A Cell-permeable Stat3 SH2 Domain Mimetic Inhibits Stat3 Activation and Induces Antitumor Cell Effects in Vitro (original) (raw)
Abstract
Given the role of constitutively active Signal Transducer and Activator of Transcription (Stat) 3 in human tumors, Stat3 inhibitors would be useful as novel therapeutics and as tools for probing Stat3-mediated tumor processes. We herein report that a 28-mer peptide, SPI, derived from the Stat3 SH2 domain, replicates Stat3 biochemical properties. Studies show SPI and Stat3 (or Stat3 SH2 domain) bind with similar affinities to known Stat3-binding phosphotyrosine (pY) peptide motifs, including those of the epidermal growth factor receptor (EGFR) and the high-affinity, IL-6R/gp130-derived pY-peptide, GpYLPQTV-NH2. Consequently, SPI functions as a potent and selective inhibitor of Stat3 SH2 domain:pTyr interactions and disrupts the binding of Stat3 to the IL-6R/gp130 peptide, GpYLPQTV-NH2. Fluorescence imaging and immunofluorescence staining/laser-scanning confocal microscopy show SPI is cell membrane-permeable, associates with the cytoplasmic tail of EGFR in NIH3T3/hEGFR, and is present in the cytoplasm, but strongly localized at the plasma membrane and in the nucleus in malignant cells harboring persistently active Stat3. Moreover, SPI specifically blocks constitutive Stat3 phosphorylation, DNA binding activity, and transcriptional function in malignant cells, with little or no effect on the induction of Stat1, Stat5, and Erk1/2MAPK pathways, or on general pTyr profile at the concentrations that inhibit Stat3 activity. Significantly, treatment with SPI of human breast, pancreatic, prostate, and non-small cell lung cancer cells harboring constitutively active Stat3 induced extensive morphology changes, associated with viability loss and apoptosis. Our study identifies SPI as a novel molecular probe for interrogating Stat3 signaling and that functions as a selective inhibitor of Stat3 activation with antitumor cell effects.
Keywords: Growth Factors, Peptides, SH2 Domains, Signal Transduction, STAT Transcription Factor, S3I-201, Stat3, Antitumor Cell Effects
Introduction
The binding of cytokines or growth factors to cognate receptors initiates a cascade of molecular events that culminate in the activation of the Signal Transducer and Activator of Transcription (STAT) family of proteins (1, 2). Among these is the recruitment of STATs, via the SH2 domain, to the receptor phosphotyrosine (pTyr)2 peptide motifs, which brings them into close proximity for phosphorylation on a key tyrosyl residue by growth factor receptor tyrosine kinases, Janus kinases (Jaks), and the Src family kinases. Consequently, dimerization between two STAT monomers is promoted through a reciprocal pTyr-SH2 domain interaction, and the active STAT dimers in the nucleus bind to specific DNA-response elements in the promoters of target genes and regulate gene expression. In response to growth factors and cytokines, normal STAT signaling promotes cell growth and differentiation, development, inflammation, and immune responses.
The STAT proteins are modular in structure and contain N-terminal domain, coiled-coil domain, DNA-binding domain, SH2 domain, and a transcriptional activation domain, with each domain engaging in important molecular events for promoting STAT functions. In particular, the SH2 domain mediates crucial interactions with specific pTyr peptide motifs, including promoting the association with receptors and holding up two activated STAT monomers together in a reciprocal SH2 domain:pTyr interactions in STAT:STAT dimerization. Among the STAT family members, Stat3 and Stat5 have been strongly implicated in malignant transformation and tumorigenesis (3–7) and have become valid targets for anticancer drug design. In general, given the role of the SH2 domain as an important motif in signal transduction, in relation to engaging in interactions with pTyr peptide modules (8), there are considerable efforts to design probes that disrupt these interactions for potential application as drugs (9–12). For Stat3, pTyr peptide mimetics have been shown to suppress its functions. Thus, the Stat3 SH2 domain:pTyr peptide interaction has become an attractive target in many drug design strategies intended to identify small molecule inhibitors as new therapeutics for cancers in which aberrant Stat3 activity is implicated (4, 5, 13–22).
Whereas the focus of the existing Stat3 drug discovery efforts have been on disrupting the Stat3 SH2 domain:pTyr peptide interactions for a good reason, the approaches have largely been directed at SH2 domain antagonists, which are pTyr peptide mimics that compete for the binding to the Stat3 SH2 domain (4, 5, 22). One of the major limitations of this approach has been finding a membrane-permeable, optimum pTyr substitute that retains the high binding affinities of the native pTyr peptide motifs, against which these antagonists will be competing for the binding to the Stat3 SH2 domain. To eliminate this issue, we have taken the converse approach of identifying a suitable Stat3 SH2 domain-mimic. Key structural information from the computational modeling of the native pTyr peptide, PpYLKTK bound to the Stat3 SH2 domain, per the crystal structure of Stat3β (23) facilitated the design of a 28-mer peptide, SPI from the Stat3 SH2 domain. Studies presented herein show that SPI retains the binding characteristics of the SH2 domain. In vitro biochemical and biophysical studies indicate SPI, like Stat3 binds to cognate pTyr-peptide motifs with a similar affinity. Accordingly, SPI blocks the binding of Stat3 (or Stat3 SH2 domain) to cognate pTyr peptide motifs, and hence functions as a selective inhibitor of constitutive Stat3 activation in human breast, prostate, pancreatic, and non-small cell lung cancer cells, with antitumor cell effects.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Normal mouse fibroblasts (NIH3T3) and counterparts transformed by v-Src (NIH3T3/v-Src) or overexpressing the human epidermal growth factor (EGF) receptor (NIH3T3/hEGFR), and the human breast (MDA-MB-231, MDA-MB-435, and MCF-7), pancreatic (Colo-357), prostate (DU145 and LNCaP), non-small cell lung (A549) cancer, and TE-71 mouse thymus epithelial stromal cells have all been previously reported (14, 24–28). The Stat3-dependent reporter, pLucTKS3 and the Stat3-independent reporter, pLucSRE, and the v-Src transformed mouse fibroblasts that stably express pLucTKS3 (NIH3T3/v-Src/pLucTKS3) or pLucSRE (NIH3T3/v-Src/pLucSRE), and the Stat3-independent β-casein luciferase reporter (β-Casein-Luc) driven by the Stat5-responsive β-casein promoter have been previously reported (13, 14, 16, 29, 30). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum.
Peptide Synthesis
The Stat3 SH2 domain peptide sequence, FISKERERAILSTKPPGTFLLRFSESSK was purchased from Peptide 2.0 (Fairfax, VA) at >95% purity.
Cloning and Protein Expression
The molecular cloning, expression, and the purification of His-tagged Stat3 and His-tagged Stat3 SH2 domain were carried out as we have previously reported (27). Clones were sequenced to verify the correct sequences and orientation. His-tagged recombinant proteins were expressed in BL21(DE3) cells and purified on Ni-ion Sepharose column.
Transient Transfection of Cells and Treatment with SPI
12–24 h following seeding, mouse fibroblasts overexpressing hEGFR (NIH3T3/hEGFR) in 6-well plates were transiently co-transfected with 4 μg of β-casein Luc and 500 ng of β-galactosidase (for normalizing) for 8 h using Lipofectamine plus (Invitrogen, Carlsbad, CA) and following the manufacturer's protocol. Twelve hours after transfection, cells were treated or untreated with increasing concentration of SPI (0–60 μm) for 12 h prior to stimulation with rhEGF (10 ng/μl) and allowed to culture for additional 12 h, after which cells were harvested and cytosolic extracts prepared for luciferase assay, as previously performed (16, 29, 30).
