Hypolithic Microbial Community of Quartz Pavement in the High-Altitude Tundra of Central Tibet (original) (raw)
Abstract
The hypolithic microbial community associated with quartz pavement at a high-altitude tundra location in central Tibet is described. A small-scale ecological survey indicated that 36% of quartz rocks were colonized. Community profiling using terminal restriction fragment length polymorphism revealed no significant difference in community structure among a number of colonized rocks. Real-time quantitative PCR and phylogenetic analysis of environmental phylotypes obtained from clone libraries were used to elucidate community structure across all domains. The hypolithon was dominated by cyanobacterial phylotypes (73%) with relatively low frequencies of other bacterial phylotypes, largely represented by the chloroflexi, actinobacteria, and bacteriodetes. Unidentified crenarchaeal phylotypes accounted for 4% of recoverable phylotypes, while algae, fungi, and mosses were indicated by a small fraction of recoverable phylotypes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00248-010-9653-2) contains supplementary material, which is available to authorized users.
Introduction
The study of lithic microbial communities, those inhabiting rock substrates, has been gathering momentum due to a growing appreciation of their importance in extreme desert ecosystems where higher plants are rare or absent [14, 17], and their application as model systems in ecological studies due to their simple community structure [34] and relevance to astrobiologists as analogs for possible life on the cold arid surface of Mars [5, 25]. Lithic microbial colonization can be categorized according to orientation with respect to the substrate into epiliths (on above-ground rock surfaces), hypoliths (on below-ground rock surfaces), chasmoliths (in cracks and fissures in rocks), and endoliths (within the rock matrix itself) [14, 18].
The study of hypoliths has aroused much recent interest since they tend to occur on translucent stones such as quartz, which are a near-ubiquitous substrate in arid locations worldwide including hot and cold deserts, polar regions, and high-altitude tundra. The term hypolith refers to the organisms, while hypolithon is used to describe the hypolithic community [18]. Hypolithic colonization is viewed as a stress-avoidance strategy from UV radiation and is also envisaged to confer marginal gains in moisture availability that creates a more favorable niche than the surrounding soil [34].
Early surveys of desert flora reported occurrence of hypolithic microbial colonization [10, 16]. Subsequent research identified cyanobacteria as the most common phylum observed in hypolithon from the Negev Desert [3], Antarctic Dry Valleys [7, 8], Namib Desert [9], coastal Antarctic [30], Mojave Desert [28], Arctic tundra [11], Atacama Desert [35], and Taklimakan Desert [34]. Morphological studies indicated hot arid deserts supported an apparent monoculture of Chroococcidiopsis sp. [34, 35], while polar regions supported a relatively more diverse cyanobacterial assemblage comprising various Oscillatoriales, Aphanocapsa, Aphanothece, Chroococcidiopsis, and Gloeocapsa morphotypes [8, 12]. Culture-independent molecular studies revealed greater cyanobacterial diversity including the presence of diazotrophic and filamentous taxa plus a heterotrophic bacterial component quite distinct from the surrounding soil [26–28, 30, 35].
The presence of eukaryotic algal morphotypes [8, 12] and phylotypes [26] has been recorded for the polar hypolithon, and culture-independent molecular studies have also revealed fungal phylotypes in arid Mojave Desert hypoliths [28]. Eukaryotic signatures could not, however, be detected from hypolithon in the hyper-arid Atacama and Taklimakan Deserts [34, 35]. The archaea have not been detected in polar hypoliths [26] or those from warm hyper-arid deserts [27, 35]. Whether the archaea occur as essential components of other hypolithic communities remains unresolved.
Here we identify microbial diversity across all domains of hypolithic communities in the high-altitude Tibetan tundra, and thus expand knowledge of hypolithic biodiversity into a previously uninvestigated environment and across multiple domains of microbial life. Importantly, we show that the Tibetan tundra hypolithon is distinct from that in other locations investigated to date, and comprises archaeal, algal, fungal, and moss components in addition to the cyanobacteria-dominated bacterial assemblage.
