments were plotted. SEMs are indicated. FI, form I DNA (supercoiled); FI′, form I′ DNA (covalently closed, relaxed); FII, form II DNA (nicked circular). A small amount of residual FII DNA present in the preparation of form I DNA comigrates with FI′ DNA.
In an assay measuring superhelical DNA relaxation, both enzymes reached saturation at the same concentration and their specific activities were identical (Fig. 3 C and D). Therefore, we conclude that the superhelical DNA relaxation activity of Topo IV was not affected by the ParC R705E R729A mutation. These observations indicate that ParC R705E R729A is able to reconstitute a fully active topoisomerase in vitro.
Effect of MukB on Topo IV Activity.
Upon confirming the full functionality of the ParC R705E R729A loss-of-interaction mutant as a topoisomerase constituent in vitro, we tested the effect of MukB on the activity of Topo IV.
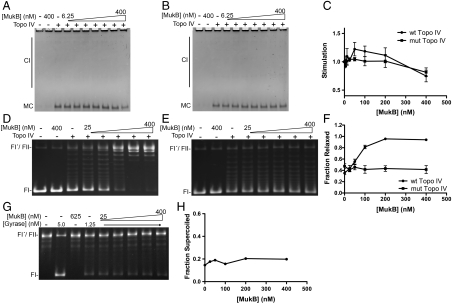
To test the effect of MukB on the decatenation activity of Topo IV, kDNA was first incubated with varying concentrations of MukB, Topo IV was then added to 0.125 nM, and the extent of stimulation of minicircles released was determined (Fig. 4 A_–_C). MukB simulated decatenation of Topo IV reconstituted with wild-type ParC by 30%, but did not stimulate Topo IV reconstituted with ParC R705E R729A. A slight inhibition was observed with both wild-type and mutant Topo IV at the highest concentration of MukB tested, possibly because increased occupancy by MukB on the DNA inhibits binding of Topo IV.
Fig. 4.
Effect of MukB on Topo IV activity. (A and B) Effect of MukB on the decatenation activity of Topo IV (0.125 nM) reconstituted with either wild-type ParC (A) or ParC R705E R729A (B). (C) The extent of stimulation of decatenation by Topo IV in A and B was determined, and the means from three independent experiments were plotted. SEMs are indicated. (D) MukB stimulates the superhelical DNA relaxation activity of wild-type Topo IV (6 nM). (E) Topo IV (6 nM) reconstituted with ParE and ParC R705E R729A is unaffected by MukB. (F) The fraction of relaxed plasmid DNA in D and E was determined, and the means from three independent experiments were plotted. SEMs are indicated. (G) MukB does not affect the supercoiling activity of DNA gyrase (0.5 nM). Reaction mixtures containing form I′ DNA and the indicated concentrations of DNA gyrase and MukB were incubated and analyzed as described. (H) The fraction of supercoiled plasmid DNA in G was determined, and the average of two independent experiments was plotted.
The effect of MukB on Topo IV-catalyzed superhelical DNA relaxation was also assessed. Negatively supercoiled DNA was first incubated with varying concentrations of MukB, Topo IV was then added to 6 nM, and the extent of DNA relaxation was determined (Fig. 4 D_–_F). In the absence of Topo IV, MukB did not affect the superhelicity of the plasmid DNA. MukB stimulated the activity of the wild-type Topo IV by 2.5-fold (Fig. 4 D and F). Interestingly, stimulation of DNA relaxation activity by MukB was abolished when Topo IV was reconstituted with ParC R705E R729A (Fig. 4 E and F). This observation implies that the physical interaction between ParC and MukB is required for stimulation, although other mechanisms are of course possible (see Discussion).
To test the specificity of the stimulation, we examined whether MukB affected the negative supercoiling activity of DNA gyrase, the other type II topoisomerase in E. coli. No effect of MukB was observed (Fig. 4 G and H), suggesting that MukB stimulation is specific for Topo IV.
Discussion
In this paper, we demonstrate that two proteins that are major determinants of rapid and efficient chromosome segregation in E. coli, MukB, a subunit of the condensin complex, and ParC, a subunit of Topo IV, interact physically and functionally in vitro. MukB stimulated the activity of wild-type Topo IV in vitro, but not of a topoisomerase comprised of ParE and ParC R705E R729A, a variant subunit that no longer physically interacted with MukB but that fully reconstituted topoisomerase activity.
