Gene Expression Profiling and Chromatin Immunoprecipitation Identify DBN1, SETMAR and HIG2 as Direct Targets of SOX11 in Mantle Cell Lymphoma (original) (raw)
Abstract
The SRY (sex determining region Y)-box 11 (SOX11) gene, located on chromosome 2p25, encodes for a transcription factor that is involved in tissue remodeling during embryogenesis and is crucial for neurogenesis. The role for SOX11 in hematopoiesis has not yet been defined. Two genes under direct control of SOX11 are the class- III β-tubulin gene (TUBB3) in neural cells and the transcription factor TEA domain family member 2 (TEAD2) in neural and mesenchymal progenitor cells. Normal, mature lymphocytes lack SOX11 but express SOX4, another member of the same group of SOX transcription factors. We and others recently identified SOX11 as aberrantly expressed in mantle cell lymphoma (MCL). Since SOX11 is variably expressed in MCL it may not be essential for tumorigenesis, but may carry prognostic information. Currently, no specific functional effects have been linked to SOX11 expression in MCL and it is not known which genes are under influence of SOX11 in lymphoma. In this study we found variable expression of SOX11, SOX4 and SOX12 mRNA in mantle cell lymphoma cell lines. Downregulation of SOX11 expression by siRNA verified that SOX11 controlled the expression of the gene TUBB3 in the MCL cell line Granta 519. Furthermore we identified, by global gene expression analysis, 26 new target genes influenced by siRNA SOX11 downmodulation. Among these genes, DBN1, SETMAR and HIG2 were found to be significantly correlated to SOX11 expression in two cohorts of primary mantle cell lymphomas. Chromatin immunoprecipitation (ChIP) analysis showed that these genes are direct targets of the SOX11 protein. In spite of almost complete downregulation of the SOX11 protein no significant effects on Granta 519 cell proliferation or survival in short term in vitro experiments was found. In summary we have identified a number of genes influenced by SOX11 expression in MCL cell lines and primary MCL. Among these genes, DBN1, SETMAR and HIG2 are direct transcriptional targets of the SOX11 protein.
Introduction
The SRY (sex determining region Y)-box 11 (SOX11) transcription factor was recently discovered as a new marker in mantle cell lymphoma (MCL), expressed in both cyclin D1 positive and negative cases [1], [2], [3], [4]. SOX11, located on chromosome 2p25, is a member of the SOX gene family and was discovered in 1995 [5]. Approximately 20 SOX genes have been identified and they are divided into eight subgroups according to the degree of similarity within and outside the high mobility domain (HMG) [6]. All SOX genes have unique and specific expression patterns and they control cell survival, proliferation and differentiation in numerous processes during embryogenesis (reviewed in [7]). SOX11, together with SOX4 and SOX12, belongs to the SOXC group, sharing a high degree of homology both in the HMG domain and the C-terminal transactivation domain [8]. In the mouse, Sox11 is important for organ development, neurogenesis, neural cell survival and neurite outgrowth [8], [9], [10] [11] while Sox4 is crucial for B lymphocyte differentiation [12]. Upregulated SOX11 has been found in brain tumors and in ovarian carcinoma, where it has been associated to tumorigenesis and clinical outcome [13], [14], [15], [16], [17]. The recently reported aberrant expression of SOX11 in most MCL [1], [2], [3], [4], [18], [19] raises the hypothesis that SOX11 may have a role in the pathogenesis of MCL.
MCL is characterized by enhanced cell proliferation, impaired cell death pathways and reduced response to DNA damaging agents and is therefore difficult to treat [20]. These features can only be partly explained by the t(11;14)(q13;q32) causing dysregulation of cyclin D1 expression. While the t(11;14) is an early event in the pathogenesis of most MCL other genetic events are necessary for lymphoma development [21], [22].
The class-III β-tubulin gene (TUBB3) is up-regulated by Sox11 in neural cells [8]. Using siRNA-mediated knockdown of SOX11 in the MCL cell lines Granta 519 and JeKo, we found that TUBB3 was influenced by SOX11 also in MCL. Additionally, 26 genes were significantly downregulated upon SOX11 silencing in Granta 519. Among these genes, drebrin 1 (DBN1), SET domain and mariner transposase fusion gene (SETMAR) and hypoxia-inducible protein 2 (HIG2) expression correlated to SOX11 expression in a series of SOX11 positive MCL [4]. We found a strong correlation between SOX11 and DBN1, SETMAR and HIG2 expression in a recently published study on SOX11 positive and negative MCL [19]. Furthermore, in MCL cell lines and primary MCL cells, DBN1 significantly correlated to SOX11 expression.
Results
Expression of SOXC Transcription Factors in MCL Cell Lines – High Expression of SOX11 in Granta 519
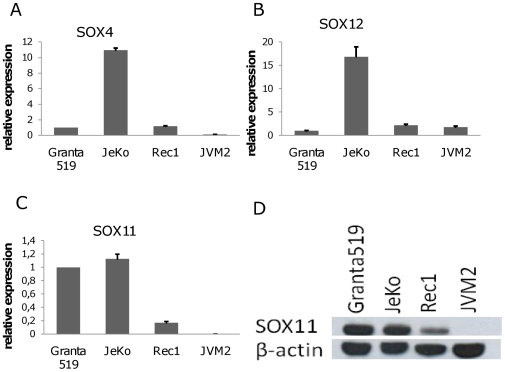
SOX4, SOX11 and SOX12 may have partially overlapping effects on transcriptional regulation [8]. We therefore investigated the expression of the three SOXC transcription factors, SOX4, SOX11 and SOX12 in the t(11;14) positive MCL cell lines Granta 519, Rec1, JeKo and JVM-2 (Figure 1). Granta 519, Rec1 and JeKo expressed SOX11 mRNA and protein while JVM-2 was negative for SOX11 expression (Figure 1). SOX4 and SOX12 protein expression could not be measured due to lack of commercially available specific antibodies. The Granta 519 cell line expressed high levels of SOX11 and very little SOX4 and SOX12 and was therefore chosen for further studies.
Figure 1. Expression of SOXC genes in MCL cell lines.