Materials and Methods
Field Sampling
The field location was a large un-named plain typical of the region’s terrain in Gyirong County, in the central southern region of the Tibet Autonomous Region of China (N29°07′, E85° 22.05′, altitude 4,638 m). The region is characterized as a cold desert [21], with an arid tundra landscape experiencing long snowbound winters and brief arid summers punctuated by moisture input from snowmelt and rainfall. Sampling was carried out in October 2005. A small-scale randomized hierarchical sampling was carried out by employing triplicate 1-m2 quadrats deployed at three separate locations across an area of 100 m2. Colonization was visually detected as a green band of microbial growth on the subsurface of stones. For each quadrat, three colonized rocks were randomly selected for molecular community analysis. All samples were aseptically transferred to sterile (gamma-irradiated) sealable plastic bags and stored at ambient temperature (at or near freezing) in darkness until processed (approx. 2 weeks travel from the remote field site). On-site measurements of photosynthetically active radiation (PAR), UV-A and UV-B radiation was carried out at solar noon using a Li-Cor LI-I400 datalogger (Li-Cor Inc, Nebraska) for PAR, and a UV UVX radiometer (UVP Inc, California) for UV irradiance. For transmittance studies through rock substrates, the sensor was placed in situ under quartz rocks, and edges were sealed using plasticine.
Microscopy
Microscopic examination was carried out using Scanning Electron Microscopy (SEM) (Stereoscan 440, Leica, Cambridge, UK). Colonized quartz surfaces were fractured from stones using a geological hammer and fixed in 2.5% glutaraldehyde for 8 h, air-dried overnight, and then gold-sputter-coated for 30 s (SCD 005, BAL-TEC, Lichtenstein) prior to visual examination.
Recovery of Environmental DNA and Target Loci
Total DNA recovery was achieved separately for each sample by lysis in CTAB with lysozyme and RNAse, followed by phenol:chloroform extraction at 60°C. Genomic DNA was checked for quality by electrophoresis in 1% agarose gels and quantified by spectrophotometry (Smartspec Plus, Bio-Rad, California). Universal PCR primers used to target small subunit rRNA genes for bacteria, archaea, and eukarya using 8F [AGA GTT TGA TCC TGG CTC AG] and 1391R [GAC GGG CGG TGW GTR CA]) [23], and 8Fa [TCY SGT TGA TCC TGC S] [13] and 1492R [GGT TAC CTT GTT ATG ACT T] [19].
Real-Time Quantitative PCR (q-PCR)
PCR amplification was quantified in real time (Applied Biosystems prism 7000, California) by flourometric monitoring with SYBR Green 1 dye (Invitrogen, California). All standard curves were constructed using plasmids from cloned rRNA genes (Qiagen, California) separately for archaea, bacteria, and eukarya.
The number of copies in standards was calculated using the Zbio.net online converter (http://www.molbiol.ru/eng/scripts/01_07.html). Slopes of the standard curves generated were−3.03, −3.21, and −3.28 for archaea, bacteria, and eukarya, respectively. All three standard curves achieved a high-correlation coefficient (>0.99). Quantification of genes in each sample was performed in triplicate. Absolute copy number of genes was obtained by interpolation from the respective standard curves generated.
Terminal Restriction Fragment Length Polymorphism (t-RFLP)
Since q-PCR revealed that the vast majority of recoverable phylotypes in all samples were bacterial, we used bacterial t-RFLP as a proxy for diversity. PCR was carried out independently on the nine samples using a ROX-labeled forward primer. Gel-purified amplicons were digested using restriction enzymes HaeIII and MspI (with MspI digest selected for further analysis). Fragment analysis was achieved by capillary electrophoresis (Applied Biosystems 3730 Genetic Analyzer), using a GeneScan ROX-labeled GS500 internal size standard. T-RFLP patterns and quality were analyzed using the freeware PeakScanner™ (version 1.0) (Applied Biosystems https://products.appliedbiosystems.com), and a data matrix comprising fragment sizes and abundance was generated. The Perl and R routines [1] were then used to identify true peaks and binned fragments of similar size. The relative abundance of a true terminal restriction fragment within a given t-RFLP pattern was generated as a ratio of the respective peak area. A virtual digest using MspI was carried out on the sequences retrieved from the bacterial clone library. This allowed the assignment of phylogenetic identity to individual peaks. All t-RFLP GeneScan reads were repeated in triplicate.