Cytosolic Extracts and Luciferase Assay
Cytosolic extract preparation from mammalian cells for luciferase assay has been described previously (29, 30). Luciferase assays were carried out according to the supplier's (Promega, Madison, WI) manual and measured with a luminometer (Lumat LB 9507, EG&G Berthold, Germany).
Nuclear Extract Preparation and Electrophoretic Mobility Shift Assay
Nuclear extract preparation and electrophoretic mobility shift assay (EMSA) were carried out as previously described (25, 30). The 32P-labeled oligonucleotide probes used were hSIE (high affinity sis-inducible element from the c-fos gene, m67 variant, 5′-AGCTTCATTTCCCGTAAATCCCTA) that binds Stat1 and Stat3 (31) and MGFe (mammary gland factor element from the bovine β-casein gene promoter, 5′-AGATTTCTAGGAATTCAA) for Stat1 and Stat5 binding (32, 33). Where appropriate, cells in culture were pretreated with SPI for 12 h, prior to treatment with sodium orthovanadate for 12 h or stimulation with EGF (10 ng/μl) for 12 min and then harvested for nuclear extract preparation.
SDS-PAGE/Western Blotting Analysis
SDS/PAGE and Western blotting analysis were performed as previously described (30, 34). Primary antibodies used were anti-Stat3, pY705Stat3, pErk1/2, and Erk1/2 (Cell Signaling).
Cell Viability, Proliferation, and Annexin V/Flow Cytometry Studies
Cells in culture in 6-well or 96-well plates were treated with or without 50 μm SPI for 24–48 h and subjected to CyQuant cell proliferation assay (Invitrogen Corp), or harvested, and the viable cells counted by trypan blue exclusion with phase contrast microscopy, or cells were processed for Annexin V and 7-AAD binding (BD Biosciences, San Jose, CA) with flow cytometry for apoptosis.
Fluorescence Imaging and Immunofluorescence with Laser-scanning Confocal Microscopy
Studies were performed as previously reported (35). Briefly, human breast cancer, MDA-MB-231, or NIH3T3/hEGFR cells were grown in multi-cell plates on slides or not, and treated with or without 5-carboxyfluorescein-labeled SPI or unlabeled SPI (30 μm) for 3 h. Cells were washed with 1× phosphate-buffered saline (PBS), fixed with ice-cold methanol, and visualized using Zeiss Axiovert 200 microscope (Zeiss, Germany) for fluorescent images, or for confocal microscopy, cells were washed three times with 1× PBS, fixed with ice-cold methanol for 15 min, washed three times in PBS, permeabilized with 0.2% Triton X-100 for 10 min, and further washed 3–4 times with PBS. Specimens were then blocked in 1% bovine serum albumin (BSA) for 1 h and incubated with anti-EGFR antibody (Santa Cruz Biotechnology) or rabbit polyclonal anti-Stat3 antibody (Cell Signaling) at 1:50 dilution at 4 °C overnight. Subsequently, cells were rinsed 4–5 times in PBS, incubated for 1 h at room temperature in the dark with AlexaFluor 546 rat secondary antibody for anti-EGFR antibody detection (Invitrogen) or AlexaFluor546 (donkey anti-rabbit) secondary antibody for anti-Stat3 antibody detection. Specimens were then washed five times with PBS, covered with cover slides with VECTASHIELD mounting medium containing DAPI, and examined immediately under a Leica TCS SP5 confocal microscope (Germany) at appropriate wavelengths. Images were captured and processed using the Leica TCS SP 5 software.
Fluorescence Polarization Assay
Fluorescence Polarization (FP) Assay was conducted as previously reported (20, 27) using the labeled phosphopeptide, 5-carboxyfluorescein-GpYLPQTV-NH2 (where pY represents phospho-Tyr) as probe and Stat3 or SPI. For saturation curves, a fixed concentration of the fluorescently labeled peptide probe (10 nm) was incubated with increasing concentration of Stat3 (0–0.8 μm) or SPI (0–400 μm) for 30 min at room temperature in the buffer, 50 mm NaCl, 10 mm HEPES, 1 mm EDTA, 0.1% Nonidet P-40, and the fluorescent polarization measurements were determined using the POLARstar Omega (BMG LABTECH, Durham, NC), with the set gain adjustment at 35 mP. For evaluating the effect of SPI as an inhibitor on Stat3 binding to pY peptide, a fixed concentration of Stat3 protein (0.8 μm) was preincubated with serial concentrations of SPI (0–150 μm), or in the case of the effect of the Stat3 inhibitor, S3I-201 on the binding of Stat3 or SPI to probe, a fixed amount of Stat3 (200 nm) or SPI (150 μm) was preincubated with increasing concentration of S3I-201 at 30 °C for 60 min in the indicated assay buffer conditions, prior to the addition of the labeled probe. Probe was then added at a final concentration of 10 nm and incubated for 30 min at room temperature following which the FP measurements were taken using the POLARstar Omega, with the set gain adjustment at 35 mP.
Surface Plasmon Resonance Analysis
SensiQ and its analysis software Qdat (ICX Technologies, Oklahoma City, OK) were used to analyze the interaction between known Stat3-binding pTyr peptide motifs (analyte) and Stat3 or the Stat3 SH2 domain (target) and to determine the binding affinity, as previously reported (27). Purified Stat3 (50 μg), Stat3 SH2 domain (30 μg), or SPI (25 μg) was immobilized on a Carboxyl Sensor Chip (for SPI) or a HisCap Sensor Chip (for Stat3 and the Stat3 SH2 domain) by injecting the peptide or protein onto the chip. Various concentrations of pTyr peptides (analyte) in running buffer (1× PBS, 0.5% DMSO) were passed over the sensor chip to produce response signals. The response signals were referenced by subtracting the response generated by passing across the unoccupied chip surface the running buffer with the analytes. The association and dissociation rate constants were calculated using the Qdat software. The ratio of the association and dissociation rate constants was determined as the affinity (KD).
Statistical Analysis
Statistical analysis was performed on mean values using Prism GraphPad Software, Inc. (La Jolla, CA). The significance of differences between groups was determined by the paired t test at p < 0.05 (*), < 0.01 (**), and < 0.001 (***).
