The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation (original) (raw)
Abstract
In the mouse, more than 16 loci are associated with mutant phenotypes that include defective pigmentation, aberrant targeting of lysosomal enzymes, prolonged bleeding, and immunodeficiency, the result of defective biogenesis of cytoplasmic organelles: melanosomes, lysosomes, and various storage granules. Many of these mouse mutants are homologous to the human Hermansky–Pudlak syndrome (HPS), Chediak–Higashi syndrome, and Griscelli syndrome. We have mapped and positionally cloned one of these mouse loci, buff (bf), which has a mutant phenotype similar to that of human HPS. Mouse bf results from a mutation in Vps33a and thus is homologous to the yeast vacuolar protein-sorting mutant vps33 and Drosophila carnation (car). This is the first found defect of the class C vacuole/prevacuole-associated target soluble _N_-ethylmaleimide-sensitive factor attachment protein receptor (t-SNARE) complex in mammals and the first mammalian mutant found that is directly homologous to a vps mutation of yeast. VPS33A thus is a good candidate gene for a previously uncharacterized form of human HPS.
Hermansky–Pudlak syndrome (HPS) is a disorder of organelle biogenesis in which oculocutaneous albinism, bleeding, and in most cases pulmonary fibrosis result from defects of melanosomes, platelet-dense granules, and lysosomes (1–4). Somewhat similar disorders, Chediak–Higashi and Griscelli syndromes, are additionally associated with severe immunodeficiency (2, 3). Important clues to the pathogenesis of these disorders have come from the mouse, in which >16 loci have been associated with mutant phenotypes similar to those of human HPS, Chediak–Higashi syndrome, and Griscelli syndrome (5, 6). Several of these genes have been identified recently and in a number of cases have been shown to result in homologous disorders in mice and humans (2–4). Although the functions of many of the corresponding gene products remain unknown, several are involved in various aspects of trafficking proteins to nascent organelles, particularly melanosomes, lysosomes, and cytoplasmic granules. In the yeast, >65 proteins have been implicated in biogenesis of the cytoplasmic vacuole, including the products of >40 vacuolar protein-sorting (vps) loci required for trafficking newly synthesized proteins from the late Golgi/_trans_-Golgi network to the vacuole (7, 8). It seems likely that at least as many proteins are associated with organellar biogenesis in mammals.
We have mapped and positionally cloned the mouse buff (bf) locus, which is characterized by recessive coat-color hypopigmentation and mild platelet-storage pool deficiency but has little if any effect on lysosomal function. We find that mouse bf results from a missense substitution in Vps33a, a homologue of yeast vps33. The bf mutation results in defective melanosome morphology and melanogenesis both in vivo and in vitro. Expression of wild-type Vps33a in transfected mouse _bf_-mutant melanocytes complements this aberrant phenotype, whereas expression of _bf_-mutant Vps33a does not. These results establish murine bf as a murine homologue to the yeast vps33 mutant and suggest VPS33A as a candidate gene for some cases of human HPS.
Materials and Methods
Mice and Backcross.
Mice carrying the spontaneous mutation bf on the C57BL/6J strain were obtained from The Jackson Laboratory and were subsequently bred at Roswell Park Cancer Institute. C57BL/6J bf/bf mice were crossed with the inbred, wild-derived Mus musculus musculus (PWK) inbred strain to obtain a high degree of polymorphism for molecular markers (9). A high-resolution genetic map of the bf critical region was generated by performing an interspecific cross between the bf/bf and PWK mice. At 6 weeks, 1,167 backcross progeny were typed for coat color and molecular markers by using microsatellite markers D5Mit188 and D5Mit212, which flank bf proximally and distally, respectively. Informative mice were typed for 20 markers between D5Mit188 and D5Mit212.
Phenotypic Analyses.
Electron microscopy of mouse eyes (10), assays of platelet morphology and function (11), and assays of kidney and urine β-glucuronidase and β-galactosidase (12) were carried out as described. Platelet aggregation was determined by the impedance method in whole blood in response to 1 μg/ml collagen in bf/bf and C57BL/6J mice (11). ATP release was determined by luminescence methods.
Mutation Analyses.
