The γTuRC Revisited: A Comparative Analysis of Interphase and Mitotic Human γTuRC Redefines the Set of Core Components and Identifies the Novel Subunit GCP8 (original) (raw)
We compared the composition of γ-tubulin ring complexes tandem-affinity purified from asynchronous and mitotic human cells by mass spectrometry. We identified various interactors including the novel core subunit GCP8. GCP8 is the first subunit with an interphase-specific role in centrosomal γ-tubulin recruitment and microtubule nucleation.
Abstract
The γ-tubulin complex is a multi-subunit protein complex that nucleates microtubule polymerization. γ-Tubulin complexes are present in all eukaryotes, but size and subunit composition vary. In Drosophila, Xenopus, and humans large γ-tubulin ring complexes (γTuRCs) have been described, which have a characteristic open ring-shaped structure and are composed of a similar set of subunits, named γ-tubulin, GCPs 2-6, and GCP-WD in humans. Despite the identification of these proteins, γTuRC function and regulation remain poorly understood. Here we establish a new method for the purification of native human γTuRC. Using mass spectrometry of whole protein mixtures we compared the composition of γTuRCs from nonsynchronized and mitotic human cells. Based on our analysis we can define core subunits as well as more transient interactors such as the augmin complex, which associates specifically with mitotic γTuRCs. We also identified GCP8/MOZART2 as a novel core subunit that is present in both interphase and mitotic γTuRCs. GCP8 depletion does not affect γTuRC assembly but interferes with γTuRC recruitment and microtubule nucleation at interphase centrosomes without disrupting general centrosome structure. GCP8-depleted cells do not display any obvious mitotic defects, suggesting that GCP8 specifically affects the organization of the interphase microtubule network.
INTRODUCTION
The γ-tubulin ring complex (γTuRC) nucleates microtubule polymerization and is a key component of microtubule organizing centers (MTOCs) such as the centrosome. γTuRCs are composed of γ-tubulin and additional subunits named _g_amma-tubulin _c_omplex _p_roteins (GCPs) in humans (Murphy et al., 1998; Fava et al., 1999; Murphy et al., 2001). Assembly of the γTuRC involves subcomplexes composed of two molecules of γ-tubulin and one of each GCP2 and GCP3 (γ-tubulin small complex, γTuSC) (Oegema et al., 1999; Kollman et al., 2008). According to a current model the additional GCPs 4-6 promote assembly of ∼13 γTuSCs into the higher order ring-shaped γTuRC by forming a stabilizing cap on one side of the ring (Moritz et al., 2000). However, in Drosophila only γ-tubulin and the orthologues of GCP2 and 3 are essential and in the absence of the GCP4-6 orthologues γ-tubulin is still recruited to centrosomes and supports microtubule nucleation (Verollet et al., 2006). Similar results were recently obtained in the fungus Aspergillus (Xiong and Oakley, 2009). It is likely that in these cases as well as in budding yeast, which naturally lacks orthologues of GCP4-6, assembly of the γTuSC into a higher order ring structure might only occur upon interaction with MTOCs. In addition to promoting assembly of cytoplasmic γTuRCs, GCPs 4-6 might have other regulatory functions, for example in controlling microtubule length and dynamics, either directly or by interaction with other proteins (Fujita et al., 2002; Venkatram et al., 2004; Bouissou et al., 2009). Orthologues of the γTuRC subunits GCP2-6 can be found in all eukaryotes containing the γTuRC, suggesting that the GCPs are critical for γTuRC structure and function. However, as the requirements for microtubule nucleation and organization vary dramatically between cells from such different organisms as yeast and humans, it is likely that with increasing cellular complexity the number of γTuRC subunits or interactors that participate in γTuRC function also increases. An example is the GCP-WD subunit (also known as NEDD1), which plays an essential role in the assembly of mitotic and meiotic spindles in animals and plants, but is not found in yeast or Aspergillus (Luders et al., 2006; Haren et al., 2006; Zeng et al., 2009; Ma et al., 2010). GCP-WD has no sequence similarity with the other GCPs and is dispensable for γTuRC assembly. Instead, GCP-WD functions as a targeting factor for the γTuRC, mediating its interaction with centrosomes and, in mitosis, also with the mitotic spindle (Luders et al., 2006; Haren et al., 2006). Spindle targeting of the γTuRC involves mitotic phosphorylation of GCP-WD, which promotes interaction with the human augmin complex (also known as HAUS complex) (Luders et al., 2006; Goshima et al., 2008; Zhu et al., 2008; Lawo et al., 2009; Uehara et al., 2009). According to the so-called amplification model augmin and γTuRC are laterally bound to spindle microtubules and cooperate in the generation of additional microtubules within the forming spindle, possibly by γTuRC-mediated nucleation (Luders and Stearns, 2007; Goshima et al., 2008). During mitosis GCP-WD also plays a role in the chromatin-dependent nucleation pathway, but the molecular details are not known (Luders et al., 2006).
Our knowledge about the composition of γTuRCs is derived from the analysis of γTuRCs from various sources such as Drosophila embryos, Xenopus oocytes, and cultured human cells (Zheng et al., 1995; Moritz et al., 1998; Murphy et al., 1998; Fava et al., 1999; Oegema et al., 1999; Murphy et al., 2001). In biochemical terms, the γTuRCs prepared from early Drosophila embryos and CSF-arrested Xenopus oocytes, respectively, most likely resemble the mitotic state, whereas the human γTuRC isolated from asynchronously growing cultured cells can be considered a nonmitotic, interphase form. A more recent large-scale analysis of mitotic protein complexes has identified additional interactors of the γTuRC in mitosis (Hutchins et al., 2010). However, whether the γTuRC undergoes cell cycle–dependent changes in subunit composition or in its interaction with other proteins has not been investigated.
Here we have performed a comparative analysis of the composition of human γTuRCs from asynchronous and mitotic cells by mass spectrometry. Our analysis reveals an interaction network comprising constitutive as well as more transient interactions and identifies a novel γTuRC core subunit.
MATERIALS AND METHODS
Molecular Biology
Full-length Fam128B was PCR amplified from a human liver cDNA library using the following primers: CCGCTCGAGCGATGGCGGCGCAGGGCGTAGG and CCGGAATTCCTAGGTGCTGCCCTGCGTAGGGCT. Full-length Fam128A was PCR-amplified from plasmid pCMV-SPORT6-Fam128A (clone # 3862861; Open Biosystems, Huntsville, AL) using the same primers.