Clone Library Construction and Sequencing
Since no significant difference between individual rRNA gene-defined rock communities could be determined using t-RFLP, equimolar mixtures of pooled PCR amplicons from each quadrat were used in the construction of three separate clone libraries for archaea, bacteria, and eukarya. Each pooled sample was gel-purified and used as template for construction of the three clone libraries (Qiagen PCR Cloningplus kit, CA, US). Plasmids were extracted from positive transformants (Mini-M™ Plasmid DNA extraction system, Viogene, Taiwan) and screened by restriction fragment length polymorphism (RFLP) using the restriction endonucleases MspI and CfoI. At least three samples from each distinct RFLP pattern were sequenced using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, California) (Applied Biosystems 3730 Genetic Analyzer). Phylotypes were delineated on the basis of 97% sequence similarity. All sequences generated by this study have been deposited in the NCBI GenBank database under accession numbers FJ790594–FJ790668. Sampling effort was assessed by the calculation of Coverage and Rarefaction curves, and estimates of library richness were made using the non-parametric estimators ACE and Chao 1. All OTU delineation was made on the basis of sequenced phylotypes.
Sequence Analysis
Approximate phylogenetic affiliations were determined by BLAST searches of the NCBI GenBank database (http://www.ncbi.nlm.nic.gov/). Screening for possible chimeric sequences used Chimera_Check (Ribosome Database Project, http://rdp.cme.msu.edu.html). Multiple alignments were created with reference to selected GenBank sequences using Clustal X v.1.81 [33]. Maximum likelihood analysis was performed using PAUP* 4.0b8 [31]. Bayesian posterior probabilities and bootstrap values (1,000 replications) are shown for branch nodes supported by more than 50% of the trees.
Results and Discussion
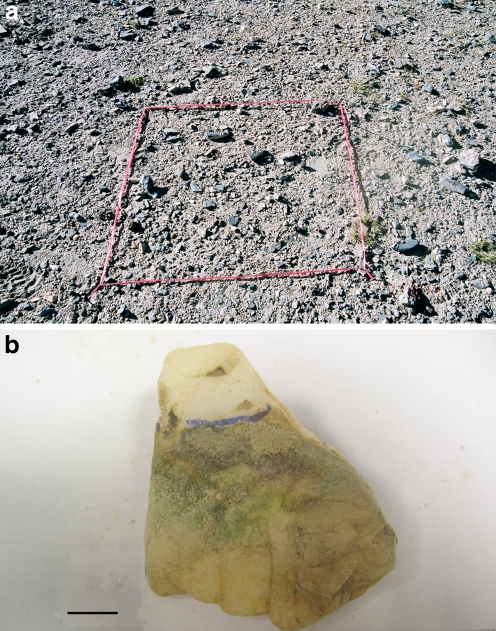
A small-scale survey of colonization at the site indicated that 35.8% (n = 455) of quartz rocks were colonized. A typical quadrat and colonized quartz rock are pictured in Fig. 1. No significant difference in colonization frequency among the nine quadrats was recorded, although we emphasize that this survey was not intended to infer landscape-scale patterns. The colonization frequency was significantly lower than that recorded in similar surveys for the warm arid Mojave Desert [28], the Arctic tundra, and the coastal Antarctic [13] where near-100% colonization occurred. It was, however, markedly higher than the values of around 5% typically reported for the hyper-arid warm and polar deserts [26, 34, 35]. The greatest similarity in colonization frequency was with snow-melt influenced soils (22%) in the Antarctic Dry Valleys [26]. Frequency of colonization has been directly linked to moisture stress in warm deserts [35], and although other stressors such as temperature and irradiance are also likely to be important in polar and high-altitude deserts, it is reasonable to assume that colonization frequency generally reflects the level of aridity. Microscopic observation of colony morphology revealed an unstructured mass of coccoid bacterial cells, plus larger morphotypes suggestive of algal cells and fungal hyphae (Supplementary Online Material, Fig. S1). Biomass was embedded in a copious extracellular polymeric substance and this likely obscured other morphotypes.