RESULTS
Computer-aided Design of SPI as a Molecular Probe and Stat3 Inhibitor
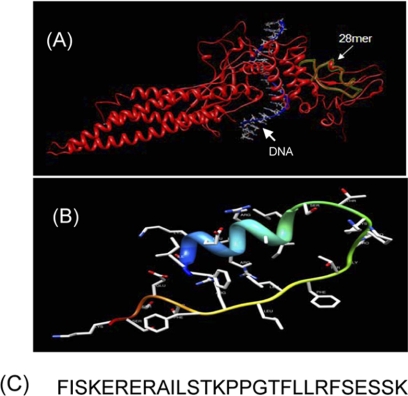
Close structural analysis of the lowest Genetic Optimization for Ligand Docking (GOLD) (36) conformation of the native pTyr peptide, PpYLKTK bound within the Stat3 SH2 domain (16), per the x-ray crystal structure of Stat3β homodimer (23), showed significant complementary interactions at the protein surface, by which a minimum SH2 domain peptide sequence was derived that retains interactions with the pTyr peptide. The lowest energy GOLD docking studies consistently showed the pTyr peptide making hydrogen bonds and electrostatic interactions with the key residues, Lys-591, Ser-611, Ser-613, and Arg-609 of the SH2 domain. The SH2 domain peptide, SPI, is composed of amino acid residues 588–615, which incorporate the aforementioned key residues (Fig. 1C). Fig. 1A shows the spatial presentation of the 28-mer peptide (28-mer, green overlay) within the context of the three-dimensional structure of Stat3 (red), per the x-ray crystal structure of Stat3β (23), and Fig. 1B shows the three-dimensional structure of the 28-mer as developed per comparative modeling method using ModWeb (University of California, San Francisco (UCSF)) (37), with a score of 0.87, and viewed by Chimera software (UCSF) (38) (because of ModWeb software requirements, of a minimum of 30 amino acids, three Gly residues were added to the N terminus of the 28-mer during the modeling, but were eliminated at the time of viewing with Chimera software). Sequence alignment analysis of SPI with SH2 motifs from other proteins reveals high homologies, 78% and 73%, respectively, to the Stat1 and Stat4 SH2 domains, and 40–57% homologies to the other proteins evaluated, including Stat2, Stat5, Stat6, Src, Fyn, Fgr, TNS3 (Tensin-3 protein), and SH2D2A (SH2 domain protein 2A) (Table 1). Molecular modeling raises the potential that SPI retains significant three-dimensional structural characteristics as in the full-length Stat3 protein. Studies were conducted for the characterization of the biochemical and biophysical properties of SPI relative to Stat3, and to determine the potential to inhibit Stat3 activation and functions.
FIGURE 1.
Computer modeling of Stat3 or the Stat3 SH2 domain peptide (SPI) and SPI amino acid sequence. A, the spatial presentation of the 28-mer peptide (28-mer, green overlay) within the context of the three-dimensional structure of Stat3 (red), per the x-ray crystal structure of DNA-bound Stat3β. B, structure of SPI, as modeled using ModWeb (University of California, San Francisco (UCSF)) and viewed by UCSF Chimera software. Model Score, 0.87; and C, amino acid sequence for residues 588–615 of the Stat3 SH2 domain.
TABLE 1.
Sequence homology between SPI and SH2 motifs of select human proteins
Alignment and homology were determined using NCBI Blast.
| Protein | Gene ID | Identity |
|---|---|---|
| % | ||
| Stat3 | 6774 | 100 |
| Stat1 | 6772 | 78 |
| Stat4 | 6775 | 73 |
| Stat2 | 6773 | 57 |
| Stat6 | 6778 | 57 |
| Stat5A | 6776 | 53 |
| Stat5B | 6777 | 53 |
| Fyn | 2534 | 53 |
| Fgr | 2268 | 50 |
| Src | 6714 | 50 |
| TNS3 | 64759 | 48 |
| SH2D2A | 9047 | 44 |
In Vitro Evidence That SPI Interacts with Cognate pTyr Peptide Motifs and Disrupts the Binding of Stat3 or the Stat3 SH2 Domain
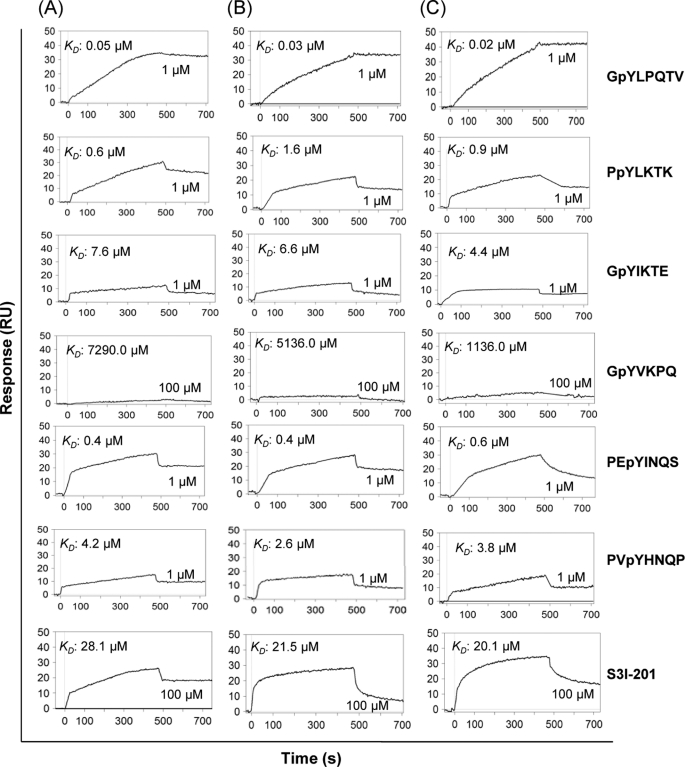
We presume that SPI would bind to pTyr peptide motifs that are known to bind to Stat3. To provide definitive evidence of direct binding of SPI to known Stat3-binding pTyr peptide motifs and to compare with the binding of Stat3 or the Stat3 SH2 domain, biophysical studies were performed. Purified, His-tagged Stat3 protein (50 μg) or His-tagged Stat3 SH2 domain (30 μg), or SPI (25 μg) was immobilized (target) on a Ni-NTA or carboxyl sensor chip surface for SPR analysis of the binding to pTyr peptides (analyte). Association and dissociation measurements were taken and the affinities were determined using Qdat software. We considered the known Stat3-binding pTyr peptide motifs, PpYLKTK (pY705Stat3 involved in Stat3:Stat3 and Stat1:Stat3 dimerization events), GpYIKTE (pY701Stat1 involved in Stat1:Stat1 and Stat1:Stat3 dimerization events), PEpYINQS and PVpYHNQP (pY1068EGFR and pY1086EGFR, respectively), and GpYLPQTV (IL-6R/gp130), and the probable binding motif, GpYVKPQ (pY694Stat5) (pTyr peptide sequences were derived from the NCBI protein data base), and the previously reported Stat3 dimerization inhibitor and the SH2 domain antagonist, S3I-201 (16).
Overall, the interactions with the aforementioned pTyr peptides (analytes) were comparable for the three targets, SPI (Fig. 2A), Stat3 SH2 domain (Fig. 2B), and Stat3 (Fig. 2C). Specifically, the interactions with the gp130-derived peptide, GpYLPQTV, were of the highest affinity, with KD values of 50.0, 30.0, and 20.0 nm, respectively, for SPI, Stat3 SH2 domain and Stat3 (Fig. 2, GpYLPQTV). Relative to these affinities, the interactions with the Stat3-derived pY705 peptide, PpYLKTK were 13–100-fold weaker, with KD values of 0.6, 1.6, and 0.9 μm, for SPI, Stat3 SH2 domain, and Stat3, respectively (Fig. 2, PpYLKTK), as were the interactions with the pY1068 of EGFR, PEpYINQS, with KD values of 0.4, 0.4, and 0.6 μm, respectively, for SPI, Stat3 SH2 domain, and Stat3 (Fig. 2, PEpYINQS). Even much weaker were the interactions of the three targets with the pY1086 peptide of EGFR, PVpYHNQP, with KD values of 4.2, 2.6, and 3.8 μm, respectively, for SPI, Stat3 SH2 domain, and Stat3, suggesting with respect to the two known Stat3-binding EGFR pTyr motifs, pY1068EGFR (PEpYINQS) and pY1086EGFR (PVpYHNQP), Stat3 and SPI display differential binding. A surprising finding is the relatively weaker binding to the Stat1 pY701 peptide, GpYIKTE, with KD values of 7.6, 6.6, and 4.4 μm, for SPI, Stat3 SH2 domain, and Stat3, respectively, which compared with the binding to Stat3 pY705 peptide (PpYLKTK) represent 3–13-fold difference in affinity, despite there being 78% sequence homology between the Stat3 and Stat1 SH2 domains (Table 1) and that when activated concurrently, Stat1 and Stat3 engage in a heterodimer formation. We deduce that overall, the binding of Stat3 (or Stat3 SH2 domain) or SPI to the Stat1 pY701 peptide is weaker compared with their binding to the pTyr peptides derived from the IL-6R/gp130, pY705Stat3, or the pY1068EGFR. The data together strongly suggest SPI and monomeric Stat3 have preference for pStat3 monomer over pStat1 monomer. SPR analysis similarly showed SPI, Stat3 SH2 domain, and Stat3 interact with the small-molecule, SH2 domain antagonist, S3I-201, with KD values of 28.1, 21.5, 20.1 μm, respectively (Fig. 2, S3I-201). Although weaker, the interactions with S3I-201 exhibit similar characteristics for SPI and Stat3 (or Stat3 SH2 domain). By contrast, data show that SPI, Stat3 SH2 domain, and Stat3 interact poorly with the pTyr peptide of Stat5, GpYVKPQ, with KD values of 7.3, 5.2, and 1.2 mm, respectively (Fig. 2, GpYVKPQ), which indicate far weaker affinities for Stat5, and which also suggest SPI, like Stat3, is more likely to interact with pStat3 than pStat5. Furthermore, the data indicate that interactions with pStat5 are rather a low probability, given the millimolar affinities.