For expression in human cells amplified GCP8 sequences were inserted into pEGFP-C1 (Clontech, Palo Alto, CA) using XhoI and EcoRI restriction sites. For the expression in Escherichia coli, the GCP8B sequence was inserted into the pGEX-4T-1 vector (GE Healthcare, Piscataway, NJ) using EcoRI and XhoI restriction sites. A plasmid expressing γ-tubulin-Myc-TAP was constructed by replacing the His-tag of γ-tubulin-Myc-His (pTS556) (Murphy et al., 1998) with the PCR-amplified TAP-tag from plasmid pBS1479 (Rigaut et al., 1999).
For RNAi-mediated depletion of GCP8A and GCP8B we designed RNA oligonucleotides targeting the following sequences within the GCP8A/B open reading frames (100% conserved between the two): CGACGUGUUCAAGAUCCUG (oligo 1, Silencer Select Pre-designed siRNA, # s198094, Ambion, Applied Biosystems, Carlsbad, CA) and CUCGCCGUCUUCCAGAUGCUCAAGU (oligo 2, Stealth version, Invitrogen, Carlsbad, CA). Sequence analysis and alignments were performed with Geneious software (Biomatters, Auckland, NewZealand).
RNAi-mediated depletion of GCP-WD was done as previously described (Luders et al., 2006).
Antibodies and Reagents
To generate anti-GCP8 antibodies, full-length GCP8B was expressed and affinity-purified as a soluble GST fusion protein in E. coli using glutathione-sepharose (GE Healthcare), according to the manufacturer's standard protocol. The protein was then used for immunization of rabbits (Antibody Production Service, Facultat de Farmàcia, Universitat de Barcelona, Spain). After passing the rabbit serum over a resin bearing immobilized GST to eliminate GST-reactive antibodies, GCP8-specific antibodies were affinity-purified using GST-GCP8 immobilized on Affi-Gel10.
The following additional antibodies were used in this study: mouse anti–γ-tubulin (GTU-88; Sigma, St. Louis, MO), mouse anti–γ-tubulin (Exbio, Prague, Czech Republic), rabbit anti–γ-tubulin (Sigma), mouse anti–α-tubulin (DM1A, Sigma), mouse anti–GCP-WD (7D10, Abnova, Walnut, CA), rabbit anti–GCP-WD (Luders et al., 2006), rabbit anti-GCP5 (H-300, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-pericentrin (Luders et al., 2006), mouse anti-GFP (3E6, Invitrogen), rabbit anti-GFP (Torrey Pines Biolabs, Houston, TX), rabbit anti-Cep215 (IHC-00063, Bethyl Laboratories, Montgomery, TX), rabbit anti-centrin (Groen et al., 2004), rabbit anti-Nek9 (Roig et al., 2002).
Alexa 488- and Alexa 568-conjugated secondary antibodies used for immunofluorescence microscopy were from Invitrogen (Carlsbad, CA), and peroxidase-coupled secondary antibodies for western blotting were from Jackson Immunoresearch Laboratories (West Grove, PA).
Cell Culture, Transfection, and Drug Treatments
U2OS cell lines were grown in DMEM containing 10% fetal calf serum.
Cells were transfected with plasmid or siRNA using Lipofectamine 2000 or Lipofectamine RNAiMAX (Invitrogen), respectively.
To generate a Hela S3 cell line stably expressing γ-tubulin-Myc-TAP cells were transfected with the γ-tubulin-Myc-TAP expression plasmid and selected in the presence of 0.4 μg/ml geneticin. Resistant clones were isolated and tested for expression of the tagged protein by Western blotting.
For interphase microtubule regrowth experiments, dishes containing coverslips with U2OS cells were incubated for 30 min in an ice-water bath to depolymerize microtubules. To allow microtubule regrowth, coverslips were then incubated in medium at 37°C, followed by methanol or 4% paraformaldehyde fixation. For mitotic microtubule regrowth experiments, cells were incubated with 250 ng/ml nocodazole for ∼16 h. Nocodazole was then washed out and microtubules were depolymerized as described above.
Cell Cycle Analysis
U2OS cells were transfected with GCP8 siRNA for 72 h and then prepared for flow cytometry by fixation in 70% ethanol.
Phosphorylated histone H3 was detected with a phosphospecific polyclonal primary antibody (Millipore, Billerica, MA) and a FITC-conjugated anti-rabbit secondary antibody (Jackson Laboratories). To measure DNA content, cells were stained with propidium iodide (Sigma). Fluorescence was measured by flow cytometry using a Coulter XL Flow Cytometer (Flow Cytometer Unit, Scientific and Technical Services Universitat de Barcelona).
Tandem-Affinity Purification of the Human γTuRC
HeLa S3 γ-tubulin-Myc-TAP cells were left untreated (for asynchronous samples) or incubated with 250 ng/ml nocodazole for ∼16 h (for mitotic samples). Cells were harvested by trypsinization or, in the case of mitotic cells, by shake-off, washed in PBS, and lysed in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.05% Triton-X100) containing Complete protease and Phostop phosphatase inhibitors (Roche, Indianapolis, IN) for 10 min on ice and sonicated with 10-s bursts at maximum power. After centrifugation for 15 min at 14,000_g_ at 4°C the γTuRC was precipitated from the cleared lysates by addition of an equal volume of 9% polyethylene glycol and incubation for 20 min on ice (Murphy et al., 2001). After centrifugation for 15 min at 12,000_g_ at 4°C the pellet was resuspended in lysis buffer and centrifuged as before. The supernatant was then incubated with IgG Sepharose beads (GE Healthcare) for 2 h at 4°C. The beads were pelleted and washed with 10 column volumes of lysis buffer and with 2 column volumes of wash buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% Triton-X100) containing phosphatase inhibitors. Beads were resuspended in 2 column volumes of wash buffer, and AcTEV protease (Invitrogen) was added to the mixture. After incubation for 2 h at 16°C beads were pelleted. The supernatant was adjusted to 1 mM imidazole and 2 mM calcium chloride and incubated with Calmodulin Sepharose beads (GE Healthcare) for 1 h at 4°C. Then the beads were washed with 10 column volumes of wash buffer containing 1 mM imidazole and 2 mM calcium chloride. Beads were pelleted and purified γTuRC was eluted by addition of 5 column volumes of elution buffer (wash buffer with 2 mM EGTA). For the analysis by SDS-PAGE the γTuRC was concentrated using Vivaspin 6 concentrators (Sartorius, Göttingen, Germany).