We observed a minor effect of MukB on the decatenation activity of Topo IV. Although the extent of stimulation was small (∼30%), stimulation was observed only with the Topo IV reconstituted with wild-type ParC and not with the Topo IV reconstituted with ParC R705E R729A. It is possible that decatenation of the singly linked kDNA network may not be an optimal assay for testing the effect of MukB on Topo IV activity or that the target activity for MukB stimulation is not decatenation.
More significant is that MukB can stimulate the superhelical DNA relaxation activity of Topo IV in a concentration-dependent manner. There are several potential mechanistic explanations for this stimulation. First, it is possible that MukB directly stimulates the catalytic activity of Topo IV and the ParC CTD acts as the regulatory domain. Because the physical interaction is required for the modulation of the activity in this case, the ParC R705E R729A mutant would lose its ability to be stimulated. Second, MukB might reconfigure DNA to produce an optimal substrate for Topo IV. Studies suggest MukB and other condensin complexes are able to reconfigure the topology of the DNA in vitro (9–12), and thus it is quite plausible that such a reconfiguration could create a preferred substrate for Topo IV. However, all the referenced studies were conducted with topoisomerases and condensins from different organisms and the use of the canonical pair (i.e., a topoisomerase and a condensin complex from the same species) can reveal a synergistic effect as seen in this paper. For example, Petrushenko and others reported that MukB inhibits superhelical DNA relaxation activity of wheat germ topoisomerase I (16); however, our result indicates that MukB stimulates the relaxation activity of Topo IV, the endogenous topoisomerase. Moreover, the observed inhibition of superhelical DNA relaxation activity in previous studies could arise from the high concentration of MukB in the reaction mixtures. Excess MukB on the DNA might inhibit the binding of topoisomerase to the DNA as seen in our decatenation assay.
It is important to note that the stimulation activity of MukB was specific to Topo IV; DNA gyrase was unaffected. This observation is consistent with the reconfiguration model, where MukB creates an optimal substrate for Topo IV: It is the CTDs of ParC and GyrA that act as the sensor for substrate specificity in Topo IV and Gyrase, respectively, and only the ParC CTD, via interaction with MukB, may be able to recognize a MukB-reconfigured substrate. Last, it is possible that MukB loads ParC onto the DNA via their physical interaction, and/or vice versa. Thus, the interaction could shift the equilibrium of Topo IV toward the DNA-bound state, increasing the effective rate of relaxation of the supercoiled substrate. Alternatively, MukB may stabilize Topo IV on the DNA via the physical interaction, and increases its processivity. These three possible mechanisms are not mutually exclusive, and the actual mechanism of stimulation is likely to involve more than one of them. It is also important to note that the activities of condensin in vitro manifest only in the presence of a topoisomerase. For instance, knotting of a circular DNA by condensin requires a type II topoisomerase. This functional interplay between condensins and topoisomerases further augments the significance of the physical interaction between MukB and ParC.
There are other proteins that are involved in the management of chromosomal DNA organization that have been found to interact with the ParC subunit of Topo IV. The C-terminal AAA+ domain of FtsK interacts with ParC and also stimulates its activity (17). FtsK is a bifunctional enzyme: The N-terminal domain forms part of the septal ring, whereas the C-terminal domain is a DNA translocase that modulates the direction of chromosomal dimer resolution by the XerC/D site-specific recombinase (18). MreB, the actin ortholog, which is required for the maintenance of cell shape (19) and has been implicated in the movement of certain regions of the chromosome (20, 21), also interacts with ParC and stimulates Topo IV activity (22). In this latter case, the stimulation is dependent on the quaternary structure of MreB: The polymerized form stimulates, whereas the monomeric form inhibits. Similar to the case in eukaryotes, these observations suggest a network of physical interactions linking the type II topoisomerase responsible for chromosome segregation and other elements of the cell responsible for chromosomal organization.