Figure 1.
a Quartz pavement at the central Tibetan field location, quadrat = 1 m2; b Typical subsurface hypolithic colonization on a quartz rock, black line drawn with marker pen indicates ground surface level, scale bar = 2 cm
Temperature (soil 5.8–28.4°C, air sub-freezing −12.8°C) and irradiance (PAR 2,072 µm/m2/s, UV-A 2.89 mw/cm2, UV-B 1,734 mw/cm2) at the time of sampling were recorded but have limited context for long-lived microbial communities such as these [35], except to reflect that UV levels were approximately fivefold greater than at sea level in southern China at the same time. Historical climate data indicates that a cold desert tundra environment has persisted in central Tibet over a geological timescale [21]. A ‘greenhouse’ warming effect for the hypolithic habitat has been observed in the Mojave Desert [28], and this was reasoned to be most advantageous during colder winter periods. It is reasonable to envisage a similar benefit to hypoliths in the cold tundra environment of Tibet. We observed that colonized quartz filtered 96.3% (±3.3) of PAR and 100% of incident UV-A and UV-B. At high altitudes where UV irradiance is high this filtering effect is likely critical in terms of community development. Although Chroococcidiopsis is also noted for its radiation-tolerance [4] it is likely that organisms with lower tolerance to radiation stress may exist within the community due to this filtering effect. The hypolithic lifestyle can therefore be viewed as generally a stress-avoidance strategy, and the term ‘refugia’ has been used to describe hypolithic niches in this regard [12].
We utilized q-PCR to establish copy number of rRNA genes as a proxy for the absolute and relative abundance of taxa. These revealed that for bacteria a mean of 8 × 106 copies/cm2 colonized rock (±7.6 × 104) occurred. This corresponded to an estimated 92.6% of the overall recoverable phylotypes for a given colonized rock. Markedly lower values were obtained for archaea (0.37 × 106 copies/cm2 colonized rock ±11.8 × 103, 4.3% of total) and eukarya (0.27 × 106 copies/cm2 colonized rock ±3.1 × 103, 3.1% of total). Other studies using an inter-domain quantitative approach recorded significantly higher copy number for bacteria and eukarya for Antarctic Dry Valleys hypoliths [26] and maritime Antarctic soils [37], although the relative abundance between domains was broadly comparable between all three studies suggesting that community structure of hypoliths varies less than standing biomass.
Since q-PCR revealed that bacteria formed the vast majority of recoverable phylotypes, we used bacterial–terminal restriction fragment length polymorphisms (t-RFLP) as a proxy for overall diversity to assess variation among colonized rocks. Analysis of t-RFLP taxon-abundance profiles revealed no significant difference between rocks (Supplementary Online Material, Fig. S2). Phylogenetic identity was assigned to t-RFLP fragments (on the basis of matching sequences from clone libraries) and revealed that the small variation in t-RFLP profiles could be largely explained by the presence/absence of plastid phylotypes and different 16S rRNA-defined strains of Chroococcidiopsis and Leptolyngbya. The t-RFLP analysis resulted in fewer recoverable phylotypes than the clone library approach using the same PCR primers (<60% the number of phylotypes recovered using clone library), and these generally corresponded to those that were most abundant in the clone library. The potential for underestimating community diversity using this technique has been acknowledged [6] and highlights the need for polyphasic studies where multiple approaches can be used to validate datasets.
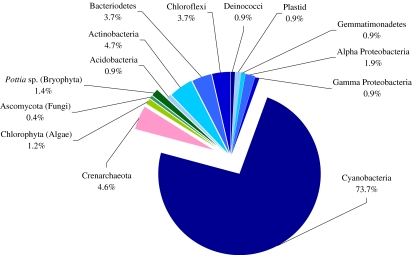
Environmental DNA was used as template for construction of three separate clone libraries using bacteria-, archaea-, and eukarya-specific PCR primers. Estimates of rarefaction, Chao 1, ACE, and Sobs indicated that sampling had approached asymptote for bacterial (n = 103), archaeal (n = 48), and eukaryal (n = 49) libraries. The relatively low diversity encountered in extreme desert environments explains how library sufficiency can be approached with relatively small libraries. Sequencing revealed a total of 62 unique phylotypes (<97% sequence similarity) and these were each assigned to a given phylum based upon phylogenetic identity. By interpolating with q-PCR data we were able to produce an estimate of average overall microbial diversity-abundance across all three domains (Fig. 2). This indicated a community dominated by cyanobacteria (73% of recoverable phylotypes across all domains). This is in general agreement with the only other quantitative estimate of hypolithic community composition, from the Antarctic Dry Valleys [26]. The most commonly recovered cyanobacterial phylotypes indicated Chroococcidiopsis (FJ790616, FJ790614) and a _Phormidium_-like taxon (FJ790627) (Fig. 3). Other cyanobacterial phylotypes belonging to Leptolyngbya, Nostoc, Oscillatoria, and some unidentified phylotypes were all present at low abundances of <10%. Most displayed phylogenetic affiliations with cyanobacterial phylotypes were recovered from hypoliths in the Taklimakan Desert, desert varnish in the Atacama Desert, or Antarctic lakes [22, 27, 32] (Fig. 3). The Chroococcidiopsis, Leptolyngbya, and Nostoc phylotypes indicate diazotrophic potential in the community in addition to photoautotrophy. Cyanobacterial (and algal) primary production appeared to support a relatively low abundance of other taxa and this probably also reflects the nutrient-limiting conditions for heterotrophy in this hypolithic niche.