Expression of SOX4 (A), SOX12 (B) and SOX11 (C) in the MCL cell lines Granta 519, JeKo, Rec1 and JVM2 by quantitative RT- PCR. Granta 519 express high levels of SOX11 mRNA but low levels of the other SOXC genes while no SOX11 mRNA expression was detected in JVM2. (D) WB showed that Granta 519, JeKo and Rec1, but not JVM2, express SOX11 protein.
Silencing of SOX11 in Granta 519 Modifies TUBB3 Expression
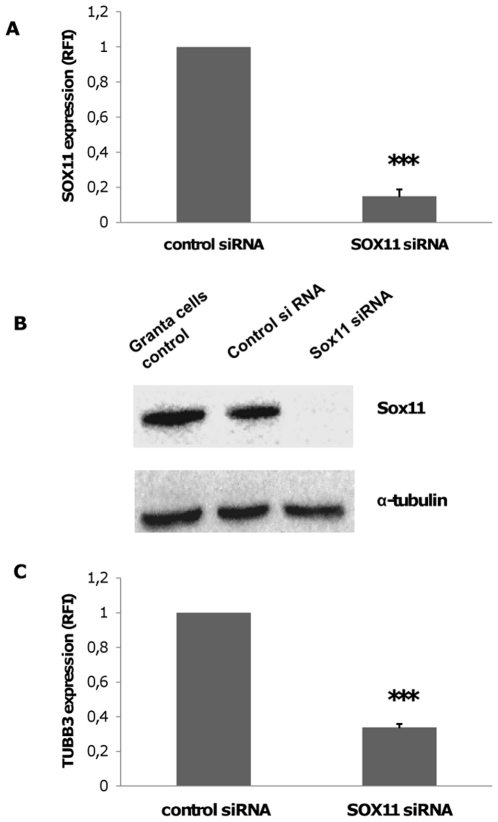
The effect of siRNA-inhibition of SOX11 in Granta 519 was investigated using the S13312 siRNA. After 20 hours, the expression of SOX11 mRNA decreased by approximately 85% compared to cells treated with control siRNA (Figure 2A). Downregulation of mRNA was stable between 12–48 hours (data not shown). The SOX11 protein could not be detected at 20 hours after SOX11 knock down (Figure 2B) and the protein levels remained undetectable for 72 hours (data not shown). Similar silencing effects were obtained by another SOX11 siRNA, S224667 (data not shown). TUBB3 has been shown to be regulated by Sox11 in non-hematopoietic cells [8]. We therefore investigated whether SOX11 is able to regulate TUBB3 in Granta 519. 20 hours after transfection with SOX11 siRNA the expression of TUBB3 was reduced by 70% compared to negative control siRNA (Figure 2C).
Figure 2. SOX11 silencing significantly reduces TUBB3 expression in the MCL cell line Granta 519.
(A) Expression of SOX11 was evaluated after transfection with either 100 pmol SOX11 siRNA or control siRNA. The SOX11 mRNA level was measured by quantitative RT-PCR 20 hours after electroporation. Error bars show standard deviation of 4 independent experiments, ***p<0.001. (B) Western blots show downregulation of SOX11 protein in Granta 519 after SOX11 siRNA knockdown but not in cells treated with control siRNA or in untransfected cells. (C) Depletion of SOX11 protein significantly reduces TUBB3 expression. Granta 519 cells were transfected with SOX11 siRNA and TUBB3 expression was investigated by quantitative RT-PCR after 20 hours. ***P<0.001.
Silencing of SOX11 Significantly Reduces the Expression of 26 Genes
We next analyzed the effect of knock down of SOX11 on global gene expression in Granta 519 cells. At the chosen time point, 20 hours after transfection, 26 genes were significantly downregulated in SOX11 siRNA treated cells compared to cells treated with control siRNA at the chosen threshold (FDR<0.002, FC≥1.5 or FC≤−1.5) (Table 1). No upregulated genes were found using this threshold.
Table 1. Differently expressed genes after SOX11 knockdown by siRNA in Granta 519 cells#.
| probe set ID | gene_assignment | p-value* | Fold-Change* |
|---|---|---|---|
| 8174189 | NM_021992//TMSB15A//thymosin beta 15a//Xq21.33-q22.3 | 6.9064e-010 | −3.94 |
| 8040070 | NM_003108//SOX11//SRY (sex determining region Y)-box 11//2p25 | 4.3570e-009 | −3.05 |
| 8110589 | NM_015455//CNOT6//CCR4-NOT transcription complex, subunit 6//5q35.3 | 7.7295e-009 | −1.52 |
| 7963054 | NM_006009//TUBA1A//tubulin, alpha 1a//12q12-q14 | 2.2166e-008 | −2.04 |
| 8117640 | NM_025231//ZSCAN16//zinc finger and SCAN domain containing 16//6p22.1 | 3.8797e-008 | −1.55 |
| 8047127 | NM_001130158//MYO1B//myosin IB//2q12-q34 | 4.4570e-008 | −2.43 |
| 8113504 | NM_004772//C5orf13//chromosome 5 open reading frame 13//5q22.1 | 1.6184e-007 | −1.78 |
| 8077370 | NM_006515//SETMAR//SET domain and mariner transposase fusion gene//3p26.1 | 2.4819e-007 | −1.58 |
| 8069880 | NM_003253//TIAM1//T-cell lymphoma invasion and metastasis 1//21q22.1|21q22 | 2.6924e-007 | −1.71 |
| 8169073 | NM_194324//TMSB15B//thymosin beta 15B//Xq22.2 | 2.7046e-007 | −1.70 |
| 7925550 | NM_001126//ADSS//adenylosuccinate synthase//1cen-q12 | 2.7195e-007 | −1.72 |
| 8005458 | NM_001040078//LGALS9C//lectin, galactoside-binding, soluble, 9C//17p11.2/ | 4.1908e-007 | −1.69 |
| 8170468 | NM_005342//HMGB3//high-mobility group box 3//Xq28 | 4.9152e-007 | −1.53 |