Mass Spectrometry
Analysis of individual protein bands: purified γTuRC was separated by SDS-PAGE and stained with Coomassie. Bands were excised from the gel and analyzed by the Proteomics Platform of the Barcelona Science Park, University of Barcelona. Briefly, excised bands were washed sequentially with 25 mM NH4HCO3 and acetonitrile. Proteins were reduced and alkylated by treatment with 10 mM DTT for 15 min at 56°C and 55 mM iodine acetamide for 15 min, respectively. After sequential washings with buffer and acetronitrile, proteins were digested overnight at 37°C with 80 ng of trypsin. Tryptic peptides were extracted from the gel matrix with 10% formic acid and acetonitrile. Mass spectrometry analysis was done by liquid-chromatography coupled to a Q-TOF mass spectrometer (capLC-ESI-Q-TOF, Micromass-Waters). The peptide digests were redissolved in 25 μl of 1% formic acid solution and injected and separated in a C18 reverse phase column (PepMap column, LC Packings, Sunnyvale, CA). Data were processed in MassLynx 4.1, and pkl files were generated using ProteinLynx software. The pkl files were submitted to database searching using the online MASCOT search engine (Matrixscience; http://www.matrixscience.com/).
For analysis of whole protein mixtures, purified γTuRC was precipitated with 20% trichloroacetic acid, in-solution digested in 5 ng/μL trypsin (in 50 mM ammonium bicarbonate, pH 8.3, 10% acetonitrile), and analyzed by liquid-chromatography coupled to tandem mass spectrometry (LC-MS/MS). Peptides were separated by reverse phase chromatography across a 90-min gradient on a C18 microcapillary column and online analyzed on an LTQ-Orbitrap hybrid mass spectrometer. For each cycle, one full MS scan acquired at high mass resolution on the Orbitrap mass analyzer was followed by 10 MS/MS spectra on the linear ion trap from the 10 most abundant ions. MS/MS spectra were searched against the IPI human protein sequence database using the Sequest algorithm. Peptide matches were filtered to <0.1% false-positives using a target-decoy database strategy (Elias and Gygi, 2007); protein false positives were <1%. Final lists of proteins interacting with γTuRCs from asynchronous and mitotic cell populations were obtained by subtracting protein matches that were also found in an untagged control sample.
Immunoprecipitation
For immunoprecipitation of EGFP-tagged GCP8 transfected U2OS cells were washed in PBS and lysed (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.5% NP-40, protease inhibitors) for 10 min on ice. After centrifugation for 15 min at 16,000_g_ at 4°C cleared lysates were incubated with anti-GFP antibodies for 1 h at 4°C. Sepharose Protein G beads (GE Healthcare) were added and the mixture was incubated for an additional hour at 4°C. The beads were pelleted and washed three times with lysis buffer. Samples were prepared for SDS-PAGE by boiling in sample buffer.
Sucrose Gradient Centrifugation
U2OS cell extracts were prepared as described above. Two hundred microliters of extract was then loaded on a 4.2 ml 10–40% sucrose gradient prepared in 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA and centrifuged in a SW-55Ti rotor (Beckman, Brea, CA) for 4 h at 55,000 rpm at 4°C. Fractions were collected and analyzed by Western blotting. Aldolase (158 K, 7S) and thyroglobulin (669 K, 19S) (GE Healthcare) were used as molecular weight standards and analyzed in parallel.
Western Blotting
Cells were washed in PBS and lysed (50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.5% NP-40, protease inhibitors) on ice. Cleared extracts were prepared by centrifugation and subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes and probed with antibodies.
Fluorescence Microscopy
U2OS cells grown on coverslips were fixed in methanol at −20°C for at least 5 min and processed for immunofluorescence. Alternatively, cells were fixed in PBS containing 4% paraformaldehyde, 0.05% glutaraldehyde, and 0.1% Triton X-100 for 15 min at RT.
Fixed cells were blocked in PBS-BT (1× PBS, 0.1% Triton X-100, and 3% BSA) and incubated with antibodies in the same buffer. Images were acquired with an Orca AG camera (Hamamatsu, Bridgewater, NJ) on a Leica DMI6000B microscope equipped with 1.4 NA 63× and 100× oil immersion objectives. AF6000 software (Leica, Wetzlar, Germany) was used for image acquisition. For further image processing and quantification of fluorescence intensities ImageJ software was used. Intensities were measured in a circular area (2–4 μm diameter) surrounding centrosomes or centrosomal microtubule asters in images acquired with constant exposure settings. For background-correction the intensity measured in an adjacent circular area of equal dimensions in the cytoplasm was subtracted.
RESULTS
Characterization of the γTuRC Interactome Identifies Constitutive and Mitosis-Specific Interactions
We developed a new method for the purification of human γTuRCs based on Hela S3 cells stably expressing γ-tubulin with a C-terminal mycTAP tag. Using this cell line and a modified tandem-affinity purification protocol we were able to produce purified human γTuRC from nonsynchronized as well as nocodazole-arrested mitotic cells. To analyze the composition of the complexes we performed mass spectrometric analysis of whole protein mixtures. In addition to the known γTuRC subunits, we identified a large number of proteins, some of which were specifically associated with mitotic γTuRCs (Supplemental Table 1). Validating our approach several of these proteins have been described previously as γTuRC interactors (Table 1) including subunits of the CCT chaperonin complex, which are involved in tubulin folding (Melki et al., 1993), CDK5RAP2 (also known as Cep215), which plays a role in centrosome targeting of γ-tubulin (Fong et al., 2008; Haren et al., 2009), the AAA+ ATPase RUVBL1, which, together with RUBVBL2, was identified as γTuRC interactor important for mitotic microtubule assembly (Ducat et al., 2008), and four proteins recently shown to interact with the γTuRC in mitosis (Hutchins et al., 2010). Of these, MOZART1 plays a role in mitotic spindle assembly (Hutchins et al., 2010), and the other three, MOZART2, NME7, and LGALS3BP, are still uncharacterized. We now demonstrate that all these proteins are associated with both interphase and mitotic γTuRCs (Table 1). As interactors enriched in the mitotic sample we identified seven of the eight subunits of the recently characterized augmin complex, which cooperates with the γTuRC in noncentrosomal microtubule formation during spindle assembly (Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009), and Polo-like kinase 1, which might regulate mitotic functions of the γTuRC (Haren et al., 2009; Zhu et al., 2009) (Table 1). In addition to previously described γTuRC interactors, we identified additional proteins that specifically copurified with γTuRCs and represent potential γTuRC interactors (Supp. Table 1). However, to eliminate false positives these interactions will have to be confirmed by alternative approaches in future studies.
Table 1.