FIGURE 2.
SPR analysis. SPR analysis of the binding of SPI (A), Stat3 SH2 domain (B), or full-length Stat3 (C) to GpYLTQTV-NH2 (high affinity IL-6R/gp-130 peptide), PpYLKTK (Stat3 pTyr peptide), GpYIKTE (Stat1 pTyr peptide), GpYVKPQ (Stat5 pTyr peptide), PEpYINQS (EGFR pTyr peptide), PVpYHNQP (EGFR pTyr peptide), and S3I-201 (Stat3 dimerization inhibitor). Data are representative of three independent determinations.
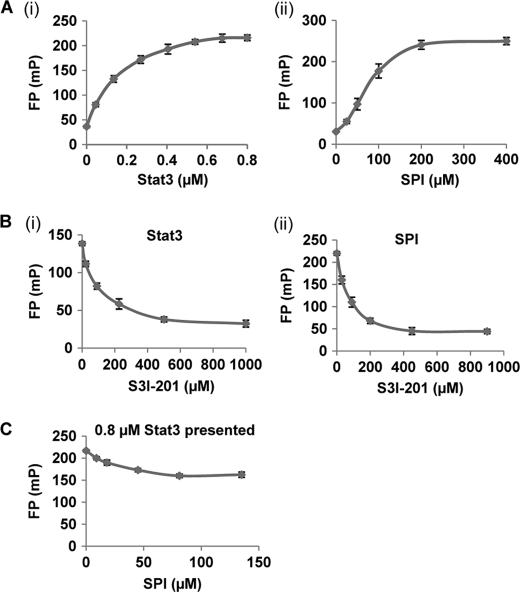
The studies so far demonstrate that SPI interacts with cognate pTyr peptide motifs recognized by Stat3 (or Stat3 SH2 domain). The interaction with SPI could block the binding of Stat3 to its cognate pTyr peptide motifs. Fluorescence polarization (FP) study has previously been used to demonstrate the binding of Stat3 (or Stat3 SH2 domain) to the high-affinity peptide, GpYLPQTV-NH2 (19, 20, 27). This assay was therefore used to verify SPI binding to the high-affinity pTyr peptide motif and to further assess the potential that such a binding could disrupt the association of Stat3 with the same peptide probe. Results of the FP assay, utilizing the 5-carboxyfluorescein-GpYLPQTV-NH2 as a probe, showed a similar binding saturation profile, measured as fluorescence polarization signal (mP), with the increasing concentration (in μm) of purified His-Stat3 (Fig. 3A, panel i) or SPI (Fig. 3A, panel ii), suggesting SPI binds similarly as Stat3 to the pTyr peptide probe. Thus, SPI behaves similarly to Stat3 in binding to cognate peptides. The results show that the binding of the fluorescent probe to Stat3 causes a much higher polarization compared with binding to SPI. This difference in the degree of polarization is not suggestive of differences in the affinities for the 5-carboxyfluorescein-GpYLPQTV-NH2 probe. Rather, it can be explained on the basis of the little ability of larger molecules to tumble, and hence, Stat3 protein being very large in size and much bigger than SPI induces a higher degree of polarization. Compared with Stat3, it would be expected that the SPI-bound probe remains sufficiently small to facilitate some degree of tumbling and therefore produces lower polarization. To further evaluate the SPI:peptide probe interaction, relative to the binding of Stat3 to the same probe, we compared the responsiveness of the interactions to the known Stat3 dimerization inhibitor, S3I-201 (16). Here, fixed concentrations of each of Stat3 (200 nm) and SPI (150 μm) were incubated with increasing concentrations of S3I-201 prior to incubation with the 5-carboxyfluorescein-GpYLPQTV-NH2 probe and subjected to FP measurements. Consistent with the observed similarities in the saturation curves (Fig. 3A), the profiles of the S3I-201-induced inhibition of the binding to the labeled peptide probe are similar, regardless of whether Stat3 or SPI is the ligand (Fig. 3B, panel i, Stat3, and B, panel ii, SPI). Next, we examined the potential that by binding to the labeled pTyr peptide probe, SPI could compete against Stat3. To address this, aliquots of fixed concentration of Stat3 (0.8 μm) were each incubated with a different concentration of SPI, prior to incubation with the fluorescein-labeled probe and subjected to FP measurements. Results show a concentration-dependent decrease in FP signal when SPI is present up to a certain concentration, when no further decreases become evident (Fig. 3C), suggesting that SPI disrupts the interaction of Stat3 with the cognate pTyr peptide probe. In this context, the FP signal decreased to the levels consistent with the displacement of Stat3 and the binding of SPI, as a ligand. The lack of complete decay of the FP signal by SPI is due to the observation in Fig. 3A, panel ii that SPI by itself behaves as a ligand in binding to the fluorescent peptide probe and induces polarization, although the FP signal is weaker than the signal induced by Stat3 binding to the same probe, as previously surmised. These findings together demonstrate that SPI, like Stat3, binds to cognate pTyr peptide motifs. By this mechanism, SPI would disrupt Stat3:Stat3 dimerization and the Stat3 binding to cognate pTyr peptide motifs. The FP assay based on the 5-carboxyfluorescein-GpYLPQTV-NH2 probe and SPI was rigorously tested and validated, with a Z factor of 0.89.
FIGURE 3.
FP assay. FP assay of the binding to the 5-carboxyfluorescein-GpYLPQTV-NH2 probe of (A) increasing concentration of (i) purified His-Stat3 or (ii) SPI; (B) a fixed amount of (i) purified His-Stat3 (200 nm) or (ii) SPI (150 μm) in the presence of increasing concentrations of S3I-201; or (C) fixed amount of Stat3 (0.8 μm) in the presence of increasing concentrations of SPI. Data are representative of four independent determinations.