| 8013450 | NM_001042685//LGALS9B//lectin, galactoside-binding, soluble, 9B//17p11.2 | 8.6597e-007 | −1.71 |
| 8177222 | NM_013230//CD24//CD24 molecule//6q21 | 9.3981e-007 | −2.20 |
| 8102311 | NM_001226//CASP6//caspase 6, apoptosis-related cysteine peptidase//4q25 | 1.1825e-006 | −1.58 |
| 8091103 | NM_006286//TFDP2//transcription factor Dp-2 (E2F dimerization partner 2) | 1.2042e-006 | −1.61 |
| 7930882 | NM_207009//FAM45A//family with sequence similarity 45, member A//10q25 | 1.2641e-006 | −1.51 |
| 8127109 | NM_016513//ICK//intestinal cell (MAK-like) kinase//6p12.1 | 3.0653e-006 | −1.67 |
| 8116051 | NM_080881//DBN1//drebrin 1//5q35.3 | 3.4139e-006 | −1.71 |
| 8005809 | NM_009587//LGALS9//lectin, galactoside-binding, soluble, 9//17q11.2//396 | 3.8368e-006 | −1.86 |
| 8135915 | NM_013332//C7orf68//chromosome 7 open reading frame 68//7q32.1 | 4.6540e-006 | −2.24 |
| 8150186 | NM_024787//RNF122//ring finger protein 122//8p12 | 6.2295e-006 | −1.85 |
| 8075263 | NM_003634//NIPSNAP1//nipsnap homolog 1 (C. elegans)//22q12.2 | 6.4654e-006 | −1.56 |
| 8074780 | NM_013313//YPEL1//yippee-like 1 (Drosophila)//22q11.2 | 2.6161e-005 | −1.99 |
| 7930181 | NM_020682//AS3MT//arsenic (+3 oxidation state) methyltransferase//10q24.32 | 0.0001 | −1.56 |
SOX11 Expression Correlates to DBN1, SETMAR and HIG2 Expression in Two Independent MCL Cohorts
To test the generality of our findings we investigated how the expression of the 26 downregulated genes correlated to SOX11 expression in 16 primary MCL, all positive for cyclin D1 and SOX11 [4]. In addition to SOX11, nine of the 26 genes were represented on the U133A chip used in this analysis. Of these, HIG2, SETMAR and DBN1 were significantly correlated to SOX11 expression values (Table 2). During the course of our study, Fernandez et al. provided evidence for lack of SOX11 expression in leukemic MCL with indolent clinical course [19]. We therefore investigated possible correlation with SOX11 among these MCL cases (GSE 16455). 19 of our 26 genes defined by SOX11siRNA were represented in this data set. The expression of 15 genes of these genes was significantly correlated to SOX11 expression, including DBN1, SETMAR and HIG2 (Table 3).
Table 2. Validation of SOX11 siRNA downregulated genes in a dataset of 16 primary MCLa).
| Probe set | Gene symbol | Spearman ratio# | p-value# |
|---|---|---|---|
| 204915_s_at *** | SOX11 | 1.00 | 0.00 |
| 206554_x_at * | SETMAR | 0.56 | 0.02 |
| 218507_at * | HIG2 | 0.57 | 0.02 |
| 202806_at * | DBN1 | 0.52 | 0.03 |
| 213996_at | YPEL1 | 0.44 | 0.08 |
| 219897_at | RNF122 | −0.37 | 0.16 |
| 208650_s_at | CD24 | −0.34 | 0.20 |
| 208651_x_at | CD24 | 0.13 | 0.63 |
| 209771_x_at | CD24 | −0.12 | 0.66 |
| 209772_s_at | CD24 | 0.03 | 0.91 |
| 216379_x_at | CD24 | −0.07 | 0.78 |
| 203236_s_at | LGALS9 | 0.19 | 0.48 |
| 201709_s_at | NIPSNAP1 | −0.16 | 0.55 |
| 201708_s_at | NIPSNAP1 | 0.11 | 0.70 |
| 204569_at | ICK | −0.11 | 0.68 |
Table 3. Validation of SOX11 siRNA downregulated genes in a series of SOX11+ and SOX11- MCLa).
| probeset | gene symbol | Spearman ratio # | p value# |
|---|---|---|---|
| 204915_s_at*** | SOX11 | 1 | 0 |
| 204914_s_at*** | SOX11 | 0.95 | 1.35E-11 |
| 204913_s_at*** | SOX11 | 0.93 | 4.46E-10 |
| 201310_s_at ** | C5orf13 | 0.84 | 1.33E-06 |
| 201309_x_at ** | C5orf13 | 0.78 | 1.97E-05 |
| 230424_at ** | C5orf13 | 0.78 | 2.16E-05 |
| 238411_x_at | C5orf13 | 0.29 | 0.18 |
| 203744_at ** | HMGB3 | 0.82 | 3.10E-06 |
| 225601_at ** | HMGB3 | 0.79 | 1.23E-05 |
| 202806_at ** | DBN1 | 0.81 | 4.12E-06 |
| 217025_s_at * | DBN1 | 0.67 | 6.06E-04 |
| 214051_at ** | MGC39900 | 0.73 | 1.15E-04 |
| 206554_x_at * | SETMAR | 0.71 | 1.95E-04 |
| 1554059_at* | SETMAR | 0.45 | 0.03 |
| 1554060_s_at* | SETMAR | 0.43 | 0.05 |
| 209790_s_at * | CASP6 | 0.70 | 2.55E-04 |
| 211464_x_at* | CASP6 | 0.46 | 0.03 |
| 209118_s_at * | TUBA1A | 0.69 | 3.63E-04 |