Composition of interphase and mitotic γTuRCs
| Protein | Interphase | Mitotic | ||||
|---|---|---|---|---|---|---|
| Control peptides | γTuRC peptides | γTuRC coverage, % | Control peptides | γTuRC peptides | γTuRC coverage, % | |
| γ-Tubulin | 3 | 42 | 61.4 | 1 | 36 | 55.4 |
| GCP2 | 1 | 61 | 52.8 | 0 | 58 | 54.5 |
| GCP3 | 3 | 63 | 46.2 | 0 | 49 | 43.7 |
| GCP4 | 0 | 27 | 53.2 | 0 | 22 | 43.6 |
| GCP5 | 0 | 53 | 45.3 | 0 | 40 | 37.4 |
| GCP6 | 0 | 61 | 37.8 | 0 | 48 | 34.4 |
| GCP-WD | 1 | 33 | 48.0 | 0 | 21 | 39.5 |
| GCP8A | 0 | 5 (2) | 49.4 | 0 | 3 (1) | 32.3 |
| GCP8B | 0 | 4 (1) | 31.0 | 0 | 3 (1) | 31.0 |
| MOZART1 | 0 | 2 | 13.4 | 0 | 2 | 52.4 |
| NME7 | 0 | 10 | 31.4 | 0 | 9 | 26.6 |
| LGALS3BP | 1 | 5 | 12.3 | 0 | 4 | 10.6 |
| CDK5RAP2 | 0 | 19 | 14.6 | 0 | 7 | 5.3 |
| RUVBL2 | 4 | 17 | 36.6 | 8 | 11 | 30.3 |
| RUVBL1 | 4 | 15 | 32.9 | 2 | 6 | 17.1 |
| TCP1 | 0 | 20 | 38.1 | 0 | 15 | 30.8 |
| CCT2 | 0 | 33 | 60.3 | 0 | 17 | 41.9 |
| CCT3 | 1 | 20 | 39.5 | 0 | 9 | 18 |
| CCT4 | 0 | 20 | 30.9 | 0 | 9 | 20.6 |
| CCT5 | 0 | 22 | 29.9 | 0 | 12 | 21.3 |
| CCT6A | 0 | 12 | 26.2 | 0 | 6 | 18.3 |
| CCT7 | 0 | 17 | 40.0 | 0 | 12 | 26.5 |
| CCT8 | 4 | 26 | 38.9 | 0 | 20 | 36.2 |
| HAUS1 | 0 | 0 | 0 | 0 | 1 | 4.0 |
| HAUS2 | 0 | 0 | 0 | 0 | 1 | 6.0 |
| HAUS3 | 0 | 0 | 0 | 0 | 5 | 10.0 |
| HAUS4 | 0 | 0 | 0 | 0 | 3 | 11.6 |
| HAUS5 | 0 | 1 | 2.5 | 0 | 7 | 13.7 |
| HAUS6 | 0 | 1 | 2.1 | 0 | 4 | 7.2 |
| HAUS8 | 0 | 0 | 0 | 0 | 3 | 10.2 |
| PLK1 | 0 | 1 | 3.2 | 0 | 4 | 9.3 |
Interphase and Mitotic γTuRCs Have a Similar Core Subunit Composition
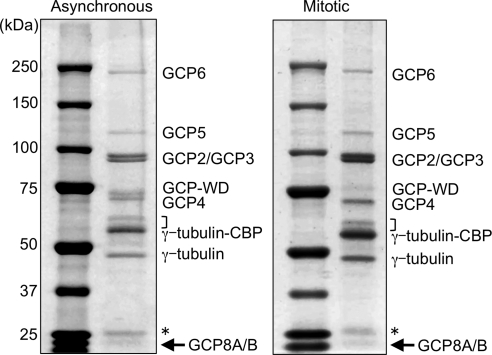
To discriminate between core components of the γTuRC and more transient interactors, we analyzed purified γTuRCs by PAGE and Coomassie staining, which revealed the most abundant polypeptides. Interphase and mitotic γTuRCs were very similar with respect to subunit composition and relative stoichiometry of subunits (Figure 1). The only obvious difference was a reduced mobility of the GCP-WD band in mitotic γTuRC, which was reported previously and shown to be caused by mitotic phosphorylation (Haren et al., 2006; Luders et al., 2006). Mass spectrometry analysis of the individual most prominent protein bands confirmed the presence of all known γTuRC subunits: γ-tubulin, GCPs 2-6, and GCP-WD (Figure 1). Interestingly, we consistently observed an additional band at ∼20 kDa which was present in both samples with an estimated stoichiometry similar to the other known subunits. Mass spectrometry identified this band as a mixture of the two closely related, uncharacterized proteins MOZART2A and MOZART2B, which were recently described as novel mitotic γTuRC interactors (Hutchins et al., 2010). We have named these proteins GCP8A and GCP8B to indicate that they are core subunits of the γTuRC (see below).
Figure 1.
Human γTuRCs purified from asynchronous and mitotic cells have a similar subunit composition. Hela S3 cells stably expressing TAP-tagged γ-tubulin were left untreated or arrested in mitosis by nocodazole treatment as indicated. γTuRCs were purified by tandem affinity chromatography and analyzed by SDS-PAGE and Coomassie staining. The position of core subunits as identified by mass spectrometry of excised protein bands are indicated. In the mitotic sample GCP-WD is present in its phosphorylated form and migrates with slightly reduced mobility. The arrow points at the novel subunit GCP8A/B. The asterisk marks contaminating IgG light chain derived from the affinity resin, the bracket indicates the position of CCT subunits.
In summary our analysis shows that γTuRCs from nonsynchronized and mitotic cells are very similar in their core subunit composition, but they differ with respect to more transient interactions.
GCP8 Is a Novel Core Subunit of the Human γTuRC
GCP8A and GCP8B polypeptides were present in all of our γTuRC preparations and this association was independent of the γTuRC cell cycle state. As GCP8A/B were readily visualized by Coomassie staining with intensities comparable to the other known subunits (Figure 1), we speculated that they might represent novel core subunits of the γTuRC. Public databases contain GCP8-related sequences from various species ranging from mammals to marine invertebrates, but we were unable to find similar sequences in plants, Drosophila, C. elegans, and yeast. The highest degree of similarity between GCP8 orthologues is found in an N-terminal region that is predicted to be of mainly helical secondary structure (Figure 2A). Human GCP8A and GCP8B are two highly related proteins with no similarity to GCPs 2-6 or GCP-WD. GCP8A and GCP8B are encoded by two distinct genes, but their amino acid sequence is more than 96% identical. Interestingly, in other species only a single GCP8 gene is present and the encoded proteins are more similar to human GCP8B (Figure 2A), suggesting that GCP8A is the result of a gene duplication in humans. For the remainder of the text we will simply use the term GCP8 when referring to both GCP8 proteins.
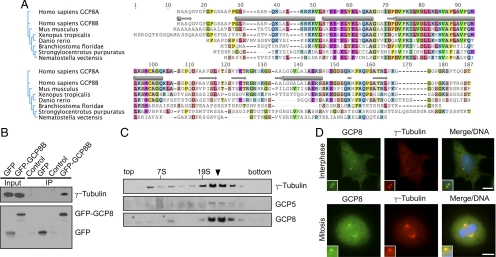
Figure 2.