Overlapping amplicons spanning the coding regions of the mouse Vps33a, Rnp24, and Atp6v0a2 genes and the murine homologues of human FLJ12975 and FLJ22471 were designed and used for RT-PCR amplification from mRNA of C57BL/6J and bf/bf mice. Kidney mRNA was prepared by using the RNeasy mini kit (Qiagen, Valencia, CA), and reverse transcription of total RNA was performed by using oligo dT primer and SuperScript II reverse transcriptase (GIBCO/BRL). Amplicons were screened for mutations by single-stranded conformational polymorphism/heteroduplex analysis (13), and a Vps33a amplicon, which showed an aberrant pattern, was reamplified and sequenced directly.
To screen for VPS33A mutations in human HPS patients, primers derived from intervening and noncoding sequences adjacent to the 13 genomic exons were used to amplify PCR products from genomic DNA of 26 HPS patients who lacked mutations in the HPS1, HPS2 (AP3B1), HPS3, and HPS4 loci. VPS33A amplicons were screened for mutations by single-stranded conformational polymorphism/heteroduplex analysis (13), and the one amplicon containing the Ile-256 → Leu variant was screened in DNA of 27 unrelated Puerto Rican controls.
Expression Plasmid Construction, Cell Culture, and Transformation.
To construct Vps33a and Vps33abf expression plasmids, we first prepared full-length wild-type and Asp-251 → Glu mouse Vps33a cDNAs by nested RT-PCR using C57BL/6J and bf/bf mouse kidney cDNA, respectively, as described above and initial primers 5′-CGGGTGCCCGGGCAAGATG-3′/5′-GTGGTGACCTGTCACGTCTC-3′ and secondary primers 5′-GACTAGTGCCCCATGGCGGCGCACCTGTCGTACGG-3′ (containing an _Spe_I linker)/5′-CCGCTCGAGGTGGTGACCTGTCACGTCTCCTCCGA-3′ (containing an _Xho_I linker). The amplified cDNA fragments were cloned in pCR2.1 (Invitrogen) and sequenced to verify accuracy. The Vps33a and Vps33abf cDNAs were subcloned in-frame in the _Kpn_I and _Xho_I sites of GATEWAY pENTR3C (Invitrogen). The cDNAs then were transferred via site-specific recombination into pDEST26 (Invitrogen) and subsequently into the _Sna_BI and _Eco_RV sites of pIREShyg2 (CLONTECH).
Wild-type Melan-a (14) and Melan-bf (15) mouse melanocytes were cultured as described (10). Melan-bf cells were transfected with pIREShyg2-m_Vps33a_, pIREShyg2-m_Vps33abf_, and pIREShyg2 by using FuGENE 6 transfection reagent (Roche Diagnostics, Basel), and stably transformed cell lines were selected in medium containing 300 μg/ml hygromycin B (Invitrogen).
Results
Characterization of the _bf_-Mutant Phenotype.
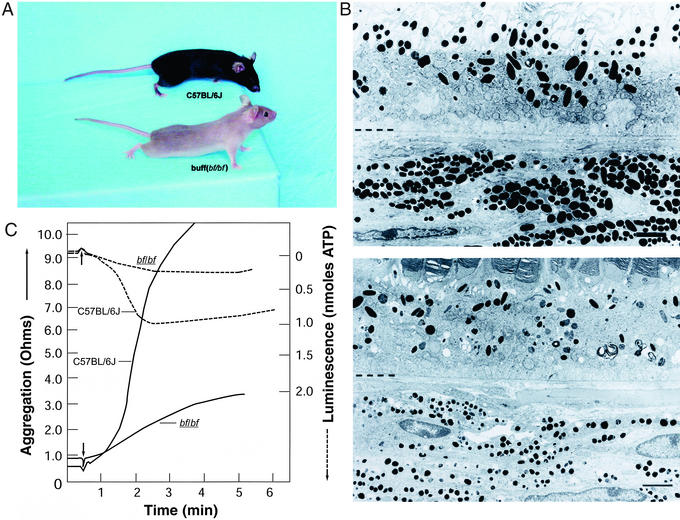
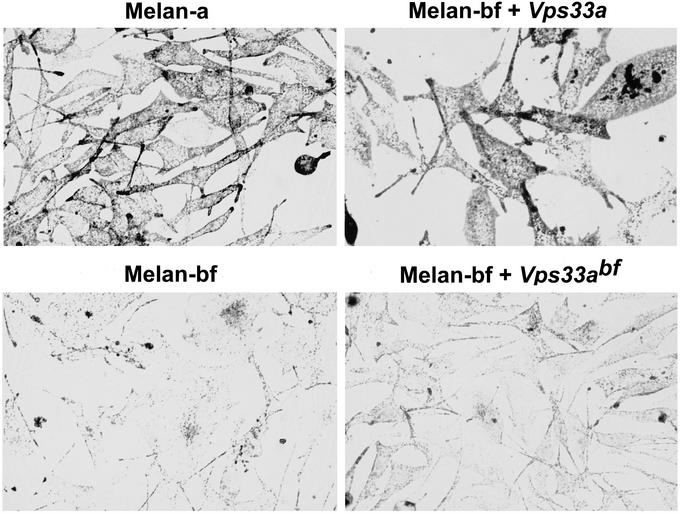