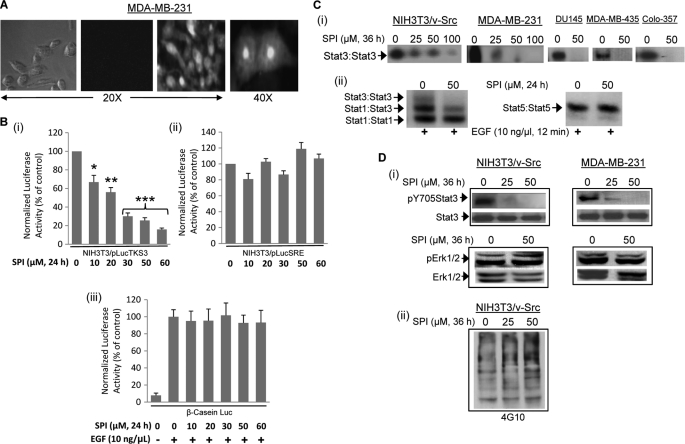
Evidence of Intracellular Accumulation of SPI and Inhibition of Intracellular Stat3 Activation
Stat3 is constitutively activated in a variety of malignant cells, including human breast and pancreatic cancer cells (3–5). Given the effect against the binding of Stat3 (or Stat3 SH2 domain) to cognate pTyr peptide motifs, we asked the question whether SPI could inhibit Stat3 activation in malignant cells. To address this, we first evaluated the extent of intracellular uptake of SPI by creating a 5-carboxyfluorescein-labeled version in which the fluorescence tag is attached to the N terminus of SPI. In contrast to cells treated with non-fluorescent SPI (negative control) (Fig. 4A, left two panels), fluorescent microscopy analysis showed strong evidence of intracellular fluorescence (Fig. 4A and data not shown), indicating significant cellular uptake of SPI following treatment of cells, including the human breast cancer line, MDA-MB-231 and the NIH3T3/hEGFR, with 30 μm SPI. Observation under higher magnification (40×) showed a wide intracellular distribution of SPI, with nuclear accumulation (Fig. 4A, right panel), demonstrating SPI is membrane-permeable and can localize in the nucleus. The mechanisms of cellular uptake and nuclear translocation remain to be determined. Dose-response studies reveal increasing intracellular uptake with increasing dose, while time course studies suggest fluorescent signal is decreased, but still evident by 22 h (supplemental Fig. S1). Malignant cells were then treated with SPI to assess biochemical and biological effects. Treatment of v-Src-transformed mouse fibroblasts harboring aberrant Stat3 activity (NIH3T3/v-Src/pLucTKS3) (13, 14) and that stably over-express the Stat3-dependent luciferase reporter, pLucTKS3 (13, 14, 29, 30) showed a dose-dependent inhibition of Stat3-mediated luciferase reporter induction (Fig. 4, B(i)), demonstrating that SPI inhibits Stat3 transcriptional activity. By contrast, a similar treatment of the v-Src-transformed mouse fibroblasts (NIH3T3/v-Src/pLucSRE) (13, 14) that stably over-express the Stat3-independent luciferase reporter, pLucSRE (29, 30), which is driven by the serum response element (SRE) of the c-fos promoter, or of EGF-stimulated mouse fibroblasts transiently-transfected with the Stat5-dependent β-casein-promoter-driven luciferase (β-casein-Luc) (16) had minimal to no effect on the induction of these reporters (Fig. 4B, panels ii and iii).
FIGURE 4.
Intracellular uptake and effect of SPI on Stat3 activation. A, phase-contrast and fluorescence imaging of MDA-MB-231 cells following treatment with 30 μm fluorescently labeled (right two panels) or unlabeled (left two panels) SPI for 3 h; Image was captured using Zeiss Axiovert 200 microscope; B, cytosolic extracts of equal total protein were prepared from 24-h SPI-treated or untreated (i) NIH3T3/v-Src/pLucTKS3 fibroblasts that stably express the Stat3-dependent luciferase reporter (pLucTKS3), (ii) NIH3T3/v-Src/pLucSRE that stably express the Stat3-independent luciferase reporter (pLucSRE), or (iii) NIH3T3/hEGFR transiently transfected with β-casein promoter-driven luciferase report (β-casein-Luc) and stimulated with EGF and analyzed for luciferase activity using a luminometer; (C) nuclear extracts of equal total protein were prepared from (i) the designated malignant cells or (ii) EGF-stimulated NIH3T3/hEGFR treated for 24 h with or without SPI and subjected to in vitro DNA binding assay using the radiolabeled hSIE probe (i) and (ii, left panel) that binds Stat1 and Stat3 or MGFe probe (ii, right panel) that binds Stat1 and Stat5 and analyzed by EMSA; and (D) SDS-PAGE and Western blotting analysis of whole cell lysates of equal total protein prepared from SPI-treated or untreated NIH3T3/v-Src and MDA-MB-231 cells and probing for (i) pY705Stat3, Stat3, pErk1/2, and Erk1/2, or (ii) general pY profile. Positions of STATs:DNA complexes or proteins in gel are labeled; control lanes (0) represent cytosolic or nuclear extracts, or whole cell lysates prepared from 0.05% DMSO-treated cells. Data are representative of 3–4 independent determinations or mean and S.D. of 3 independent determinations. *, p < 0.05; **, p < 0.01, and ***, p < 0.005.
Consistent with the inhibition of Stat3 transcriptional activity, v-Src-transformed mouse fibroblasts (NIH3T3/v-Src), or the human breast (MDA-MB-231 and MDA-MB-435), prostate (DU145), or pancreatic (Colo-357) cancer cells treated with SPI showed a dose-dependent inhibition of constitutive Stat3 activation, as measured by DNA binding activity in nuclear extract preparations using EMSA, with a near complete inhibition at 50 μm (Fig. 4C, panel i). The induction of the EGF receptor in fibroblasts activates Stat1, Stat3, and Stat5, which form homo- and hetero-dimers, as measured by DNA binding/EMSA analysis (Fig. 4C, panel ii). Consistent with the effects on Stat3 activation, the prior treatment of mouse fibroblasts over-expressing the human EGFR (NIH3T3/hEGFR) with 50 μm SPI completely blocked EGF-induced Stat3 activation, measured as Stat3:Stat3:DNA complex (Fig. 4C, panel ii, left panel, upper band), and suppressed the Stat1:Stat3 heterocomplex, measured as Stat1:Stat3:DNA complex (Fig. 4C, panel ii, left panel, intermediate band). By contrast, similar treatment had little or no effect on Stat1:Stat1:DNA complex (Fig. 4C, panel ii, left panel, lower band) or Stat5:Stat5:DNA complex (Fig. 4C, panel ii, right panel). Furthermore, immunoblotting analysis of whole cell lysates prepared from SPI-treated NIH3T3/v-Src and MDA-MB-231 cells showed a selective suppression of pY705Stat3 (Fig. 4D, panel i, upper panel), with no repression of pErk1/2 levels (Fig. 4D, panel i, lower panel). Total Stat3 protein levels remain unchanged (Fig. 4D, panel i, upper panel). Immunoblotting analysis further shows no significant changes in the general pTyr profile of v-Src-transformed mouse fibroblasts treated with SPI (Fig. 4D, panel ii). Thus, at concentrations up to 50 μm, SPI selectively and strongly inhibits constitutive Stat3 activation and transcriptional activity in malignant cells.