GCP8 has properties of a γTuRC subunit. (A) Amino acid sequence alignment of human GCP8A/B and putative orthologues from various species as indicated. Conserved amino acids (identical in at least 50% of the aligned sequences) are shaded in colors. The relationship between sequences is indicated by a phylogenetic tree on the left. Predicted secondary structures are indicated for human GCP8B above its sequence. Helical regions are indicated by a tube symbol, β-sheets by a block arrow symbol. (B) GFP and GFP-GCP8B, respectively, were transiently expressed in Hela cells and immunoprecipitated with anti-GFP antibody. After Western blotting the immunoprecipitates were probed with antibodies against the indicated proteins. (C) Extract of U2OS cells was fractionated by sucrose gradient centrifugation. Fractions were analyzed by Western blotting with antibodies against the indicated proteins. The arrowhead marks the γTuRC peak fraction. Aldolase (158 kDa, 7S) and thyroglobulin (669 kDa, 19S) were used as molecular weight standards. (D) U2OS cells were fixed and stained with antibodies against GCP8 and γ-tubulin as indicated. DAPI was used to stain DNA. Insets show magnified centrosome areas. Scale bar, 10 μm.
We cloned human GCP8B from a human liver cDNA library to characterize its function in detail. To test the interaction of GCP8B with the γTuRC we expressed GFP-tagged GCP8B in human cells and immunoprecipitated the protein with GFP-specific antibodies. Probing with γ-tubulin–specific antibodies demonstrated that γ-tubulin coprecipitated with GFP-GCP8B (Figure 2B). To investigate whether GCP8 has properties of a genuine γTuRC subunit we produced an antibody against GCP8, fractionated human cell extract by sucrose gradient centrifugation and probed with the GCP8 antibody. Due to the high degree of sequence similarity between GCP8A and GCP8B this antibody recognizes both forms (Supplemental Figure S1). Indeed GCP8 cofractionated with γ-tubulin and GCP5 with a major peak at a size expected for the γTuRC (Figure 2C). Interestingly, some GCP8 also cofractionated with γ-tubulin but not GCP5 at a smaller size, which could represent a γTuRC subcomplex such as the γTuSC. Costaining of GCP8 and γ-tubulin in fixed human cells revealed localization of GCP8 to centrosomes throughout the cell cycle and to spindle microtubules in mitosis (Figure 2D). Similar results were obtained for GFP-tagged GCP8 after transient expression and staining of fixed cells with GFP- and γ-tubulin–specific antibodies (Supplemental Figure S2). Together our data are consistent with the interpretation that GCP8 is a novel γTuRC subunit.
Centrosome Localization of GCP8 Requires GCP-WD
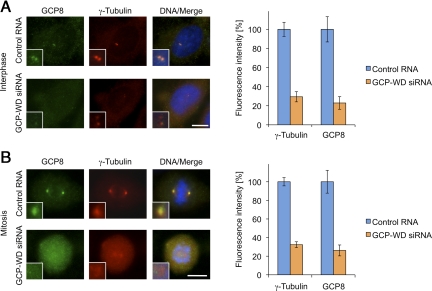
Centrosome localization of the γTuRC requires the GCP-WD subunit, which in human cells functions as a targeting factor for the γTuRC (Haren et al., 2006; Luders et al., 2006). We tested whether centrosomal targeting of GCP8 also depends on the presence of GCP-WD. In agreement with previous reports, RNAi-mediated depletion of GCP-WD strongly interfered with centrosomal recruitment of γ-tubulin both in interphase and mitosis (∼70% reduction, respectively; Figure 3, A and B). Interestingly, centrosomal GCP8 was also reduced to a similar degree in interphase and mitotic GCP-WD–depleted cells (∼75% reduction, respectively; Figure 3, A and B). Thus, also with respect to its centrosome targeting, GCP8 behaves like a true γTuRC subunit.
Figure 3.
Centrosome targeting of GCP8 requires GCP-WD. (A) Interphase U2OS cells transfected with either control RNA or siRNA against GCP-WD were stained with γ-tubulin– and GCP8-specific antibodies. DNA was stained with DAPI. The insets show magnifications of centrosomal areas. The fluorescence intensities of centrosomal γ-tubulin and GCP8 staining were quantified, and the centrosomal signal measured in control cells was set to 100%. Mean values are plotted as percentages of intensities in control cells (n > 20, error bars: SEM). (B) Mitotic U2OS cells analyzed as in A. Scale bars, 10 μm.
GCP8 Is Not Required for the Assembly of Cytoplasmic γTuRCs
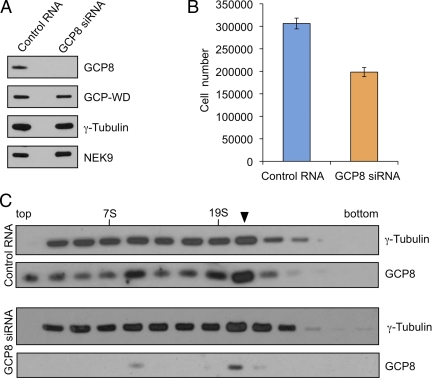
We used RNAi to deplete GCP8 from human cells. All RNAi-derived data were reproduced with two different siRNAs (see Materials and Methods) indicating the specificity of the effects. Western blotting of whole cell lysate 72 h after transfection indicated that GCP8 was efficiently depleted. Importantly, GCP8 depletion did not significantly affect the total levels of other γTuRC subunits (Figure 4A). We noticed that GCP8-depleted cells had a slightly slower proliferation rate compared with control cells (Figure 4B). Surprisingly, however, FACS analysis indicated that the cell cycle profile after GCP8 depletion was similar to control cells (Supplemental Figure S3). There was no increase in the mitotic index or in the subG1 population of cells, suggesting that the reduced proliferation was not due to mitotic progression defects or cell death (Supplemental Figure S3). To investigate whether the inhibitory effect on cell proliferation might be related to an impaired γTuRC we tested whether the γTuRC was present in GCP8-depleted cells. We analyzed extracts from control and GCP8-depleted cells by sucrose gradient centrifugation. γ-Tubulin in control-transfected extracts fractionated mostly at the size of the large γTuRC (Figure 4C) and the profile was comparable to untreated extracts (Figure 2C). GCP8-depleted extracts showed a very similar γ-tubulin profile indicating that the γTuRC was not disrupted (Figure 4C). Interestingly, the small amount of GCP8 that was left after the depletion still comigrated with the γTuRC indicating its tight association with the complex. We concluded that unlike GCPs 2-6, which are important for γTuRC assembly (Verollet et al., 2006; Xiong and Oakley, 2009), GCP8 has no structural role in the γTuRC.
Figure 4.