Figure 2.
Relative abundance of hypolithic phylotypes across all domains from quartz in Tibetan tundra
Figure 3.
Phylogenetic relationships among cyanobacterial 16S rRNA phylotypes recovered from hypolithon in Tibetan tundra. Phylotypes recovered during this study are shown in bold type. Tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar represents 0.1 nucleotide changes per position
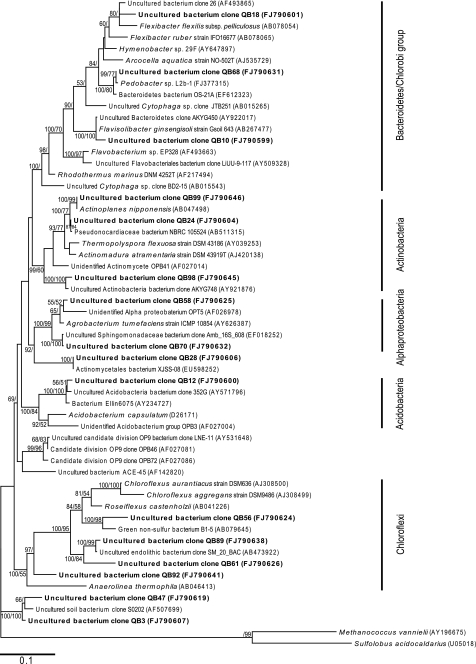
Non-cyanobacterial phyla accounted for approximately 20% of total recoverable phylotypes. Among these the Actinobacteria, Bacteriodetes, and Chloroflexi phylotypes were most commonly encountered (Fig. 4). These had highest affiliation to the Alpine, Antarctic, and Atacama Desert phylotypes and so probably also represent cold/arid-adapted phylotypes. The Chloroflexi have also been recovered from tundra soils in Tibet [13] and hypolithon of warm hyper-arid deserts in China [27]. A likely ecological role for these photoheterotrophs in arid environments is therefore emerging, in addition to their well-studied role in hot spring microbial mats (e.g. [24]). Additional phyla previously recovered from the hypolithon in other deserts were present in the library with relatively low abundance (∼1%); including the Acidobacteria, Deinococci, and Gamma Proteobacteria. This low diversity and abundance may reflect the lack of available organic substrates in addition to environmental stress, since hypolithic communities are thought to rely to a significant degree by photo-excretory products from the primary producers in nutrient-poor desert soils [28].
Figure 4.
Phylogenetic relationships among bacterial 16S rRNA phylotypes recovered from hypolithon in Tibetan tundra. Phylotypes recovered during this study are shown in bold type. Tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar represents 0.1 nucleotide changes per position
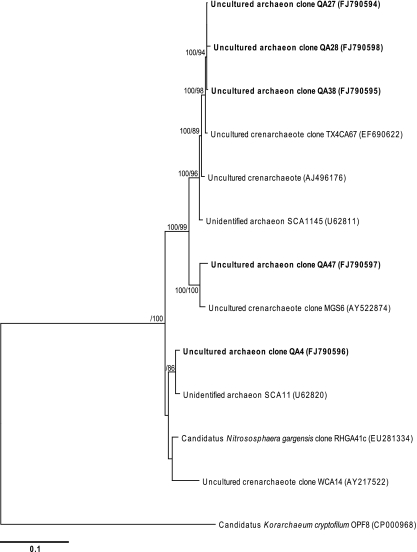
The second most abundant domain after the bacteria was the crenarchaeota (4.3% of recoverable phylotypes). A single phylotype (FJ790594) with phylogenetic affiliations to uncultivated soil crenarchaeotes from non-extreme environments accounted for 90% of archaeal phylotypes (Fig. 5). The archaea remain a relatively under-studied domain in environmental microbiology and almost nothing is known of their ecological role. It is apparent that at the rRNA gene level they appear phylogenetically less diverse in the environment than bacteria [2]. Few studies have targeted archaea in lithic niches, but they were regarded as absent from hypolithon in hyper-arid warm deserts [27, 34] and Antarctic Dry Valleys hypoliths [26], and endoliths [15], after repeated attempts to amplify archaeal rRNA genes using archaea-specific PCR primers.