bf arose spontaneously in mice of the C57BL/6J strain (16) and is represented by a single mutant allele. Homozygous bf/bf mice exhibit reduced coat-color pigmentation (Fig. 1A). Lysosomal enzyme activities have been reported to be increased in kidneys of bf mice (17), although we have not been able to confirm these findings (see below). To characterize the bf phenotype further, we carried out electron microscopy of the eyes of bf/bf and C57BL/6J mice. As shown in Fig. 1B, melanosomes are markedly reduced in both size and number in the retinal pigment epithelium and choroid of bf mice. Similar findings were observed on analysis of a permanent line of mouse melanocytes derived from bf/bf mice (Fig. 2, Melan-bf) (15).
Figure 1.
Phenotype of _bf_-mutant mice. (A) Coat-color hypopigmentation in adult bf/bf versus C57BL/6J mice. (B) Reduced size and number of melanosomes in the eyes of bf/bf (Lower) versus wild-type C57BL/6J (Upper) mice. The dotted lines represent demarcation between retinal pigment epithelium (above) and choroid (below). (Scale bar, 2 μm.) (C) Reduced collagen-mediated platelet aggregation (solid line) and dense granule ATP secretion (dotted line) in bf/bf versus C57BL/6J mice. The arrows indicate the time of addition of collagen.
Figure 2.
Complementation of _bf_-mutant mouse melanocytes by Vps33a. Bright-field microscopy of wild-type (Melan-a) mouse melanocytes (14), _bf_-mutant (Melan-bf) mouse melanocytes (15), and _bf_-mutant melanocytes stably transformed by expression vectors containing wild-type (Melan-bf + Vps33a) or _bf_-mutant mouse Vps33a (Melan-bf + Vps33abf) cDNAs. Pigmented cytoplasmic granules are melanized melanosomes.
To assess platelet function in bf mice, we first tested bleeding time, which was prolonged significantly, to 9.7 ± 1.2 min in bf/bf homozygotes, versus 4.7 ± 0.7 min in bf/+ heterozygous controls (P < 0.02). Collagen-mediated platelet aggregation, secretion of dense-granule ATP (Fig. 1C), and platelet serotonin (not shown) all were reduced significantly in bf/bf mice. Furthermore, the number of dense granules per platelet, determined by whole-mount electron microscopy, was reduced greatly in bf/bf (5.07 ± 0.37) versus C57BL/6J (12.2 ± 0.79) mice (P < 0.001). These results establish that the _bf_-mutant phenotype includes significant platelet-storage pool defect, although this defect is less pronounced than in other mouse HPS-like mutants (6).
In contrast, we observed much less effect of the bf mutation on lysosomal enzyme activity and secretion. Kidney β-glucuronidase and β-galactosidase activities were not altered significantly in bf/bf versus C57BL/6J mice (not shown), and whereas secretion of β-galactosidase into urine was reduced by 53% (P < 0.02), the slight reduction we observed in β-glucuronidase secretion was not significant. Thus, the principal physiological effect of the bf mutation on organelles seems to be on melanosomes, with lesser effects on platelet-dense granules and little or no effect on lysosomes.
Fine Mapping and Identification of the Murine bf Locus.