Interestingly, it has been shown that a condensin subunit and type II topoisomerase (Topo II) also interact in Drosophila embryonic extracts (23). These proteins are known to be constituents of the scaffold of condensed metaphase chromosomes, display a barber pole-like localization pattern (24), and they are essential for proper chromosome organization and segregation (23, 25, 26). There is debate in the literature over the precise roles of these proteins in chromosome condensation from prophase to metaphase (27, 28); however, it is evident that their functions are essential to create suitable chromosomal structures for the eventual separation of the sister chromatids in anaphase. Considering the evolutionary conservation of important biological functions and the coexistence of condensin complex and type II topoisomerases, especially Topo IV, in prokaryotes, it is conceivable that the same mechanism exists in bacteria. Clearly the bacterial condensin complex is required for proper organization of the chromosome. And although Topo IV is currently thought to be the decatenating enzyme, additional functions as either a structural component of the chromosome or the nucleoid are also conceivable. These additional functions might even be exclusive to ParC. Our observation that MukB was able to stimulate the relaxation activity of Topo IV, an intramolecular reaction, is consistent with this idea. Topo IV might be involved in chromosome organization in cooperation with MukB, possibly creating right-handed knots as previously reported (16). Although Topo IV can remove supercoils, a coordinated strand passage reaction could bring together distal DNA segments, thus condensing the chromosome.
In rapidly growing bacteria, geometric and topological organization of the nascent DNA is essential for efficient and faithful chromosome segregation, and the condensin complex and Topo IV may weave a suitable chromosomal structure for such a process via their interaction. Preliminary observations indicate that the parCR705ER729A allele gives rise to a partially penetrant par phenotype in vivo. Further characterization of the MukB-Topo IV interaction and the phenotype of the mutant parC allele will be necessary to elucidate the mechanism of interplay of these proteins during rapid chromosome segregation in prokaryotes.
Materials and Methods
E. coli Strains and Plasmids.
pET11a-mukB was constructed by inserting a mukB PCR fragment copied from C600 genomic DNA into NdeI- and BamHI-digested pET11a plasmid DNA (Novagen). pET11a-HA-parC and pET11a-HA-gyrA were constructed by inserting HA-parC and HA-gyrA PCR fragments copied from C600 genomic DNA template into NdeI- and BamHI-digested pET11a plasmid DNA (Novagen). The following primers were used to tag ParC and GyrA with the HA epitope by PCR: for HA-parC, 5′-GCGCATATGTACCCATACGATGTTCCAGATTACGCTGAATT-CAACAACAACATGAGCGATATGGCAGAGCGCCTTGC-3′ and 5′-CGATCAGGATCCTTACTC-TTCGCTATCACCGCTGC-3′, and for HA-gyrA, 5′-GCTCATATGTACCCATACGATGTTCCAGATTACGCTGAATTCAACAACAACATGAGCGACCTTGCGAGAGAAATTACACCG-3′ and 5′-CGAGGATCCTTATTCTTCTTCTGGCTCGTCGTCAACG-3′. Plasmid DNAs carrying the mutant HA-ParC genes were created by site directed mutagenesis of pET11a-HA-parC plasmid using a commercial kit (Agilent Technologies). BL21(DE3) cells were used for all protein overproduction. Chromosomal allele replacement to create the MG1655_mukB_-HA3 was conducted in E. coli strain C600 as previously described (29, 30).
Protein Purification.
Tagged and untagged Topo IV and DNA gyrase proteins were purified as described (2, 22). For MukB, BL21(DE3)(pET11a-mukB) was grown at 37 °C in 20 L of LB medium containing 100 μg/mL ampicillin to an OD600 of 0.4 and MukB overproduction was induced by the addition of 0.4 mM IPTG for 3 h at 25 °C. Cells were harvested, resuspended at 1∶1 wt/vol in 50 mM Tris-HCl (pH 7.5 at 4 °C), 10% sucrose, and frozen in liquid nitrogen. After gradually thawing the frozen cells, the cell suspension was adjusted to 50 mM Tris-HCl (pH 8.0 at 4 °C), 150 mM NaCl, 10 mM DTT, 20 mM EDTA, and 0.2 mg/mL lysozyme, incubated on ice for 10 min, room temperature (RT) for 10 min, and ice for another 10 