Depletion of GCP8 does not affect assembly or stability of the γTuRC. (A) U2OS cells were transfected with control RNA or siRNA targeting GCP8. After 72 h whole cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. (B) Equal numbers of control-transfected cells and cells transfected with GCP8 siRNA were seeded on culture dishes and grown for 72 h. After harvesting the cell numbers were determined by counting. Mean values from four independent transfections were plotted. Error bars: SEM. (C) U2OS cells were transfected with control RNA or with siRNA targeting GCP8. After 72 h cells were lysed and extracts fractionated by sucrose gradient centrifugation. Fractions were analyzed by Western blotting and probed with antibodies against the indicated proteins. The arrowhead marks the γTuRC peak fraction. Peak fractions of aldolase (158 kDa, 7S) and thyroglobulin (669 kDa, 19S) as molecular weight standards are indicated.
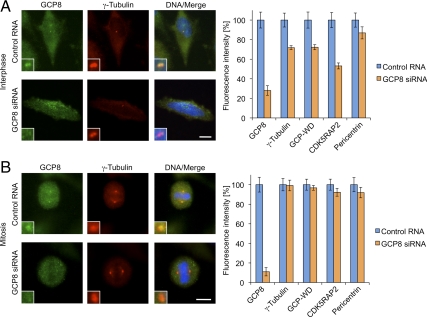
GCP8 Depletion Impairs Recruitment of γTuRC Components to Interphase Centrosomes
γTuRC recruitment and microtubule nucleation activity are essential for the microtubule organizing function of centrosomes. To investigate whether GCP8 depletion affects centrosomal γTuRC we costained control and GCP8-depleted cells with antibodies against GCP8 and various centrosome components. At interphase centrosomes with strongly reduced levels of GCP8 we observed a reduction of both GCP-WD and γ-tubulin (∼30% reduction, respectively) (Figure 5A). Interestingly, GCP8 depletion caused an even stronger reduction in the amount of centrosomal CDK5RAP2 (∼45% reduction). Importantly, the levels of pericentrin, a component of the PCM, were only slightly reduced (∼15% reduction) and staining with the centriole marker centrin revealed the presence of normal numbers of centrioles (Figure 5A and Supplemental Figure S4), indicating that GCP8 depletion did not disrupt the general centrosome structure. This conclusion is further supported by FACS analysis, which failed to detect a significant increase in the G1/S population of cells, suggesting that GCP8 depletion does not induce a specific G1/S arrest, as previously reported for RNAi treatments that compromise centrosome integrity (Supplemental Figure S3). Interestingly, depletion of GCP8 did not impair the localization of γTuRC components to centrosomes in mitosis (Figure 5B), suggesting a specific effect on interphase centrosomes. In mitotic cells depleted of GCP8 localization of γ-tubulin to spindle microtubules was also unaffected (Figure 5B).
Figure 5.
Depletion of GCP8 interferes with the recruitment of γTuRC components to interphase centrosomes. U2OS cells transfected with either control RNA or siRNA against GCP8 were stained with antibodies against various centrosome proteins. As an example, interphase (A) and mitotic (B) cells stained with GCP8 and γ-tubulin antibodies are shown. DNA was visualized with DAPI. Scale bars, 10 μm. The fluorescence intensities of centrosomal signals were quantified for each of the detected proteins in interphase (A) and mitotic (B) cells, respectively. Mean values are plotted as percentages of intensities in control cells, which were set to 100% (n > 20, error bars: SEM).
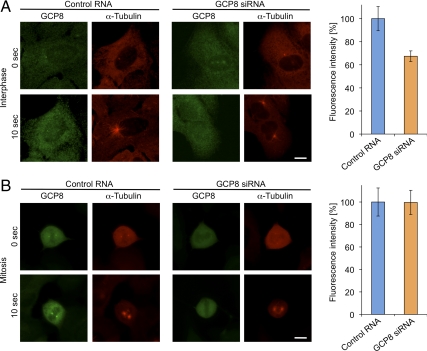
GCP8 Depletion Inhibits Microtubule Nucleation at Interphase Centrosomes
We next tested whether centrosomes in GCP8-depleted cells were able to nucleate microtubules. We performed a cell-based microtubule regrowth assay, in which cells were cold-treated to completely depolymerize microtubules and then incubated at 37°C to allow microtubule nucleation and regrowth. After 10 s of regrowth, small microtubule asters had formed at centrosomes in both control and GCP8-depleted cells (Figure 6A). However, centrosomal microtubule asters in GCP8-depleted cells were slightly smaller than in control cells and quantification of the microtubule signal surrounding the centrosomes indicated a ∼30% reduction (Figure 6A). Again, the effect was specific to interphase centrosomes as we could not detect differences in nucleation activity between control and GCP8-depleted centrosomes in mitosis (Figure 6B). In summary, GCP8 depletion impairs the nucleation activity of centrosomes in interphase by interfering with the centrosomal recruitment of γTuRC components specifically during this phase of the cell cycle.
Figure 6.
Depletion of GCP8 interferes with interphase centrosomal microtubule nucleation. (A) Nonsynchronized U2OS cells were transfected either with control RNA or GCP8 siRNA and were subjected to a microtubule regrowth assay. Microtubules were depolymerized on ice (time point 0) and after warming microtubules were allowed to regrow (time point 10 s) before fixation and staining with antibodies against GCP8 and α-tubulin. Scale bar, 10 μm. The intensities of the microtubule asters that had formed around centrosomes in interphase after 10 s of regrowth were quantified and mean values were plotted as percentages of intensities in control cells (n > 20, error bars: SEM). (B) U2OS cells transfected either with control RNA or GCP8 siRNA were arrested in mitosis with nocodazole. Cells were subjected to a microtubule regrowth assay and analyzed as in A. Scale bar, 10 μm. The intensities of the microtubule asters that had formed around centrosomes in mitotic cells after 10 s of regrowth were quantified, and mean values were plotted as percentages of intensities in control cells (n > 20, error bars: SEM).
DISCUSSION
We have developed a tandem affinity-based method that allows the efficient purification of γTuRC from human cells while favoring protein–protein interactions. Our protocol includes a polyethylene glycol precipitation step to selectively enrich γTuRC (Murphy et al., 2001). Even though we cannot rule out that in addition to γTuRC other γ-tubulin containing protein complexes were copurified, the most abundant polypeptides in our preparations were almost exclusively γTuRC subunits, and their relative amounts were very similar to those of immunoaffinity-purified γTuRCs in previous studies (Zheng et al., 1995; Moritz et al., 1998; Murphy et al., 1998; Fava et al., 1999; Oegema et al., 1999; Murphy et al., 2001), suggesting that our method is well suited to purify γTuRC. Here we have used purified γTuRCs for a comprehensive analysis of their composition and interactions at distinct cell cycle stages by mass spectrometry.