Mouse bf was mapped previously to 65.0 centimorgans on mouse chromosome 5, although not with high precision (18). To refine localization of the bf gene, we carried out a backcross of C57BL/6J mice homozygous for bf with mice of the PWK strain and typed 1,167 progeny for recombination events near bf using markers D5Mit188 and D5Mit282, which flank bf proximally and distally, respectively. Fifty-six recombinant mice were then phenotyped for coat color and genotyped for 20 additional microsatellite markers previously mapped to this region of mouse chromosome 5: D5Mit319, D5Mit368, D5Mit408, D5Mit137, D5Mit96, D5Mit160, D5Mit65, D5Mit369, D5Mit139, D5Mit279, D5Mit138, D5Mit95, D5Mit140, D5Mit161, D5Mit212, D5Mit244, D5Mit30, D5Mit162, D5Mit29, and D5Mit165. These data localized bf to the 0.7-centimorgan interval between D5Mit137 proximally and markers D5Mit138, D5Mit139, and D5Mit279 distally; within this interval, four markers were nonrecombinant with bf: D5Mit96, D5Mit160, D5Mit65, and D5Mit369.
By analysis of the United Kingdom's Mouse Genome Centre database, we identified a sequence-tagged site within the bf interval, AI120173, which represents a mouse orthologue of human CD36L1. This indicated that the bf region of mouse chromosome 5 is homologous to distal human chromosome 12q24. The NCBI and Celera human genome databases indicated that this region is ≈2.1 megabases in size, containing ≈71 known and predicted genes, of which VPS33A, RNP24, and ATP6V0A2 had the greatest apparent biological relevance. We therefore screened for mutations in the mouse homologues of these three genes as well as the murine homologues of the predicted genes FLJ12975 and FLJ22471 by RT-PCR of kidney mRNA from bf/bf versus wild-type C57BL/6J mice and single-stranded conformational polymorphism/heteroduplex analysis (13) of the RT-PCR products. We detected an obvious abnormality in mouse Vps33a, and DNA sequence analyses of both the RT-PCR product and the genomic DNA defined a missense substitution of glutamic acid for aspartic acid at codon 251 (GAT → GAG). This substitution was not present in the wild-type C57BL/6J background on which the bf mutation arose. As shown in Fig. 3, Asp-251 has been conserved among all Vps33 homologues for which sequences are available and is located only three residues distal to the glycine disrupted by the carnation (car) mutation of Drosophila (19).
Figure 3.
Alignment of Vps33a-related amino acid sequences. Mouse Vps33a amino acid residues 220–260 are shown aligned to all known Vps33 orthologues and paralogues. Residues absolutely conserved among all available Vps33-related sequences (Vps33 unless indicated) are shown in red, and residues conserved among the three mammalian Vps33a paralogues are shown in yellow. <1>, mouse Vps33a bf Asp-251 → Glu (GAT → GAG) substitution; <2>, Drosophila car 1 Gly-249 → Val substitution (19); <3>, human VPS33A Ile-256 → Leu (ATT → CTT) substitution.
Characterization and Complementation of Mouse _bf_-Mutant Melanocytes.
To confirm that the bf gene is indeed Vps33a, we stably transformed a line of melanocytes derived from homozygous _bf_-mutant mice (15) by using both wild-type and _bf_-mutant (Asp-251 → Glu) Vps33a cDNAs. As shown in Fig. 2, _bf_-mutant melanocytes contain a reduced number of undermelanized melanosomes, which is similar to what is seen in vivo (Fig. 1B). Stable transformation by a vector expressing wild-type Vps33a cDNA completely complemented this abnormal phenotype, restoring normal melanosomal number and pigmentation (Fig. 2). In contrast, stable transformation by a plasmid expressing _bf_-mutant (Asp-251 → Glu) Vps33a cDNA or by the empty vector did not alter the mutant phenotype (Fig. 2). These results confirm that Vps33a is the mouse bf gene and that the Asp-251 → Glu substitution pathologically affects function of the Vps33a protein in vivo.
Analysis of VPS33A in Human Patients with HPS.
We screened human VPS33A as a candidate HPS gene in 26 unrelated patients with HPS who lack mutations in the HPS1, HPS2, HPS3, and HPS4 loci. One patient, of mixed Puerto Rican/Dominican origin, was found to be heterozygous for a missense substitution, Ile-256 → Leu (ATT → CTT). Ile-256 has been conserved among all known Vps33-related homologues (Fig. 3). Nevertheless, no other mutation was found in this patient. Although this substitution was not observed among 27 Puerto Rican controls, at present we cannot be certain whether this substitution has pathological significance or is a rare nonpathological polymorphism.