| 226157_at* | TFDP2 | 0.62 | 0.00 |
| 203588_s_at* | TFDP2 | 0.62 | 0.00 |
| 203589_s_at* | TFDP2 | 0.51 | 0.01 |
| 217970_s_at * | CNOT6 | 0.50 | 0.02 |
| 222476_at | CNOT6 | 0.11 | 0.64 |
| 205347_s_at * | TMSB15A | 0.46 | 0.03 |
| 209772_s_at* | CD24 | 0.46 | 0.03 |
| 208650_s_at* | CD24 | 0.44 | 0.04 |
| 208651_x_at | CD24 | 0.39 | 0.07 |
| 216379_x_at | CD24 | 0.36 | 0.10 |
| 209771_x_at | CD24 | 0.36 | 0.10 |
| 227310_at* | ADSS | 0.46 | 0.03 |
| 221761_at | ADSS | -0.29 | 0.19 |
| 218507_at * | HIG2 | 0.44 | 0.04 |
| 1554452_a_at * | HIG2 | 0.44 | 0.04 |
| 213135_at * | TIAM1 | 0.43 | 0.04 |
| 206409_at | TIAM1 | 0.17 | 0.44 |
| 225351_at * | FAM45A | 0.44 | 0.04 |
| 1563919_a_at | FAM45A | 0.17 | 0.46 |
| 1563920_at | FAM45A | -0.06 | 0.79 |
| 204569_at | ICK | 0.40 | 0.06 |
| 1552837_at | ICK | 0.18 | 0.43 |
| 212364_at | MYO1B | -0.30 | 0.17 |
| 212365_at | MYO1B | -0.12 | 0.58 |
| 203236_s_at | LGALS9 | 0.11 | 0.64 |
| 219676_at | ZSCAN16 | 0.22 | 0.34 |
Expression of SETMAR and DBN1 are Influenced by SOX11 in vitro and SOX11 Expression Correlated to DBN1 in Primary MCL Cells
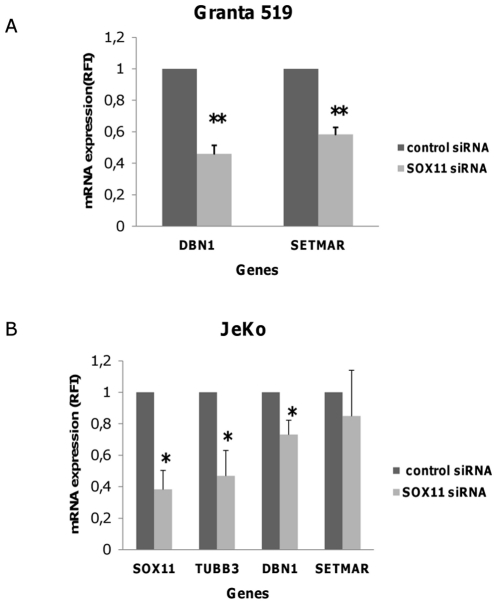
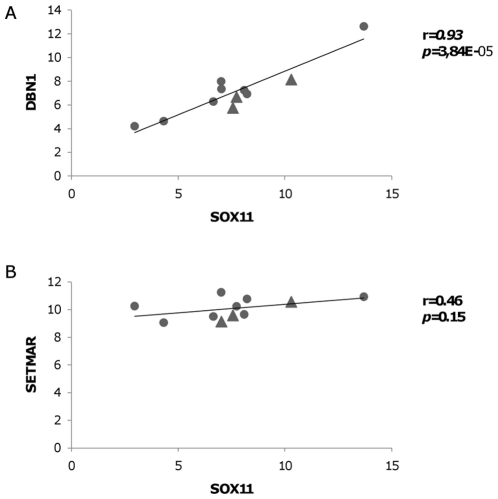
To validate the results of the gene expression analysis, SETMAR and DBN1 were analysed by quantitative RT-PCR in Granta 519 after SOX11 siRNA treatment. Both SETMAR and DBN1 mRNA levels were significantly (p<0.001) reduced by SOX11 downregulation in Granta 519 cells (Figure 3A). In another MCL cell line, JeKo, SOX11 downregulation caused similar reduction of TUBB3 and DBN1 levels but not of SETMAR (Figure 3B). We also found a significant correlation between the expression of SOX11 and DBN1 in primary MCL cells (8 samples from 6 patients as described in Materials and Methods and Table 4) and in the MCL cell lines Granta 519, Rec1 and JeKo (Figure 4).
Figure 3. Reduced expression of SETMAR and DBN1 in response to SOX11siRNA.
Analysis by qRT-PCR confirms downregulation of DBN1 and SETMAR (A) in SOX11 siRNA treated Granta 519 cells. (B) Expression of SOX11, TUBB3, SETMAR and DBN1 in the JeKo cell line was evaluated after transfection with either 100 pmol SOX11 siRNA or control siRNA. mRNA levels were measured by quantitative RT-PCR 20 hours after electroporation. Error bars show standard deviation of 4 independent experiments. * P<0.05, **P<0.01.
Table 4. Isolated primary MCL cells from 6 patientsa).
| Sample number | Patient age | sex | tissue | tumor cell content# |
|---|---|---|---|---|
| A1 | 82 | M | blood | 86% |
| A2 | 82 | M | pleural exudate | 95% |
| B1 | 77 | F | blood | 89% |
| B2 | 77 | F | bone marrow | 92% |
| C | 69 | M | spleen | 80% |
| D | 73 | M | spleen | 88% |
| E | 77 | M | lymph node | 85% |
| F | 66 | M | blood | 69% |
Figure 4. Expression of DBN1 correlates to SOX11 expression in primary MCL cells and MCL cell lines.
The expression values of SOX11, DBN1 or SETMAR were analyzed by quantitative RT-PCR. The threshold cycle (Ct) of the gene against the Ct of β actin was determined by subtraction. All values were first tested for normal distribution, and then the Pearson correlation coefficients and P value were calculated. ▴ MCL cell lines Granta 519, Rec1 and JeKo; • primary MCL patients.
DBN1, SETMAR, and HIG2 are Direct Targets of SOX11 Protein
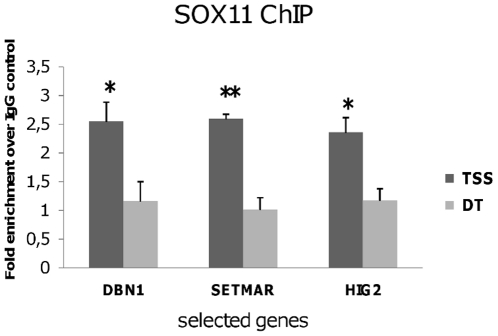
To investigate whether DBN1, SETMAR and HIG2 are direct targets of SOX11 protein, we performed chromatin immunoprecipitation (ChIP) assays on Granta 519 cells using two pairs of primers for each gene as described in Material and Methods. Granta 519 cells were cross-linked, and protein-DNA complexes were immunoprecipitated using antibodies recognizing normal rabbit IgG or anti-SOX11 antibody. We found that SOX11 was significantly recruited to DBN1, SETMAR and HIG2 promoter regions close to the transcription start site (TSS), but not to regions 2 kb distal to the TSS (DT) (Figure 5).