Biochemical Mechanism of Inhibition of Intracellular Stat3 Activation
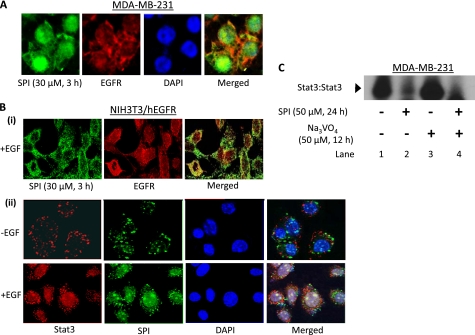
We next examined the mode of inhibition of Stat3 activation. Per the SPR and FP studies, SPI will bind to pTyr peptide motifs, thereby disrupt Stat3:Stat3 dimerization, as observed for small molecule or peptidomimetic dimerization disruptors of Stat3 (13, 14). Per the SPR data showing that both Stat3 (or Stat3 SH2 domain) and SPI bind to the known Stat3 binding pY1068EGFR and pY1086EGFR motifs with comparable affinities, presumably, SPI, like the Stat3 SH2 domain will associate with receptor pTyr motifs (Fig. 2). The binding to receptor motifs will in turn obstruct Stat3 binding and thereby block de novo Stat3 phosphorylation and activation. To verify this and to determine the intracellular localization and colocalizations of SPI and EGFR, fluorescence microscopy, and immunofluorescence staining with laser-scanning confocal microscopy analyses were conducted. Cells were treated with the fluorescently labeled SPI and subjected to immunostaining for EGFR (red), as described under “Experimental Procedures.” Results from the microscopy study showed that in MDA-MB-231 cells treated with fluorescently labeled SPI (green), the 28-mer is widely distributed throughout cells, with a strong nuclear localization (Fig. 5A, SPI, and merged, blue (DAPI), and green (SPI)). Moreover, SPI and EGFR are colocalized at the plasma membrane in MDA-MB-231 and in mouse fibroblasts overexpressing EGFR (NIH3T3/hEGFR) and treated with fluorescently labeled SPI (Fig. 5, A and B, panel i, merged, yellow). Moreover, to verify that SPI associates with Stat3, NIH3T3/hEGFR cells were treated with the fluorescently labeled SPI (green) and subjected to immunostaining for Stat3 (red), as described under “Experimental Procedures.” Laser-scanning confocal microscopy data showed that in unstimulated cells, Stat3 and SPI are predominantly at the plasma membrane, with weak presence in the cytoplasm and the nucleus, and with little evidence of co-localization (Fig. 5B, panel ii, upper panel), given that Stat3 is not phosphorylated. By contrast, upon cell stimulation by EGF, the fluorescently labeled SPI (green) and Stat3 (red) are universally distributed in the cells, with a strong localization at the perinuclear region and in the nucleus (Fig. 5B, panel ii, lower panel, green, red, and blue (DAPI)), with evidence of colocalization (Fig. 5B, panel ii, merged, green, red, and blue).
FIGURE 5.
Studies of colocalization of SPI with EGF receptor or Stat3 and the effect on Stat3 activation. A, fluorescence imaging of MDA-MB-231 cells treated with fluorescently labeled SPI (green; 30 μm, 3 h), fixed, and stained with rabbit anti-EGFR antibody (red) or DAPI nuclear staining (blue), or (B) immunofluorescence with laser-scanning confocal microscopy of unstimulated (−EGF) or EGF-stimulated (+EGF, 10 ng/ml, 10 min) NIH3T3/hEGFR fibroblasts growing in culture and pretreated with fluorescently labeled SPI (green; 30 μm, 3 h), fixed and stained with (i) rabbit anti-EGFR antibody (red) or (ii) rabbit anti-Stat3 antibody (red), or DAPI nuclear staining (blue). Images were captured using Zeiss Axiovert 200 fluorescent microscope or Leica TCS SP5 laser-scanning confocal microscope; or (C) Nuclear extracts were prepared from MDA-MB-231 cells pretreated with or without SPI prior to treatment with or without sodium orthovanadate and subjected to in vitro DNA binding assay using the radiolabeled hSIE probe and analyzed by EMSA. Positions of STATs:DNA complexes in gel are labeled; control (−), no treatment. Data are representative of three independent studies.
The observed colocalization indicates SPI associates with EGFR. To test the assertion that SPI association with receptors would compete against the binding of Stat3, thereby blocking de novo activation, MDA-MB-231 cells were treated with or without SPI for 12 h prior to treatment with sodium orthovanadate (protein phosphatase inhibitor) for an additional 12 h. Analysis by in vitro DNA binding activity assay/EMSA of nuclear extract preparations showed, as expected that the treatment with SPI alone inhibits Stat3 DNA binding activity (Fig. 5C, lane 2, compared with lane 1), while the treatment with orthovanadate alone increased Stat3 activity above the existing levels (Fig. 5C, lane 3, compared with lane 1), the latter, which is in part due to the blockade of the pStat3 turnover by the inhibition of protein phosphatases. By contrast, unlike cells treated with orthovanadate alone, in cells treated first with SPI and then with orthovanadate, Stat3 activation was completely inhibited (Fig. 5C, lane 4). This can be explained on the basis of the blockade of de novo Stat3 activation (by SPI), concomitant with the physiological turnover (elimination) of pre-existing pStat3 during the period prior to the addition of orthovanadate. Thus, by the time of the orthovanadate addition, there were no residual pStat3 levels. The microscopy and the gel shift data together demonstrate that SPI associates with Stat3 binding motifs of receptors, consequently preventing de novo Stat3 phosphorylation.
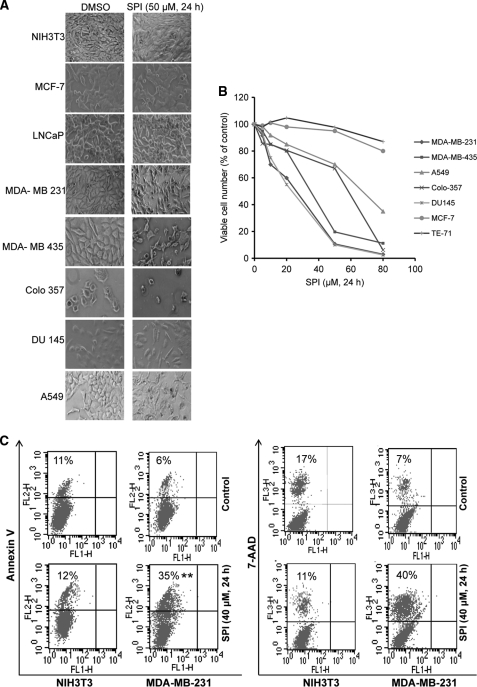
SPI Blocks Cell Viability and Growth, and Induces Apoptosis of Malignant Cells Harboring Constitutively Active Stat3
Aberrantly active Stat3 promotes malignant cell proliferation and survival and malignant transformation (4, 5, 39). We asked the question whether SPI is able to selectively decrease the viability and growth of malignant cells that harbor aberrant Stat3 activity. The human breast (MDA-MB-231 and MDA-MB-435), pancreatic (Colo-357), prostate (DU145), and non-small cell lung cancer (A549) lines that harbor constitutively-active Stat3 in culture were treated with or without an increasing concentration of SPI for 24 h and visualized under phase-contrast microscope for morphology changes, or analyzed for viable cell numbers by CyQuant cell proliferation/viability kit or by trypan blue exclusion with phase-contrast microscopy. Compared with the control (DMSO treatment), the human tumor cells harboring aberrant Stat3 activity (MDA-MB-231, MDA-MB-435, DU145, Colo-357, and A549) and treated with SPI showed significant morphology changes and had reduced viable cell numbers (Fig. 6, A and B, and data not shown). Importantly, the Stat3-dependent tumor cell lines showed dose-dependent decreases in viability and growth following 24-h treatment with increasing concentration of SPI (Fig. 6B, and data not shown). By contrast, the morphology, viability, and growth of cells that do not harbor aberrant Stat3 activity (normal NIH3T3, human breast cancer, MCF-7, murine thymus stromal epithelial cells, TE-71 and prostate cancer, LNCaP) were not significantly affected by similar treatment with SPI (Fig. 6, A and B, and data not shown). Furthermore, cultured MDA-MB-231 cells that harbor aberrant Stat3 activity and treated with SPI for 24 h and subjected to Annexin V binding/flow cytometry analysis showed evidence of significant apoptosis (35%), compared with DMSO-treated control (6%), while the normal NIH3T3 fibroblasts similarly treated showed little evidence of apoptosis, compared with DMSO-treated control (Fig. 6C, left panel). These data together indicate that SPI selectively represses constitutive Stat3 activation in malignant cells, thereby inducing antitumor cell effects that are dependent on the presence of constitutively active Stat3 in cells.