min, followed by centrifugation at 100,000 × g for 1 h at 4 °C. The supernatant (fraction 1) was collected, brought to 35% saturation of [NH4]2SO4, and stirred for 1 h at 4 °C. The collected precipitate was resuspended in Buffer A (50 mM Hepes-KOH (pH 7.6), 25 mM KCl, 0.1 mM EDTA, 2 mM DTT, 10% glycerol, fraction 1′). Fraction 1′ (340 mg) was diluted with buffer A to a conductivity equivalent to 39 mM NaCl in buffer A, and loaded onto a 100-mL Q Sepharose (Amersham) column equilibrated with buffer A. The column was washed with two column volumes (CVs) of buffer A and protein was eluted with a 10-CV gradient of 0 to 1 M NaCl in buffer A. Peak fractions (eluting at 230 mM NaCl) were pooled according to SDS-PAGE analysis for the MukB protein band (fraction 2). Fraction 2 was dialyzed overnight against buffer A (fraction 2′). Half of fraction 2′ (23 mg) was loaded onto a 10-mL Heparin-agarose column (Sigma) equilibrated with buffer A. After washing with 2 CVs of buffer A, protein was eluted with a 10-CV gradient of 0 to 1 M NaCl in buffer A. The peak fractions (eluting at 140 mM NaCl) were pooled according to SDS-PAGE analysis. Peak fractions from two separate runs were combined (fraction 3), [NH4]2SO4 was added to 50% saturation, and the precipitated protein was collected and resuspended in buffer A (fraction 3′, 17 mg). Fraction 3′ was gel filtered through a 125-mL Superdex 200 column (GE Healthcare, 2.3 cm × 58 cm) equilibrated with buffer A* [50 mM Hepes-KOH (pH 7.6), 500 mM NaCl, 0.1 mM EDTA, 2 mM DTT, 10% glycerol]. The column was developed at 1 mL/ min. Peak fractions were pooled (fraction 4, 7.5 mg), dialyzed against storage buffer [50 mM Hepes-KOH (pH 7.6), 150 mM NaCl, 0.1 mM EDTA, 2 mM DTT, 40% glycerol], frozen in liquid nitrogen, and stored at -80 °C. Fraction 4 had no detectable endonuclease activity at 37 °C.
MukB-ParC Interaction Assay.
Monoclonal anti-HA antibody-conjugated agarose beads (Sigma) were activated with 0.1 M glycine (pH 2.5) as described by the manufacturer, washed twice with immunoprecipitation buffer (IP) buffer [50 mM Hepes-KOH (pH 7.6), 150 mM NaCl, 10 mM magnesium acetate, 10 mM DTT, 0.05% NP-40], and resuspended in an equal volume of IP buffer. To conjugate HA-tagged proteins, the activated beads were mixed with cleared lysates from cells overproducing HA-tagged proteins. The extracts were prepared as described above. Beads (50 μL) were mixed with extract (400 μL) containing 4 to 7 mg of protein for 2 h at 4 °C on a rotator, the beads were collected by centrifugation, washed with 1 mL IP buffer four times, and resuspended in 50 μL of IP buffer. For the MukB pull-down experiments, 10 μL of protein beads were mixed with 0.5 μM MukB in 100 μL IP buffer for 2 h at 4 °C on a rotator, the beads were collected, washed with 400 μL IP buffer four times, and resuspended in 30 μL Laemmli SDS-PAGE loading buffer. After heating to 100 °C for 5 min, samples were analyzed by 8% SDS-PAGE, and proteins were visualized by staining with Coomassie brilliant blue.
Coimmunoprecipitation Experiments.
Crude extracts were prepared from MG1655_mukB_-HA3 cells as for purification of MukB. Anti-HA antibody-conjugated agarose beads were prepared as described above. For the immunoprecipitation of ParC protein, polyclonal anti-ParC rabbit antibody was conjugated to Protein A/G coupled agarose beads (Thermo Scientific) and resuspended in IP buffer. For the experiment in Fig. 1B, 20 μL of bead suspension was mixed with 280 μL of crude extract (12 mg/mL) from MG1655 _mukB_-HA3 and the suspension was incubated for 2 h at 4 °C on a rotator. The beads were pelleted, washed four times with 400 μL IP buffer, resuspended in 40 μL Laemmli SDS-PAGE loading buffer, and bound protein analyzed by 8% SDS-PAGE. Proteins were transferred from the gel to nitrocellulose membranes and visualized by Western blotting with appropriate antibodies. All the antibodies—monoclonal mouse anti-HA (Sigma), polyclonal rabbit anti-ParC, goat anti-Rabbit-HRP, and anti-mouse IgG-HRP conjugates (Bio-Rad)—were diluted by 10,000-fold in 25 mM Tris-HCl (pH 8.0 at RT), 125 mM NaCl, 0.1% Tween 20.
Relaxation, Decatenation, and Supercoiling Assays.