Figure 5. SOX11 protein directly targets DBN1, SETMAR and HIG2.
ChIP assay of SOX11 protein binding to the selected three genes. Immunoprecipitated DNA was analyzed by quantitative RT-PCR for two different primers as described in material and method. TSS: primer designed close to transcription start site (TSS) and DT: primer distal (∼2 kb) to the TSS. Columns represent the mean of fold enrichment of DBN1, SETMAR or HIG2 relative to IgG control. Error bars show standard deviation of 3 independent experiments.* P<0.05, ** P<0.01 compared to the IgG control.
Discussion
The main finding of this study is the identification of 26 genes influenced by SOX11 in MCL. SOX11 has critical roles in embryonic neurogenesis and tissue remodeling [5], [9], [23], [24]. It is also required for neuron survival and neurite growth [25] but no defined role in hematopoiesis has yet been shown.
The SOX11 protein belongs to the HMG box super family of DNA-binding proteins which are highly conserved between animals. They are involved in the regulation of such diverse development processes as specification of early embryonic germ layers [26], [27], organ development and neurogenesis [28]. SOX11, together with two other SOX family members SOX4 and SOX12, belong to the same subgroup, SOXC. SOX4 is so far the only gene in this family that has been found to have an important role in lymphopoiesis. SOX4–null hematopoietic cells grafted into wild type mice remain blocked at the pro-B cell stage [12]. Although SOX11 is highly homologous to SOX4, its role in lymphopoiesis remains unclear.
Recently, SOX11 was found to be aberrantly expressed in MCL [1], [2], [3], [4], [18]. Importantly, approximately 10% of cyclin D1 positive MCL lack nuclear expression of SOX11 [2], [4], [19]. SOX11 is also expressed in subsets of Burkitt lymphomas, lymphoblastic leukemias and hairy cell leukemias [1], [3], [18]. Thus, expression of SOX11 in lymphoma seems independent of the t(11;14) cyclin D1 translocation.
The possible role of the aberrant SOX11 expression in lymphoma is unknown. Others have shown that SOX11 is needed for the expression of pan-neuronal genes, including the Class-Ш β-tubulin gene TUBB3 (also named Tuj1). TUBB3 was the first gene to be identified as a potential direct target of SOXC proteins. In this study, we showed that SOX11 can modify the expression of TUBB3 also in MCL.
By gene expression analysis using a very stringent threshold, we identified 26 genes, all significantly downregulated, after SOX11 knockdown in MCL cells. Three of these genes were also found to be significantly correlated to SOX11 expression levels in 16 SOX11 positive primary MCL samples from various tissues. During the course of our study, Fernandez et al. published results on MCL with leukemic, non-nodal presentation and a very indolent clinical course [19]. Many of these tumors had in fact been misdiagnosed as other types of lymphomas. 15 of the 26 SOX11siRNA downregulated genes in Granta 519 were found to be significantly correlated to SOX11 expression in the Fernandez cohort, confirming our observations.
Prior to this study, Tubb3 and Tead2 were the only genes proven to be directly regulated by Sox11 [8], [9]. By chromatin immunoprecipitation we showed direct binding of the SOX11 protein to the promoter regions of the DBN1, SETMAR and HIG2 genes in MCL cells.
DBN1 and three other genes downregulated by SOX11 silencing, TMSB15A, TMSB15B and C5orf13 are actin binding and involved in cell shape and motility. DBN1 encodes for the actin binding protein drebrin 1 (_d_evelopmentally _re_gulated br_ain protei_n), cloned in 1988 [29], [30]. DBN1 was later shown to be is involved in neurite formation and synaptic signalling (reviewed in [31]) and to be downregulated in brains of Alzheimer patients [32], [33]. Drebrin is also expressed in epithelial cells of the stomach, kidney, colon and in cell lines from fibroblasts and astrocytoma [34]. Interestingly, drebrin is cleaved by caspase-6 [35], encoded by another of the SOX11siRNA downregulated genes.TMSB15A and TMSB15B encode for different isoforms of the actin binding protein thymosin beta 15. Thymosin beta 15 promotes cell motility and was discovered as highly upregulated in human breast, prostate and colon cancer [36], [37], [38], [39]. According to NCBI, C5orf13 is coding for the protein P311, highly expressed in glioma and involved in glioma cell migration through the reorganization of the actin cytoskeleton [40]. Thus a number of genes involved in cell shape and motility were downregulated by SOX11 siRNA treatment of Granta 519 cells. We could, however, not detect any significant change in shape or growth pattern of the cells after siRNA treatment (data not shown).
SETMAR, HIG2 and HMGB3 could potentially influence cell division and response to cytostatic treatment. SETMAR, also called METNASE, is cooperating with topoisomerase II alpha in decoiling of chromosomes during mitosis. Recent evidence in acute myeloid leukemia suggests that SETMAR may confer resistance to the topoisomerase II alpha inhibitor VP-16 [41], [42]. The HMGB3 protein belongs to the high mobility group of proteins and may, as shown for other HMG proteins, play a role in DNA replication and transcription. Hmgb3 is highly expressed in hematopoietic stem cells [43]. Recent experiment in mice indicate that Hmgb3 downregulation is associated with increased Wnt-signalling, more rapid renewal of stem cells and fewer lymphoid and myeloid progenitor cells [44]. We therefore investigated a possible influence on cell proliferation or cell survival in vitro after SOX11 knock-down by siRNA. In these short term experiments, we did however not detect any significant changes in cell proliferation or viability after SOX11 downregulation (manuscript in preparation), in contrast to what has been reported for neural cells [25]. However, since the effect on gene expression in siRNA treated cells is transient, we cannot exclude possible effects of SOX11 expression on cell survival or proliferation in other experimental settings.