Figure 5.
Phylogenetic relationships among archaeal 18S rRNA phylotypes recovered from hypolithon in Tibetan tundra. Phylotypes recovered during this study are shown in bold type. Tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar represents 0.1 nucleotide changes per position
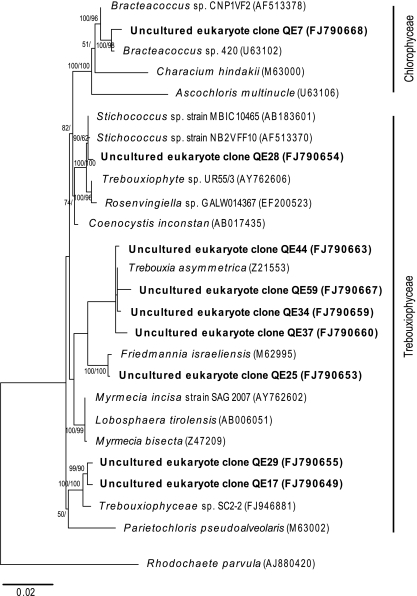
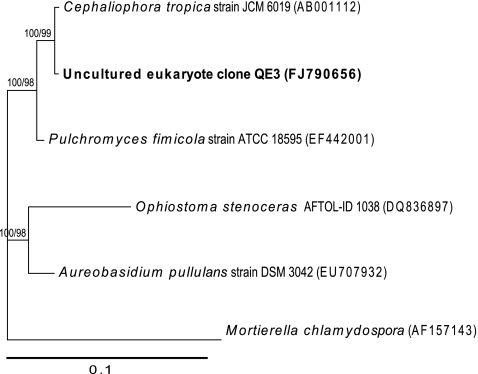
Moss phylotypes accounted for almost half of the recoverable phylotypes from the eukaryal clone library but accounted for only around 1.5% of overall phylotypes in the hypolithon. Moss phylotypes indicated a _Pottia_-like organism that is known as a common colonist in polar locations [29]. No visual evidence of moss growth was observed, and this suggests that the moss phylotypes may have represented resting structures awaiting more favorable conditions for germination. Presence of the known endolithic lichen phycobiont Trebouxia sp. and related taxa [36] was indicated (Fig. 6) although the only fungal phylotype recovered (Elutherascus sp.) (Fig. 7) is not a previously recorded lichen mycobiont. The nature of the lichen relationship in lithic environments is poorly understood and so this may reflect a previously unappreciated lichen association and/or free living eukaryotes.
Figure 6.
Phylogenetic relationships among algal 18S rRNA phylotypes recovered from hypolithon in Tibetan tundra. Phylotypes recovered during this study are shown in bold type. Tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar represents 0.1 nucleotide changes per position
Figure 7.