Discussion
Organelle biogenesis is a complex series of processes that organize trafficking of newly synthesized “cargo” proteins to the nascent organelles. In yeast, some 65 different genes have been implicated in biogenesis of the lysosome-like vacuole including >40 so-called vps loci required for assembly of the vacuolar ATPase and selective transport of certain soluble and membrane-bound proteins from the late Golgi/_trans_-Golgi network to the vacuole (7, 8). In mice and humans, fewer genes are known, but the diversity of organelles virtually assures that an even larger number of proteins are required for organellar biogenesis in mammals. The constellation of HPS-, Chediak–Higashi syndrome-, and Griscelli syndrome-like disorders of humans and mice indicates that melanosomes, lysosomes, and certain cytoplasmic granules share common pathways of organellar protein trafficking, although these pathways are as yet largely unknown. Here we have shown that the mouse bf locus corresponds to Vps33a, encoding a homologue of yeast vps33. This is the first mammalian mutant shown to involve a direct homologue of a classical yeast vps mutant, and the large amount known about protein trafficking to the yeast vacuole makes bf particularly instructive.
Vps33, a homologue of Sec1p-like regulators of membrane fusion, interacts with the Vps11, Vps16, Vps18, and probably also the Vps39 and Vps41 proteins to form the class C Vps complex (20), which in yeast is required for soluble _N_-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-mediated vesicle docking and fusion with the vacuole (21–23). Vps41, in turn, interacts with the AP-3 adaptor complex in a clathrin-independent pathway of vacuolar protein trafficking (24, 25). There is weak genetic interaction between vps33 and a synthetic deletion of apm3, encoding the μ chain of AP-3 (26), although the nature of this interaction has not been defined. In Drosophila, the car eye color locus likewise corresponds to vps33 (19), the _car_1 allele involving a Gly-249 → Val substitution, only three residues proximal to that homologous to Asp-251 in mouse Vps33A (Fig. 3).
In mammals (mouse, rat, and human), there are two vps33 paralogues, Vps33a and Vps33b (27, 28), and the respective functions of the corresponding two Vps33-like proteins are not known. The relative specificity of the _bf_-mutant phenotype to melanosomes and platelet granules versus virtual sparing of lysosomal function suggests that a Vps33a-containing class C Vps complex might be specifically involved in biogenesis of melanosomes and granules, whereas a Vps33b-containing class C Vps complex might be specifically involved in biogenesis of lysosomes. Because expression of both Vps33a and Vps33b seems to be ubiquitous (28, 29), any such functional specificity of these two Vps33-related proteins must reside at the protein level.
The identification of Vps33a as the murine bf locus suggests human VPS33A and perhaps also VPS33B as candidate HPS loci in man. Our data indicate that VPS33A, at least, is not a common HPS gene in humans. Nevertheless, rare cases of HPS may result from pathological mutations. The close functional association in yeast between the Vps33 protein and Vps11, Vps16, Vps18, Vps39, Vps41, and the AP-3 complex suggests these genes as additional candidates for the HPS-like loci of mouse and humans. Indeed, the mouse pearl (pe) and human HPS2 loci have been found to correspond to Ap3b1, and the mouse mocha (mh) and Drosophila garnet (g) loci correspond to Ap3d1 (2–4). Together, these findings strongly implicate a clathrin-independent AP-3 adaptor-class C Vps complex–soluble _N_-ethylmaleimide-sensitive factor attachment protein receptor pathway in mammalian melanosome, lysosome, and granule biogenesis.
Acknowledgments
We thank M. Reddington, S. Y. Jiang, L. Zhen, and D. Tabaczynski for expert technical assistance. This work was supported by National Institutes of Health Grants AR39892 (to R.A.S.), HL51480, HL31698, and EY12104 (to R.T.S.), and EY10199 (to P.G.S.), Roswell Park Cancer Institute Cancer Center Support Grant CA16056, and Welch Foundation Grant I-1300 (to P.G.S.).
Abbreviations
HPS
Hermansky–Pudlak syndrome
Vps
vacuolar protein sorting
bf
buff
car
carnation
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF439858 (mouse Vps33a cDNA and amino acid sequences) and AF439857 (VPS33A cDNA and amino acid sequences)].