SOX11 is aberrantly expressed in MCL but the molecular mechanism(s) responsible for its upregulation in lymphoid and hematopoietic cells are not yet defined, in contrast to other cell types. In chicken neural cells overexpression of Ngn2 and Ascl1 upregulates Sox11 while the transcriptional repressors Id1 and REST/NRSF downregulates its expression [11]. In the mouse embryonal neuronal plate Sox11 was upregulated by FoxD5 and Notch signalling [45], [46]. SOX11 mRNA expression was also rapidly (within 3 hours) upregulated after exposure of retinal microglia to 17beta-estradiol [47]. Even though SOX11 is highly expressed in most MCL, expression of SOX11 seems not to be dependent on the t(11;14) since it is a feature of both cyclin D1 positive and cyclin D1 negative cases [3]. Furthermore, SOX11 may be expressed in other aggressive B cell lymphomas that lack cyclin D1.
In summary, we have used selective siRNA targeting and downregulation of SOX11 to identify a set of SOX11 responsive genes. Our results have been validated in primary MCL tumors and freshly isolated lymphoma cells. Three of the identified genes, DBN1, SETMAR and HIG2, were found to be directly targeted by the SOX11 protein in MCL.
Materials and Methods
Ethical Permission
This study was approved by the Ethical Committe at the Karolinska Institutet, and performed according to the principles expressed in the Declaration of Helsinki. All the patients gave their written informed consent to participate in the study.
Patient Samples
Eight samples of tumor cells were collected from 6 patients diagnosed with cyclin D1+ MCL at the Department of Pathology (Karolinska University Hospital Huddinge, Sweden) (Table 4). Tumor cells were isolated from different tissues (bone marrow, blood, lymph node, spleen, pleural exudate), viability frozen in RPMI 1640 medium containing 10% DMSO and 40% FBS and stored at -135°C.
Cell Lines
Granta 519, Rec1, JeKo, JVM2 cell lines and primary cells were obtained and cultured as previously described [48]. In short, cell lines were maintained in culture medium (RPMI 1640, supplemented with 10% fetal bovine serum (FBS) and 50 µg/ml gentamicin (all from Invitrogen, Carlsbad, CA)) under standard conditions (humidified atmosphere, 95% air, 5% CO2, 37°C). The cells were maintained between 3×105 and 1.5×106/ml. Culture medium was changed twice weekly.
siRNA and Transient Transfection
Two silence ® select pre-designed SOX11 siRNAs, S13312 and S224667, and one non-targeting silence® select Negative Control #1 siRNA were obtained from Ambion (Ambion, Austin, TX). siRNA S13312, used in most experiments had the sense targeting sequence (5′-3′): GACCUGAUGUUCGACCUGAtt. Results were confirmed using S224667 with the sense targeting sequence (5′-3′): GGAGCUGAGCGAGAUGAUCtt. Freeze-dried siRNAs were suspended in nuclease-free water (Ambion, Applied Biosystem, Austin, TX) in a final concentration of 50 µM. Transient transfection were performed on an Amaxa Nucleofection Device (Lonza Cologne AG, Cologne, Germany) according to the manufacturer's instruction. In brief, Granta 519 cells were split at a density of 5×105/ml in the medium 48 hours before transfection. Thereafter, 4×106 cells were collected and resuspended in 100 µl human cell line nucleofector solution C with 100 pmol of either SOX11 siRNA or control siRNA using the X-01 electroporation program. The two SOX11 siRNAs were tested for their individual ability to knock down SOX11 expression. Both siRNA sequences were able to knock down SOX11 expression by 80%-85% in Granta 519 cells.
RNA Isolation and Affymetrix GeneChip Human Gene 1.0 ST Array
Cells were harvested 20 hours after siRNA transfection. Total RNA from individual transfection experiments in Granta 519 cells (n = 4) or patient samples (n = 8) was extracted with Qiagen RNeasy Plus Mini kit according to the instructions of the manufacturer (Qiagen, Valencia, CA). RNA quantity was measured using a NanoDrop (NanoDrop Technologies, Wilmington, DE) set for RNA measurement (A260/A280 ratio). RNA quality was assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA) and samples with high-quality RNA were hybridized to Affymetrix GeneChip Human Gene 1.0 ST arrays (Affymetrix Inc. Santa Clara, CA) in accordance with the Affymetrix standard protocol at the Bioinformatics and Expression Analysis Core facility (BEA, Department of Biosciences and Nutrition, Karolinska Institutet).
Gene Expression Data Analysis
The microarray image data were processed in the BEA core facility according to standard procedures. CEL data was analyzed with Partek Genomic Suite vs 6.5 (Partek Inc., St. Louis, MO) which can normalize and process multiple datasets simultaneously. Data normalization was performed by Robust Multiarray Analysis algorithm (RMA). Statistical differences were examined using one way ANOVA. Significant probesets were defined with a p-value cutoff significant with a False Discovery Rate (FDR) of <0.002, and a fold-change (FC) equal or greater than 1.5(FC≥1.5) for upregulated genes and equal or less than −1.5(FC≤1.5) for downregulated genes, respectively.
Gene expression values for the selected genes were retrieved from two primary array databases. Our previously published gene expression analysis included 16 MCL tumor samples, all expressing cyclin D1 and SOX11 [49]. We also retrieved data from a recently published study from [19] which included 7 indolent, SOX11 negative MCL and 15 conventional, SOX11 positive MCL (http://www.ncbi.nlm.nih.gov/geo/with the GSE accession number GSE16455).