Phylogenetic relationships among fungal 18S rRNA phylotypes recovered from hypolithon in Tibetan tundra. Phylotypes recovered during this study are shown in bold type. Tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar represents 0.1 nucleotide changes per position
The occurrence and multi-domain composition of this community may have important ecological implications. Primary productivity estimates for Tundra soils have not previously taken into account the hypolithic community, and this is clearly a significant subsurface category of biomass in otherwise biologically relatively depauperate tundra soils. All three components (cyanobacteria, putative lichen, and moss) are known individually as colonists of cold environments [14]. The major cyanobacterial component can be envisaged as being a significant driver of carbon and nitrogen input. The lichenized part of the community may become important under severe xeric conditions since lichens can exploit niches with relatively low-water activities [20]. Conversely, the mosses can be envisaged to provide a significant contribution to productivity in times of moisture sufficiency. This presents a picture of a cold-adapted community with partitioned activity among co-colonists that creates the potential for hypolithic productivity over a range of moisture availability regimes encountered in the high-altitude Tibetan tundra.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are grateful to the Tibet Ministry of Geology for their fieldwork assistance. This work was supported by the Hong Kong Research Grants Council (grant number HKU 7733/08M).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Abdo Z, Schüette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- 2.Aller JY, Kemp PF. Are archaea inherently less diverse than bacteria in the same environments? FEMS Microbiol Ecol. 2008;65:74–87. doi: 10.1111/j.1574-6941.2008.00498.x. [DOI] [PubMed] [Google Scholar]
- 3.Berner T, Evanari M. The influence of temperature and light penetration on the abundance of the hypolithic algae in the Negev Desert of Israel. Oecologia. 1978;33:255–260. doi: 10.1007/BF00344852. [DOI] [PubMed] [Google Scholar]
- 4.Billi D, Friedmann IE, Hofer KG, Grilli-Caiola M, Ocampo-Friedmann R. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl Environ Microbiol. 2000;66:1489–1492. doi: 10.1128/AEM.66.4.1489-1492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billi D, Ghelardini P, Onofri S, Cockell CS, Rabbow E, Horneck G. Desert cyanobacteria under simulated space and Martian conditions. EPSC Abstracts. 2008;3:EPSC2008-A-00474. [Google Scholar]
- 6.Blackwood CB, Hudleston D, Zak DR, Buyer JS. Interpreting ecological diversity indices applied to t-RFLP data: insights form simulate microbial communities. Appl Environ Microbiol. 2007;73:5276–5283. doi: 10.1128/AEM.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broady PA. The ecology of sublithic terrestrial algae at the Vestfold Hills, Antarctica. British Phycol J. 1981;16:231–240. doi: 10.1080/00071618100650241. [DOI] [Google Scholar]
- 8.Broady PA. The distribution of terrestrial and hydro-terrestrial algal associations at three locations in southern Victoria Land, Antarctica. Algalogical Studies. 2005;118:95–112. doi: 10.1127/1864-1318/2006/0118-0095. [DOI] [Google Scholar]
- 9.Budel B, Wessels DCJ. Rock inhabiting blue green algae from hot arid regions. Archiv Fur Hydrobiologie. 1991;92:385–398. [Google Scholar]
- 10.Cameron RE, Blank GB. Soil studies—microflora of desert regions VIII. Distribution and abundance of microorganisms. Space Programs Summary. 1965;4:193–202. [Google Scholar]
- 11.Cockell CS, Stokes MD. Widespread colonization by polar hypoliths. Nature. 2004;431:414–415. doi: 10.1038/431414a. [DOI] [PubMed] [Google Scholar]
- 12.Cockell CS, Stokes MD. Hypolithic colonization of opaque rocks in the Arctic and Antarctic polar desert. Arct Antarct Alp Res. 2006;38:335–342. doi: 10.1657/1523-0430(2006)38[335:HCOORI]2.0.CO;2. [DOI] [Google Scholar]
- 13.Costello EK, Schmidt SK. Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ Microbiol. 2006;8:1471–1486. doi: 10.1111/j.1462-2920.2006.01041.x. [DOI] [PubMed] [Google Scholar]
- 14.Cowan D, Ah Tow L. Endangered Antarctic environments. Ann Rev Microbiol. 2004;58:649–690. doi: 10.1146/annurev.micro.57.030502.090811. [DOI] [PubMed] [Google Scholar]