References
- 1.Hermansky F, Pudlak P. Blood. 1959;14:162–169. [PubMed] [Google Scholar]
- 2.Spritz R A. Clin Genet. 1999;55:309–317. doi: 10.1034/j.1399-0004.1999.550503.x. [DOI] [PubMed] [Google Scholar]
- 3.Spritz R A. Trends Genet. 1999;15:337–340. doi: 10.1016/s0168-9525(99)01785-0. [DOI] [PubMed] [Google Scholar]
- 4.Spritz R A. Pigm Cell Res. 1999;13:15–20. doi: 10.1034/j.1600-0749.2000.130104.x. [DOI] [PubMed] [Google Scholar]
- 5.Bennett D. Int Rev Cytol. 1993;146:191–260. doi: 10.1016/s0074-7696(08)60383-6. [DOI] [PubMed] [Google Scholar]
- 6.Swank R T, Novak E K, McGarry M P, Rusiniak M E, Feng L. Pigm Cell Res. 1998;11:60–80. doi: 10.1111/j.1600-0749.1998.tb00713.x. [DOI] [PubMed] [Google Scholar]
- 7.Wendland B, Emr S D, Riezman H. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- 8.Pelham H R B. Philos Trans R Soc London B. 1999;354:1471–1478. doi: 10.1098/rstb.1999.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien E P, Novak E K, Keller S A, Poirier C, Guenet J L, Swank R T. Mamm Genome. 1994;5:356–360. doi: 10.1007/BF00356554. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T, Li W, Zhang Q, Novak E K, Sviderskaya E V, Wilson A, Bennett D C, Roe B A, Swank R T, Spritz R A. Genomics. 2001;78:30–37. doi: 10.1006/geno.2001.6644. [DOI] [PubMed] [Google Scholar]
- 11.Swank R T, Sweet H O, Davisson M T, Reddington M, Novak E K. Genet Res. 1991;58:51–62. doi: 10.1017/s0016672300029608. [DOI] [PubMed] [Google Scholar]
- 12.Brandt E J, Elliott R W, Swank R T. J Cell Biol. 1975;67:774–788. doi: 10.1083/jcb.67.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S-T, Park S-K, Lee K-H, Holmes S A, Spritz R A. Mol Cells. 1995;5:668–672. [Google Scholar]
- 14.Bennett D C, Cooper P J, Hart I R. Int J Cancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 15.Samaraweera P, Donatien P D, Qazi S, Kobayashi T, Hearing V J, Panthier J J, Orlow S J. Eur J Biochem. 1999;266:924–934. doi: 10.1046/j.1432-1327.1999.00930.x. [DOI] [PubMed] [Google Scholar]
- 16.Dickie M M. Mouse Newslett. 1964;30:30. [Google Scholar]
- 17.Hakansson E M, Lundin L G. Biochem Genet. 1977;15:75–85. doi: 10.1007/BF00484549. [DOI] [PubMed] [Google Scholar]
- 18.Kerscher S, Glenister P H, Favor J, Lyon M F. Genomics. 1996;36:17–21. doi: 10.1006/geno.1996.0420. [DOI] [PubMed] [Google Scholar]
- 19.Sevrioukov E A, He J-P, Moghrabi N, Sunio A, Kramer H. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- 20.Rieder S E, Emr S D. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T K, Rehling P, Peterson M R, Emr S D. Mol Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 22.Peterson M R, Emr S D. Traffic. 2001;2:476–486. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- 23.Wurmser A E, Sato T K, Emr S D. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehling P, Darsow T, Katzmann D J, Emr S D. Nat Cell Biol. 1999;1:346–353. doi: 10.1038/14037. [DOI] [PubMed] [Google Scholar]
- 25.Darsow T, Katzmann D J, Cowles C R, Emr S D. Mol Biol Cell. 2001;12:37–51. doi: 10.1091/mbc.12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stepp J D, Huang K, Lemmon S K. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pevsner J, Hsu S C, Hyde P S, Scheller R H. Gene. 1996;183:7–14. doi: 10.1016/s0378-1119(96)00367-8. [DOI] [PubMed] [Google Scholar]
- 28.Huizing M, Didier A, Walenta J, Anikster Y, Gahl W A, Krämer H. Gene. 2001;264:241–247. doi: 10.1016/s0378-1119(01)00333-x. [DOI] [PubMed] [Google Scholar]
- 29.Carim L, Sumoy L, Andreu N, Estivill X, Escarceller M. Cytogenet Cell Genet. 2000;89:92–95. doi: 10.1159/000015571. [DOI] [PubMed] [Google Scholar]