cDNA Synthesis and Real-time Quantitative RT-PCR
cDNA was generated according to the protocol for Omniscript Reverse Transcription (Qiagen, GmbH, Hilden, Germany). 2 µg of RNA was used in conjunction with Oligo(dT) primer and 20 µl of cDNA was generated. 1 µl of cDNA was added to qPCR Kit Platinum SYBR Green qPCR SuperMix-UGD with FITC (Invitrogen) according to the manufacturer's instructions, run in triplicate on a CFX96 Real-time System (C1000 Thermal Cycler, BIO-RAD, city, CA). The following primers were used: SOX11, 5′-CATGTAGACTAATGCAGCCATTGG-3′ and 5′-CACGGAGCACGTGTCAATTG-3′; SOX4, 5′- CAGCCCCTAATTTCTCCATGTT and 5′- GGTGGCAGGTTAAGGGATACTG; SOX12, 5′- CCCAGGTCCACCCTCAGTAC and 5′- CGAGAGTCTTCCTGCCATCAC; SETMAR, 5′-GCGGAAGCGGCAAAGAC-3′ and 5′-GCCTCAGGCTTCTCCTTAAACTC-3′; DBN1, 5′-GCCCCACCTGCTAACCAA-3′ and 5′-GTGATTGACTGAAGTACCCCTCACT-3′; TUBB3,5′- GGAGCGGATCAGCGTCTACT-3′ and 5′-GCTCGAGGCACGTACTTGTG-3′; β-actin, 5′-AAAGACCTGTACGCCAACACA-3′ and 5′-AGTACTTGCGCTCAGGAGGA-3′. The cycle parameters used was: 50°C 2 min, 95°C 2 min, followed by 40 cycles of 95°C for 15 seconds and 59°C for 30 seconds. Ct values (Threshold cycles) were obtained from amplification of SOX11, SOX4, SOX12, SETMAR, DBN1, TUBB3 and β-actin. The ΔCt value was calculated by subtracting the Ct value of β-actin from the Ct value of target gene. A ΔΔCt value was then calculated by subtracting the ΔCt value for siRNA condition from the ΔCt value for control condition with relative fold increase (RFI) reported as 2−ΔΔCt.
Western blotting
Western blotting was performed as described previously [4]. The blotted membranes were probed with rabbit anti-human SOX11 (HPA000536, Atlas Antibodies, Uppsala, Sweden), and antibodies to tubulin (SC 8035, Santa Cruz Biotechnology, Santa Cruz, CA) or actin (A4700, Sigma, Saint Louis, MO) was used as loading controls. Antibody binding was detected by enhanced chemoluminescence using SupersignalWest Pico (Pierce Biotechnology, Shelton) CTchemiluminescent substrate.
Chromatin immunopreciption (ChIP)
Granta 519 cells were split at a density of 5×105/ml in the medium and cultured for 72 hours. Thereafter, 2×107 cells were collected and crosslinked with 1% formaldehyde for 10 min. Cross-linking was quenched by adding 125 mM glycine and cells were washed with cold PBS, harvested and resuspended in lysis buffer containing protease inhibitors cocktail (Roche, Mannheim, Germany) and sonicated 15 min for three times. The soluble chromatin was collected by centrifugation and the supernatants were incubated with 30 µl protein A/G Sepharose (50% slurry; GE Healthcare Bio-Sciences Corp., Uppsala, Sweden) under gentle agitation over night at 4°C. The supernatant was transferred to a new microcentrifuge tube, followed by immunoprecipitation with 1 µg of anti-SOX11 antibody and non-immune rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) as control at 4 C overnight. Protein A/G Sepharose (20 µl of a 50% slurry) was then added and incubated for 1.5–2 h. The pellets were successively washed with different buffer as previously described [50]. Protein-DNA crosslinks were reversed by overnight incubation at 65°C in 120 µl elution buffer [TE; 1% SDS]. DNA was purified using a PCR purification kit (QIAGEN, Valencia, CA) and eluted in 50 µl of elution buffer. The immunoprecipitated DNA was amplified by real-time PCR using Fast SYBR Green Master Mix (Applied Biosystems, Warrington,UK) Two pairs of primers were designed for DBN1, SETMAR and HIG2 with one primer designed close to transcription start site (TSS) and one distal (∼2 kb) to the TSS (DT).
The primer pairs used are as follows:
DBN1 TSS forward: 5′-TGAGGTGGAAGGATGTTTGCT-3′; Reverse: 5′-CGGCGGTAAGGGAGTCACT-3′.
DBN1 DT forward: 5′-CCCTGCCGTGGGAGTCT-3′; Reverse: 5′-TCCCAGAGGAGTCCCAAGTAGA-3′.
SETMAR TSS forward: 5′-GGGAGCCAGACCCAAAAAGT-3′; Reverse: 5′-TTCTCAGGAGTGGCCTGGAA-3′.
SETMAR DT forward: 5′-AGAAAGATACAGAGAAGGGACTACTTGAG-3′; Reverse: 5′-TTCTTGTATCTCCAGTGCCTTTACC-3′.
HIG2 TSS forward: 5′-TCACTCCAGAACACAATGACTCAA-3′; Reverse: 5′-CGCGGAGCTGTTTCCAAA-3′.
HIG2 DT forward: 5′-TTTCCAACTGGCATGACCTTT-3′; Reverse: 5′-GCACCTCTCCAACAATTCTTTCTC-3′.
Statistical Analysis
Quantitative RT-PCR data were statistically analyzed using paired t -test with P values <0.05 as statistical significance. The correlation between SOX11 and other genes as well as the expression differences among MCL were analyzed with the Spearman's rank correlation coefficient test or Pearson correlation coefficient for qRT-PCR results using Origin 8 software (OriginLab, MA USA) using Origin 8 software. 2-tailed test for significance was used.