FIGURE 6.
SPI induces morphology change, suppresses viability, and induces apoptosis of malignant cells that harbor persistently active Stat3. A, photomicrographs of human breast (MDA-MB-231, MDA-MB-435, and MCF-7), prostate (DU145 and LNCaP), non-small cell lung (A549), and pancreatic (Colo-357) cancer cells, murine thymus stromal epithelial (TE-71) cells, and normal mouse fibroblasts (NIH3T3) in culture were treated once or untreated with 50 μm SPI for 24 h; (B) indicated cells were untreated or treated once with increasing concentration of SPI for 24 h and assayed for viability using CyQuant cell proliferation kit; or (C) human breast cancer cells (MDA-MB-231) or normal mouse fibroblasts (NIH3T3) in culture were untreated (control) or treated once with SPI (40 μm, 24 h) and subjected to Annexin V and 7-AAD binding and analyzed by flow cytometry for apoptosis. Data are representative of 3–4 independent determinations.
DISCUSSION
Protein-protein interactions are a common molecular event important in signal transduction and many other physiological processes. In the case of Stat3, the recruitment via the SH2 domain to cognate receptor pTyr peptide motifs is a key initial step for phosphorylation. Data herein show that the Stat3 SH2 domain-derived 28-mer peptide, SPI alone is sufficient to reproduce some aspects of the biochemical properties of Stat3 (or the Stat3 SH2 domain), thereby serving as a critical motif that engages in the inter-molecular interactions with the key residues of the cognate pTyr peptides to which Stat3 binds. Using biophysical analysis, such as SPR, we demonstrate similarities in the binding characteristics of Stat3 (or Stat3 SH2 domain) and SPI to known cognate pTyr peptides, including the native IL-6R/gp-130 derived peptide, GpYLPQTV-NH2, the Stat3 peptide, pY705Stat3, and the EGFR motif, pY1068EGFR, which Stat3 and SPI interact relatively stronger, and to the EGFR motif, pY1086EGFR and the Stat1 peptide, pY701Stat1, which they bind to with rather low affinities. The similarity in the binding characteristics of SPI and Stat3 is further evident by the SPR analysis that further suggests rather unfavorable interactions with the native Stat5 phosphopeptide, pY694Stat5, with affinities that are in milli-molar concentrations (KD of 1–7 mm). Together, present studies indicate SPI, like Stat3, shows preferential binding to different cognate pTyr peptide motifs, specifically showing stronger binding to the Stat3 phosphopeptide, compared with weaker binding to the Stat1 phosphopeptide. Given the observed differences in the affinities, we surmise that the type and number of binding partners to which Stat3 (or Stat3 SH2 domain) or SPI would interact with would be strongly influenced by their intracellular concentrations. Fluorescence polarization analysis based on the binding to the gp130-derived peptide (as 5-carboxyfluorescein-GpYLPQTV-NH2), (40), further support the similarities in the binding characteristics between SPI and Stat3.
The ability to mimic the SH2 domain and interact with pTyr peptide motifs raises the possibility that SPI could compete against Stat3. This assertion is supported by the fluorescence polarization studies that SPI strongly competes against Stat3 for the binding to IL-6R/gp13-derived pTyr peptide probe. Furthermore, evidence is presented that SPI exhibits selectivity at certain concentrations in the inhibition of intracellular Stat3 phosphorylation, DNA-binding and transcriptional activities. The inhibition of intracellular Stat3 activation may be explained in part by the ability to disrupt Stat3:Stat3 dimerization, as has been observed for other Stat3 dimerization inhibitors (13, 14, 16, 17), and on the basis of the potential that by associating with receptor pTyr motifs, SPI could block Stat3-binding to receptors, and hence prevented de novo phosphorylation. Thus, while the treatment of cells with the protein phosphatase inhibitor, sodium orthovanadate alone, increased the levels of activated Stat3 above constitutive levels, due to the blockade of pStat3 turnover by protein phosphatases, the prior exposure of cells to SPI squelched any subsequent orthovanadate-induced accumulation of activated Stat3. Moreover, the observation that SPI exhibits preferential inhibition of Stat3 activation relative to Stat1, despite the physiological occurrence of a Stat1:Stat3 heterodimer, when the two STAT family members are concurrently activated by EGF, suggests not only does SPI have preference for Stat3 over Stat1, but also that activated Stat3 protein may prefer to form a homodimer when concurrently activated with Stat1.
The SPI-mediated inhibition of Stat3 activation, via binding to cognate pTyr peptide motifs, is in direct converse to the mechanisms of Stat3 inhibition by many of the existing Stat3-inhibitory modalities, which are pY peptide mimetics and bind to the Stat3 SH2 module (4, 5, 13–15, 19, 22, 40, 41), or to the approaches that have been reported for the inhibition of other SH2 domain-containing proteins, such as the adapter protein, Grb2 (12), although the biochemical outcome of the inhibition would be the same, which is to disrupt pY:SH2 domain interactions. Whereas there are many protein entities with an SH2 module that are involved in promoting signal transduction and other biochemical processes (8, 9), present findings provides evidence of specificity for SPI action, which is demonstrated in the lack of effect on EGF-induced Stat5 activation and transcriptional activity, ErkMAPK activation, the c_-fos_-promoter-driven luciferase reporter, or on Stat1 activation at concentrations that inhibit Stat3 activity. Accordingly, antitumor cell effects of SPI are observed at concentrations that inhibit Stat3 activity and are consistent with the blockade of aberrant Stat3 activation (4, 5, 13–15, 40, 41). Specifically, human breast, pancreatic, prostate, and non-small cell lung cancer cells harboring aberrant Stat3 activity are more sensitive to SPI. We note an increased nuclear accumulation of SPI in malignant cells, which we speculate will promote a stronger degree of inhibition of nuclear activated Stat3 and Stat3 transcriptional activity. By contrast, in mouse fibroblasts overexpressing the EGFR in which Stat3 is not aberrantly-activated, SPI is predominantly localized to the cell membrane. The mechanism(s) for the cellular uptake and the nuclear translocation, and whether there are specific factors that promote the increased nuclear localization in malignant cells remain to be determined. Herein is reported a Stat3 SH2 domain-mimetic that functions as an inhibitor of Stat3 activation. The approach to the inhibition of Stat3 activation by SPI avoids the known challenges of mimicking the pTyr functionality for the existing SH2 domain-binding inhibitors. The significant in vitro activity of SPI makes it a suitable candidate for developing a modality for further evaluation of in vivo effects. Furthermore, SPI serves as a valuable molecular probe for interrogating aberrant Stat3 functions in tumor processes and for designing in vitro pY peptide binding assays.
Supplementary Material
Supplemental Data
Acknowledgments
We thank all colleagues and members of our laboratory for stimulating discussions.
*
This work was supported, in whole or in part, by NCI, National Institutes of Health Grants CA106439 and CA128865 (to J. T.).