Core Components of the Human γTuRC
Based on our analysis we can redefine the set of human γTuRC core subunits, which account for the most abundant polypeptides present in purified γTuRC and are found in both interphase and mitotic complexes. This set of subunits comprises γ-tubulin, GCPs 2-6, GCP-WD, and the novel previously uncharacterized protein GCP8. In a previous study published during the preparation of this manuscript, GCP8 was termed MOZART2 (Mitotic spindle organizing protein associated with a ring of γ-tubulin) (Hutchins et al., 2010). In agreement with this study our results suggest that GCP8/MOZART2 does not play an important role in spindle assembly. In addition, we show that GCP8 has properties of a γTuRC core subunit. We would like to propose the uniform “GCP” nomenclature for all core components of the human γTuRC, as identified in our study (Murphy et al., 1998). Accordingly, we suggest the term GCP8 for MOZART2 (counting GCP-WD/NEDD1 as GCP7). Even though GCP-WD and GCP8 are not sequence-related to GCPs 2-6, this simplified nomenclature appreciates that all these proteins qualify as true γTuRC subunits by standard assays.
Our study shows that MOZART1, another mitotic interactor of γTuRCs (Hutchins et al., 2010), is also present in interphase γTuRCs. However, due to its small size (8.5 kDa) we did not detect this protein in our initial analysis by PAGE and Coomassie staining and cannot assess its abundance in the γTuRC. Future studies will show whether MOZART1 qualifies as an additional core subunit of the γTuRC.
γTuRC Interactors
Apart from the γTuRC core subunits we have also identified additional proteins present in purified γTuRCs. As these proteins are less abundant than the core subunits, their interaction with the γTuRC might be of lower affinity or be subject to regulation. Examples of such interactors present in both interphase and mitotic γTuRCs are the nucleoside-diphosphate kinase NME7, subunits of the CCT chaperonin, the centrosomal protein CDK5RAP2, and the AAA+ ATPase RUVBL1. NME7 belongs to a family of nucleoside diphosphate kinases, which play an important role in the generation of nucleoside triphosphates such as ATP and GTP. Interestingly, a Chlamydomonas protein with homology to NME7 was identified as a component of basal bodies and of the flagellum (Keller et al., 2005; Pazour et al., 2005), and the characterization of NME7 knockout mice suggested a function in ciliary motility (Vogel et al., 2010). Whether human NME7 also has a function in ciliary motility and what role the interaction with the γTuRC might play is currently unclear. The CCT chaperonin complex binds to unfolded γ-tubulin and promotes its proper folding (Melki et al., 1993). As the γTuRC purification via tagged γ-tubulin might result in the copurification of some partially folded γ-tubulin, the presence of chaperonin subunits in our γTuRC preparations is not surprising. Our γTuRC preparations also contained CDK5RAP2, a structural component of the PCM participating in γTuRC recruitment (Fong et al., 2008; Haren et al., 2009), whereas pericentrin, for which a similar function has been described, was not detected (Zimmerman et al., 2004; Haren et al., 2009). One interpretation would be that the interaction between pericentrin and the γTuRC is restricted to the centrosome (Dictenberg et al., 1998), whereas CDK5RAP2 might also associate with noncentrosomal γTuRC. In this context it is interesting to note that both CDK5RAP2 and γTuRC have recently been implicated in the centrosome-independent regulation of microtubule plus end dynamics (Fong et al., 2009; Bouissou et al., 2009). RUVBL1 and RUVBL2 have both been described as γTuRC interactors, and RUVBL1 was shown to have a role in mitotic spindle assembly (Ducat et al., 2008). However, we find that both proteins are also associated with interphase γTuRCs suggesting that these proteins might also play a role in nonmitotic functions of the γTuRC. Addressing this question will be a major challenge as RUVBL1 and RUVBL2 have been implicated in multiple essential cellular processes such as chromatin remodeling, DNA repair, and transcription. Interestingly, CDK5RAP2, RUVBL1 and RUVBL2 were not identified as γTuRC interactors in the large-scale analysis of mitotic protein complexes (Hutchins et al., 2010). The different tagging strategy and method used for the purification of the γTuRCs might explain these differences.
Mitosis-Specific γTuRC Interactors
Another category of transient interactors are proteins that interact with γTuRCs in a cell cycle–dependent manner. In preparations of mitotic γTuRC we identified seven of the eight subunits of the human augmin complex, most of which could not be detected in γTuRCs obtained from interphase cells. A mitosis-specific interaction with γTuRC components has been demonstrated previously for the augmin subunit HAUS6 (Zhu et al., 2008). Our results indicate that this interaction most likely involves the entire augmin complex and highlights the importance of this interaction for the function of the γTuRC in noncentrosomal microtubule formation during mitotic spindle assembly (Luders et al., 2006; Luders and Stearns, 2007; Goshima et al., 2008). In addition to augmin we also identified other proteins preferentially associated with mitotic γTuRCs including the mitotic kinase Plk1, for which an interaction with the γTuRC subunit GCP-WD has been demonstrated previously (Haren et al., 2009; Zhu et al., 2009). It will be interesting to test whether Plk1 also binds and potentially regulates other components of the γTuRC.
The Novel Subunit GCP8
GCP8, the novel γTuRC subunit described in this study, is not sequence-related to GCPs 2-6 and does not seem to have a structural role in the γTuRC. GCP8 shares this property with the recently described GCP-WD subunit, suggesting that both proteins might have regulatory functions. In agreement with this interpretation GCP-WD was recently shown to function as a centrosomal attachment factor for the γTuRC and to be regulated by mitotic phosphorylation (Luders et al., 2006; Haren et al., 2006). Our results indicate that GCP8 depletion reduces centrosomal microtubule nucleation by interfering with the recruitment of both GCP-WD and γ-tubulin specifically during interphase. This interphase-specific role of GCP8, which is unique among the characterized γTuRC subunits, might explain why GCP8-depleted cells did not display obvious mitotic defects. We also did not observe centriole duplication defects after GCP8 depletion. It is possible that the remaining nucleation activity at GCP8 depleted centrosomes is sufficient to support centriole duplication or that GCP8 depletion selectively depletes a centrosomal fraction of γ-tubulin that might not be involved in centriole duplication. Indeed, distinct subpopulations of centrosomal γ-tubulin have been described (Khodjakov and Rieder, 1999).