The kDNA decatenation assay with Topo IV was conducted as described (22). For the superhelical DNA relaxation assay, reaction mixtures (20 μL) containing negatively supercoiled pUCO DNA (4,521 base pairs, 200 ng), 50 mM Hepes-KOH (pH 7.6), 20 mM KCl, 100 μg/mL BSA, 10 mM DTT, 2 mM ATP, and the indicated concentrations of Topo IV were incubated at 37 °C for 5 min. Reactions were terminated by the addition of EDTA and NaCl to 20 mM and 300 mM, respectively, and the incubation continued at 37 °C for 5 min. SDS and Proteinase K were then added to 0.5% and 100 μg/mL, respectively, and the incubation was continued at 37 °C for 15 min. Samples were analyzed by electrophoresis at 25 V for 15 h through vertical 1% agarose gels using 50 mM Tris-HCl (pH 7.8 at 23 °C), 40 mM NaOAc, and 1 mM EDTA as the electrophoresis buffer. Gels were stained with ethidium bromide and photographed.
To assay MukB stimulation, reaction mixtures for decatenation or relaxation assay were first incubated in the absence of Topo IV with the indicated concentration of MukB at 37 °C for 10 min, either wild-type or mutant Topo IV (final concentration of 0.125 nM for decatenation and 6 nM for relaxation) was then added and the incubation continued at 37 °C for 5 min for both assays. Samples were then analyzed as above.
To prepare the relaxed circular DNA substrate for DNA gyrase supercoiling assays, negatively supercoiled pUCO DNA (10 μg) was treated with Topo IV (36 nM) at 37 °C for 45 min. The relaxed DNA was recovered by ethanol precipitation after extraction with phenol chloroform and resuspended in 100 μL 10 mM Tris-HCl (pH 7.5 at 4 °C), 1 mM EDTA. Reaction conditions for supercoiling by DNA gyrase were the same as for DNA relaxation by Topo IV. The final concentration of DNA gyrase was 0.5 nM.
Acknowledgments.
We thank Chong Lee, Pearl Nurse, and Georgiy Teverovskiy for assistance. These studies were supported by NIH Grant GM34558.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 18749.
References
- 1.Kato J, et al. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 2.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 3.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graumann PL, Knust T. Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 2009;17:265–275. doi: 10.1007/s10577-008-9014-x. [DOI] [PubMed] [Google Scholar]
- 5.Woo JS, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 6.Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: Architecture and functional relationship to kleisins. EMBO J. 2005;24:1921–1930. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- 9.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 10.Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: Implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 11.Stray JE, Crisona NJ, Belotserkovskii BP, Lindsley JE, Cozzarelli NR. The Saccharomyces cerevisiae Smc2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J Biol Chem. 2005;280:34723–34734. doi: 10.1074/jbc.M506589200. [DOI] [PubMed] [Google Scholar]
- 12.Petrushenko ZM, Lai CH, Rai R, Rybenkov VV. DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J Biol Chem. 2006;281:4606–4615. doi: 10.1074/jbc.M504754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Y, Petrushenko ZM, Rybenkov VV. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol. 2008;15:411–418. doi: 10.1038/nsmb.1410. [DOI] [PubMed] [Google Scholar]
- 14.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Rauch CA, White JH, Englund PT, Cozzarelli NR. The topology of the kinetoplast DNA network. Cell. 1995;80:61–69. doi: 10.1016/0092-8674(95)90451-4. [DOI] [PubMed] [Google Scholar]
- 16.Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J Biol Chem. 2006;281:34208–34217. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espeli O, Lee C, Marians KJ. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J Biol Chem. 2003;278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
- 18.Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 19.Wachi M, Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Madabhushi R, Marians KJ. Actin homolog MreB affects chromosome segregation by regulating topoisomerase IV in Escherichia coli. Mol Cell. 2009;33:171–180. doi: 10.1016/j.molcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 24.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell. 2003;4:467–480. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 25.Uemura T, et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 26.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 27.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol. 2003;160:645–655. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coelho PA, Queiroz-Machado J, Sunkel CE. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J Cell Sci. 2003;116:4763–4776. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- 29.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espeli O, Nurse P, Levine C, Lee C, Marians KJ. SetB: An integral membrane protein that affects chromosome segregation in Escherichia coli. Mol Microbiol. 2003;50:495–509. doi: 10.1046/j.1365-2958.2003.03736.x. [DOI] [PubMed] [Google Scholar]