2
The abbreviations used are:
pTyr or pY
phosphorylated tyrosine
SH2
Src homology 2
EMSA
electrophoretic mobility shift assay
EGFR
epidermal growth factor receptor
ErkMAPK
extracellular signal-regulated kinase-mitogen-activated protein kinase
GOLD
genetic optimization for ligand docking
SPR
surface plasmon resonance
FP
fluorescence polarization.
REFERENCES
- 1.Bromberg J. (2000) Breast Cancer Res. 2, 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darnell J. E., Jr. (2002) Nat. Rev. Cancer 2, 740–749 [DOI] [PubMed] [Google Scholar]
- 3.Yu H., Jove R. (2004) Nat. Rev. Cancer 4, 97–105 [DOI] [PubMed] [Google Scholar]
- 4.Yue P., Turkson J. (2009) Expert Opin. Investig. Drugs 18, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turkson J. (2004) Expert Opin. Ther. Targets 8, 409–422 [DOI] [PubMed] [Google Scholar]
- 6.Turkson J., Jove R. (2000) Oncogene 19, 6613–6626 [DOI] [PubMed] [Google Scholar]
- 7.Darnell J. E. (2005) Nat. Med. 11, 595–596 [DOI] [PubMed] [Google Scholar]
- 8.Yaffe M. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 177–186 [DOI] [PubMed] [Google Scholar]
- 9.Sawyer T. K. (1998) Biopolymers 47, 243–261 [DOI] [PubMed] [Google Scholar]
- 10.Burke T. R., Jr., Luo J., Yao Z. J., Gao Y., Zhao H., Milne G. W., Guo R., Voigt J. H., King C. R., Yang D. (1999) Bioorg. Med. Chem. Lett. 9, 347–352 [DOI] [PubMed] [Google Scholar]
- 11.Burke T. R., Jr., Smyth M. S., Otaka A., Nomizu M., Roller P. P., Wolf G., Case R., Shoelson S. E. (1994) Biochemistry 33, 6490–6494 [DOI] [PubMed] [Google Scholar]
- 12.Dharmawardana P. G., Peruzzi B., Giubellino A., Burke T. R., Jr., Bottaro D. P. (2006) Anticancer Drugs 17, 13–20 [DOI] [PubMed] [Google Scholar]
- 13.Turkson J., Kim J. S., Zhang S., Yuan J., Huang M., Glenn M., Haura E., Sebti S., Hamilton A. D., Jove R. (2004) Mol. Cancer Ther. 3, 261–269 [PubMed] [Google Scholar]
- 14.Turkson J., Ryan D., Kim J. S., Zhang Y., Chen Z., Haura E., Laudano A., Sebti S., Hamilton A. D., Jove R. (2001) J. Biol. Chem. 276, 45443–45455 [DOI] [PubMed] [Google Scholar]
- 15.Song H., Wang R., Wang S., Lin J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4700–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiquee K., Zhang S., Guida W. C., Blaskovich M. A., Greedy B., Lawrence H. R., Yip M. L., Jove R., McLaughlin M. M., Lawrence N. J., Sebti S. M., Turkson J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7391–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiquee K. A., Gunning P. T., Glenn M., Katt W. P., Zhang S., Schrock C., Schroeck C., Sebti S. M., Jove R., Hamilton A. D., Turkson J. (2007) ACS Chem. Biol. 2, 787–798 [DOI] [PubMed] [Google Scholar]
- 18.Coleman D. R., 4th, Ren Z., Mandal P. K., Cameron A. G., Dyer G. A., Muranjan S., Campbell M., Chen X., McMurray J. S. (2005) J. Med. Chem. 48, 6661–6670 [DOI] [PubMed] [Google Scholar]
- 19.Ren Z., Cabell L. A., Schaefer T. S., McMurray J. S. (2003) Bioorg Med. Chem. Lett. 13, 633–636 [DOI] [PubMed] [Google Scholar]
- 20.Schust J., Sperl B., Hollis A., Mayer T. U., Berg T. (2006) Chem. Biol. 13, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 21.Gunning P. T., Glenn M. P., Siddiquee K. A., Katt W. P., Masson E., Sebti S. M., Turkson J., Hamilton A. D. (2008) Chembiochem. 9, 2800–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher S., Turkson J., Gunning P. T. (2008) Chem. Med. Chem. 3, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker S., Groner B., Müller C. W. (1998) Nature 394, 145–151 [DOI] [PubMed] [Google Scholar]
- 24.Johnson P. J., Coussens P. M., Danko A. V., Shalloway D. (1985) Mol. Cell. Biol. 5, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C. L., Meyer D. J., Campbell G. S., Larner A. C., Carter-Su C., Schwartz J., Jove R. (1995) Science 269, 81–83 [DOI] [PubMed] [Google Scholar]
- 26.Garcia R., Bowman T. L., Niu G., Yu H., Minton S., Muro-Cacho C. A., Cox C. E., Falcone R., Fairclough R., Parsons S., Laudano A., Gazit A., Levitzki A., Kraker A., Jove R. (2001) Oncogene 20, 2499–2513 [DOI] [PubMed] [Google Scholar]
- 27.Zhang X., Yue P., Fletcher S., Zhao W., Gunning P. T., Turkson J. (2010) Biochem. Pharmacol 79, 1398–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farr A. G., Hosier S., Braddy S. C., Anderson S. K., Eisenhardt D. J., Yan Z. J., Robles C. P. (1989) Cell. Immunol. 119, 427–444 [DOI] [PubMed] [Google Scholar]
- 29.Turkson J., Bowman T., Adnane J., Zhang Y., Djeu J. Y., Sekharam M., Frank D. A., Holzman L. B., Wu J., Sebti S., Jove R. (1999) Mol. Cell. Biol. 19, 7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turkson J., Bowman T., Garcia R., Caldenhoven E., De Groot R. P., Jove R. (1998) Mol. Cell. Biol. 18, 2545–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner B. J., Hayes T. E., Hoban C. J., Cochran B. H. (1990) EMBO J. 9, 4477–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouilleux F., Moritz D., Humar M., Moriggl R., Berchtold S., Groner B. (1995) Endocrinology 136, 5700–5708 [DOI] [PubMed] [Google Scholar]
- 33.Seidel H. M., Milocco L. H., Lamb P., Darnell J. E., Jr., Stein R. B., Rosen J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3041–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Turkson J., Carter-Su C., Smithgall T., Levitzki A., Kraker A., Krolewski J. J., Medveczky P., Jove R. (2000) J. Biol. Chem. 275, 24935–24944 [DOI] [PubMed] [Google Scholar]
- 35.Jaganathan S., Yue P., Turkson J. (2010) J. Pharmacol. Exp. Ther. 333, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones G., Willett P., Glen R. C., Leach A. R., Taylor R. (1997) J. Mol. Biol. 267, 727–748 [DOI] [PubMed] [Google Scholar]
- 37.Eswar N., John B., Mirkovic N., Fiser A., Ilyin V. A., Pieper U., Stuart A. C., Marti-Renom M. A., Madhusudhan M. S., Yerkovich B., Sali A. (2003) Nucleic Acids Res. 31, 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 39.Siddiquee K., Turkson J. (2008) Cell Res. 18, 254–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Bai L., Bernard D., Nikolovska-Coleska Z., Gomez C., Zhang J., Yi H., Wang S. (2010) ACS Med. Chem. Lett. 1, 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhasin D., Cisek K., Pandharkar T., Regan N., Li C., Pandit B., Lin J., Li P. K. (2008) Bioorg Med. Chem. Lett. 18, 391–395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data