- 15.de la Torre JR, Goebel BR, Friedmann EI, Pace NR. Microbial diversity of cryptoendolithic communities from the McMurdo dry valleys, Antarctica. Appl Environ Microbiol. 2003;69:3858–3867. doi: 10.1128/AEM.69.7.3858-3867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedmann I, Galun M. Desert algae, lichens and fungi. Desert Biology Vol II. New York: Academic; 1974. pp. 165–212. [Google Scholar]
- 17.Friedmann IE. Endolithic microorganisms in the Antarctic cold desert. Science. 1982;215:1045–1053. doi: 10.1126/science.215.4536.1045. [DOI] [PubMed] [Google Scholar]
- 18.Golubic S, Friedmann I, Schneider J. The lithobiontic ecological niche, with special reference to microorganisms. J Sediment Petrol. 1981;51:475–478. [Google Scholar]
- 19.Harris JK, Kelley ST, Pace NR. New perspectives on uncultured bacterial phylogenetic division OP11. Appl Environ Microbiol. 2004;70:845–849. doi: 10.1128/AEM.70.2.845-849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kranner I, Beckett R, Hochman A, Nash TH. Desiccation-tolerant lichens: a review. The Bryologist. 2008;111:576–593. doi: 10.1639/0007-2745-111.4.576. [DOI] [Google Scholar]
- 21.Kuhle M (1990) The cold deserts of high Asia (Tibet and contiguous mountains). GeoJournal:319–323
- 22.Kuhlman KR, Venkat P, La Duc MT, Kuhlman GM, McKay CP. Evidence of a microbial community associated with rock varnish at Yungay, Atacama Desert, Chile. J Geophys Res, Biogeosciences. 2008;113:G04022. doi: 10.1029/2007JG000677. [DOI] [Google Scholar]
- 23.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester: Wiley; 1991. pp. 115–148. [Google Scholar]
- 24.Lau MCY, Pointing SB. Vertical partitioning and expression of primary metabolic genes in a thermophilic microbial mat. Extremophiles. 2009;13:533–540. doi: 10.1007/s00792-009-0240-8. [DOI] [PubMed] [Google Scholar]
- 25.McKay CP. Relevance of Antarctic microbial ecosystems to exobiology. In: Friedmann EI, editor. Antarctic microbiology. New York: Wiley; 1993. pp. 593–601. [Google Scholar]
- 26.Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens J, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pointing SB, Warren-Rhodes K, Lacap DC, Rhodes KL, McKay CP. Hypolithic community shifts occur as a result of liquid water availability along environmental gradients in China’s hot and cold hyperarid deserts. Environ Microbiol. 2007;9:414–424. doi: 10.1111/j.1462-2920.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 28.Schlesinger WH, Pippen JS, Wallenstein MD, Hofmockel KS, Klepeis DM, Mahall BE. Community composition and photosynthesis by photoautotrophs under quartz pebbles, southern Mojave Desert. Ecology. 2003;84:3222–3231. doi: 10.1890/02-0549. [DOI] [Google Scholar]
- 29.Seppelt RD, Green TGA, Schwartz A-MJ, Frost A. Extreme southern locations for moss sporophytes in Antarctica. Antarct Sci. 1992;4:37–39. doi: 10.1017/S0954102092000087. [DOI] [Google Scholar]
- 30.Smith MC, Bowman JP, Scott FJ, Line MA. Sublithic bacteria associated with Antarctic quartz stones. Antarct Sci. 2000;12:177–184. [Google Scholar]
- 31.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Version 4.0b8. Sunderland: Sinauer; 2001. [Google Scholar]
- 32.Taton A, Grubisic S, Balthasart P, Hodgson DA, Laybourn-Parry J, Wilmotte A. Biogeographical distribution and ecological ranges of cyanobacteria in East Antarctic lakes. FEMS Microbiol Ecol. 2006;57:272–289. doi: 10.1111/j.1574-6941.2006.00110.x. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren-Rhodes K, Rhodes KL, Pointing SB, Boyle L, Dungan J, Liu S, Zhou P, McKay CP. Lithic cyanobacterial ecology across environmental gradients and spatial scales in China’s hot and cold deserts. FEMS Microbiol Ecol. 2007;61:470–482. doi: 10.1111/j.1574-6941.2007.00351.x. [DOI] [PubMed] [Google Scholar]
- 35.Warren-Rhodes K, Rhodes KL, Pointing SB, Ewing S, Lacap DC, Gómez-Silva B, Amundson R, Friedmann IE, McKay CP. Hypolithic cyanobacteria, dry limit of photosynthesis and microbial ecology in the hyperarid Atacama Desert, Chile. Microb Ecol. 2006;52:389–398. doi: 10.1007/s00248-006-9055-7. [DOI] [PubMed] [Google Scholar]
- 36.Wong FKY, Lau MCY, Aitchison JC, Cowan DA, Pointing SB. Endolithic microbial communities of limestone niches in a high-altitude arid environment. Microb Ecol. 2010 doi: 10.1007/s00248-009-9607-8. [DOI] [PubMed] [Google Scholar]
- 37.Yergeau E, Bokhorst S, Huiskes AHL, Boshker HTS, Aerts R, Kowlachuk GA. Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microb Ecol. 2007;59:436–451. doi: 10.1111/j.1574-6941.2006.00200.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.