What are the consequences of the defects in centrosomal γ-tubulin recruitment and microtubule nucleation in GCP8-depleted cells? It is tempting to speculate that changes in the organization of the interphase microtubule network might play a role in the reduced proliferation rate of GCP8-depleted cells. However, a recent study has demonstrated that cells with a compromised or even completely depolymerized microtubule network can progress through interphase with near-normal kinetics (Uetake and Sluder, 2007). Microtubules were present in GCP8-depleted cells, but we cannot exclude more subtle defects such as changes in microtubule dynamics. GCP8 might also play a role in noncentrosomal γTuRC functions. For example, the γTuRC was recently shown to localize along interphase microtubules and to regulate microtubule plus end dynamics in Drosophila S2 cells (Bouissou et al., 2009).
We could not identify GCP8-related sequences in yeast, nematodes, flies, or plants, however we cannot rule out the existence of functional homologues with a low degree of conservation at the primary amino acid sequence level. Alternatively, GCP8 might be less important for cells that do not actively nucleate microtubules at centrosomes in interphase. In Drosophila, for example, interphase centrosomes do not recruit detectable amounts of γ-tubulin and do not function as MTOCs (Rogers et al., 2008).
In this study we have identified and extended the set of γTuRC core subunits and provided a comprehensive list of γTuRC interactors. Together with previous studies our work lays grounds for detailed future analyses of the roles of distinct γTuRC interactors in microtubule organization in both interphase and mitosis.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We thank Aaron Groen and Ryoma Ohi for the kind gift of the anti-centrin antibody. This work was supported by the Plan Nacional of I+D+I grants BFU-2007-62087 (to C.C.), BFU2008-03441 (to J.R.) and BFU2009-08522 (to J.L.) (Ministerio de Ciencia e Innovación, Spain). In addition, J.L. acknowledges support from a Marie Curie International Reintegration Grant (FP7-PEOPLE-2007-4-3-IRG, project #224835) and from IRB intramural funds. J.R. is supported by the Ramón y Cajal Program (MICINN, Spain), N.T. is the recipient of an IRB Barcelona Interprogram Postdoctoral Fellowship, and M.T.B. is a recipient of a FPU fellowship (MICINN, Spain).
Abbreviations used:
PCM
Pericentriolar material
γTuRC
γ-tubulin ring complex.
Footnotes
REFERENCES
- Bouissou A., Verollet C., Sousa A., Sampaio P., Wright M., Sunkel C. E., Merdes A., Raynaud-Messina B. {gamma}-Tubulin ring complexes regulate microtubule plus end dynamics. J. Cell Biol. 2009;187:327–334. doi: 10.1083/jcb.200905060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg J. B., Zimmerman W., Sparks C. A., Young A., Vidair C., Zheng Y., Carrington W., Fay F. S., Doxsey S. J. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat D., Kawaguchi S., Liu H., Yates J. R., 3rd, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol. Biol. Cell. 2008;19:3097–3110. doi: 10.1091/mbc.E07-11-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J. E., Gygi S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Fava F., Raynaud-Messina B., Leung-Tack J., Mazzolini L., Li M., Guillemot J. C., Cachot D., Tollon Y., Ferrara P., Wright M. Human 76p: A new member of the gamma-tubulin-associated protein family. J. Cell Biol. 1999;147:857–868. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the {gamma}-tubulin ring complex. Mol. Biol. Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K. W., Hau S. Y., Kho Y. S., Jia Y., He L., Qi R. Z. Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol. Biol. Cell. 2009;20:3660–3670. doi: 10.1091/mbc.E09-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Vardy L., Garcia M. A., Toda T. A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol. Biol. Cell. 2002;13:2360–2373. doi: 10.1091/mbc.02-01-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R. D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A. C., Cameron L. A., Coughlin M., Miyamoto D. T., Mitchison T. J., Ohi R. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Haren L., Remy M. H., Bazin I., Callebaut I., Wright M., Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Stearns T., Luders J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 2009;4:e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J. R., et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., 3rd, Marshall W. F. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman J. M., Zelter A., Muller E. G., Fox B., Rice L. M., Davis T. N., Agard D. A. The Structure of the {gamma}-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell. 2008;19:207–215. doi: 10.1091/mbc.E07-09-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., et al. HAUS, the 8-subunit human augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- Luders J., Patel U. K., Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Luders J., Stearns T. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Ma W., Baumann C., Viveiros M. M. NEDD1 is crucial for meiotic spindle stability and accurate chromosome segregation in mammalian oocytes. Dev. Biol. 2010;339:439–450. doi: 10.1016/j.ydbio.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Melki R., Vainberg I. E., Chow R. L., Cowan N. J. Chaperonin-mediated folding of vertebrate actin-related protein and gamma-tubulin. J. Cell Biol. 1993;122:1301–1310. doi: 10.1083/jcb.122.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M. B., Guenebaut V., Heuser J., Agard D. A. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moritz M., Zheng Y., Alberts B. M., Oegema K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. M., Preble A. M., Patel U. K., O'Connell K. L., Dias D. P., Moritz M., Agard D., Stults J. T., Stearns T. GCP5 and GCP 6, two new members of the human gamma-tubulin complex. Mol. Biol. Cell. 2001;12:3340–3352. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. M., Urbani L., Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J. Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Wiese C., Martin O. C., Milligan R. A., Iwamatsu A., Mitchison T. J., Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Peifer M., Rogers S. L. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J., Mikhailov A., Belham C., Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R. S., Tomioka A., Petry S., Vale R. D., Obuse C., Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Sluder G. Cell-cycle progression without an intact microtuble cytoskeleton. Curr. Biol. 2007;17:2081–2086. doi: 10.1016/j.cub.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram S., Tasto J. J., Feoktistova A., Jennings J. L., Link A. J., Gould K. L. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell. 2004;15:2287–2301. doi: 10.1091/mbc.E03-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verollet C., Colombie N., Daubon T., Bourbon H.M., Wright M., Raynaud-Messina B. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 2006;172:517–528. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P., Read R., Hansen G. M., Freay L. C., Zambrowicz B. P., Sands A. T. Situs inversus in Dpcd/Poll−/−, Nme7−/−, and Pkd1l1−/− mice. Vet. Pathol. 2010;47:120–131. doi: 10.1177/0300985809353553. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Oakley B. R. In vivo analysis of the functions of gamma-tubulin-complex proteins. J. Cell Sci. 2009;122:4218–4227. doi: 10.1242/jcs.059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C. J., Lee Y. R., Liu B. The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell. 2009;21:1129–1140. doi: 10.1105/tpc.109.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wong M. L., Alberts B., Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- Zhu H., Coppinger J. A., Jang C. Y., Yates J. R., 3rd, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Fang K., Fang G. FAM29A, a target of Plk1 regulation, controls the partitioning of NEDD1 between the mitotic spindle and the centrosomes. J. Cell Sci. 2009;122:2750–2759. doi: 10.1242/jcs.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman W. C., Sillibourne J., Rosa J., Doxsey S. J. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]