Acknowledgments
We are grateful to the personnel at the Flow Cytometry Unit at Karolinska University Hospital Huddinge for the help with collection of patients' samples and flow cytometry. We also thank Jianguang Ji at Lund University for statistical advice and David Brodin at BEA core facility for help with gene expression analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a grant from the Swedish Cancer Society, the Swedish Research Council, the Cancer Society in Stockholm, the Karolinska Institutet Funds, and the Stockholm Country Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chen YH, Gao J, Fan G, Peterson LC. Nuclear expression of sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol. 2010;23:105–112. doi: 10.1038/modpathol.2009.140. [DOI] [PubMed] [Google Scholar]
- 2.Ek S, Dictor M, Jerkeman M, Jirstrom K, Borrebaeck CA. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood. 2008;111:800–805. doi: 10.1182/blood-2007-06-093401. [DOI] [PubMed] [Google Scholar]
- 3.Mozos A, Royo C, Hartmann E, De Jong D, Baro C, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94:1555–1562. doi: 10.3324/haematol.2009.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Asplund AC, Porwit A, Flygare J, Smith CI, et al. The subcellular Sox11 distribution pattern identifies subsets of mantle cell lymphoma: correlation to overall survival. Br J Haematol. 2008;143:248–252. doi: 10.1111/j.1365-2141.2008.07329.x. [DOI] [PubMed] [Google Scholar]
- 5.Jay P, Goze C, Marsollier C, Taviaux S, Hardelin JP, et al. The human SOX11 gene: cloning, chromosomal assignment and tissue expression. Genomics. 1995;29:541–545. doi: 10.1006/geno.1995.9970. [DOI] [PubMed] [Google Scholar]
- 6.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, et al. The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haslinger A, Schwarz TJ, Covic M, Chichung Lie D. Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur J Neurosci. 2009;29:2103–2114. doi: 10.1111/j.1460-9568.2009.06768.x. [DOI] [PubMed] [Google Scholar]
- 11.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 13.Brennan DJ, Ek S, Doyle E, Drew T, Foley M, et al. The transcription factor Sox11 is a prognostic factor for improved recurrence-free survival in epithelial ovarian cancer. Eur J Cancer. 2009;45:1510–1517. doi: 10.1016/j.ejca.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Weigle B, Ebner R, Temme A, Schwind S, Schmitz M, et al. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol Rep. 2005;13:139–144. [PubMed] [Google Scholar]
- 15.Hide T, Takezaki T, Nakatani Y, Nakamura H, Kuratsu J, et al. Sox11 prevents tumorigenesis of glioma-initiating cells by inducing neuronal differentiation. Cancer Res. 2009;69:7953–7959. doi: 10.1158/0008-5472.CAN-09-2006. [DOI] [PubMed] [Google Scholar]
- 16.de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10:648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57:201–214. doi: 10.1023/a:1015773818302. [DOI] [PubMed] [Google Scholar]
- 18.Dictor M, Ek S, Sundberg M, Warenholt J, Gyorgy C, et al. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt's lymphoma. Haematologica. 2009;94:1563–1568. doi: 10.3324/haematol.2009.008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez V, Salamero O, Espinet B, Sole F, Royo C, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70:1408–1418. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- 20.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–762. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 21.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, et al. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. Embo J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. Embo J. 1994;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hargrave M, Wright E, Kun J, Emery J, Cooper L, et al. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev Dyn. 1997;210:79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, et al. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankowski MP, Cornuet PK, McIlwrath S, Koerber HR, Albers KM. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143:501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Klymkowsky MW. The Sox axis, Nodal signaling, and germ layer specification. Differentiation. 2007;75:536–545. doi: 10.1111/j.1432-0436.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- 27.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, et al. Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 29.Kojima N, Kato Y, Shirao T, Obata K. Nucleotide sequences of two embryonic drebrins, developmentally regulated brain proteins, and developmental change in their mRNAs. Brain Res. 1988;464:207–215. doi: 10.1016/0169-328x(88)90027-7. [DOI] [PubMed] [Google Scholar]
- 30.Shirao T, Kojima N, Kato Y, Obata K. Molecular cloning of a cDNA for the developmentally regulated brain protein, drebrin. Brain Res. 1988;464:71–74. doi: 10.1016/0169-328x(88)90020-4. [DOI] [PubMed] [Google Scholar]
- 31.Dun XP, Chilton JK. Control of cell shape and plasticity during development and disease by the actin-binding protein Drebrin. Histol Histopathol. 25:533–540. doi: 10.14670/HH-25.533. [DOI] [PubMed] [Google Scholar]
- 32.Harigaya Y, Shoji M, Shirao T, Hirai S. Disappearance of actin-binding protein, drebrin, from hippocampal synapses in Alzheimer's disease. J Neurosci Res. 1996;43:87–92. doi: 10.1002/jnr.490430111. [DOI] [PubMed] [Google Scholar]
- 33.Hatanpaa K, Isaacs KR, Shirao T, Brady DR, Rapoport SI. Loss of proteins regulating synaptic plasticity in normal aging of the human brain and in Alzheimer disease. J Neuropathol Exp Neurol. 1999;58:637–643. doi: 10.1097/00005072-199906000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Keon BH, Jedrzejewski PT, Paul DL, Goodenough DA. Isoform specific expression of the neuronal F-actin binding protein, drebrin, in specialized cells of stomach and kidney epithelia. J Cell Sci. 2000;113(Pt 2):325–336. doi: 10.1242/jcs.113.2.325. [DOI] [PubMed] [Google Scholar]
- 35.Klaiman G, Petzke TL, Hammond J, Leblanc AC. Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteomics. 2008;7:1541–1555. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao L, Zetter BR. Molecular cloning and structural characterization of the rat thymosin beta15 gene. Gene. 2000;260:37–44. doi: 10.1016/s0378-1119(00)00452-2. [DOI] [PubMed] [Google Scholar]
- 37.Bao L, Loda M, Zetter BR. Thymosin beta15 expression in tumor cell lines with varying metastatic potential. Clin Exp Metastasis. 1998;16:227–233. doi: 10.1023/a:1006540824969. [DOI] [PubMed] [Google Scholar]
- 38.Gold JS, Bao L, Ghoussoub RA, Zetter BR, Rimm DL. Localization and quantitation of expression of the cell motility-related protein thymosin beta15 in human breast tissue. Mod Pathol. 1997;10:1106–1112. [PubMed] [Google Scholar]
- 39.Bao L, Loda M, Janmey PA, Stewart R, Anand-Apte B, et al. Thymosin beta 15: a novel regulator of tumor cell motility upregulated in metastatic prostate cancer. Nat Med. 1996;2:1322–1328. doi: 10.1038/nm1296-1322. [DOI] [PubMed] [Google Scholar]
- 40.McDonough WS, Tran NL, Berens ME. Regulation of glioma cell migration by serine-phosphorylated P311. Neoplasia. 2005;7:862–872. doi: 10.1593/neo.05190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wray J, Williamson EA, Royce M, Shaheen M, Beck BD, et al. Metnase mediates resistance to topoisomerase II inhibitors in breast cancer cells. PLoS One. 2009;4:e5323. doi: 10.1371/journal.pone.0005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wray J, Williamson EA, Sheema S, Lee SH, Libby E, et al. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–1858. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemeth MJ, Curtis DJ, Kirby MR, Garrett-Beal LJ, Seidel NE, et al. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102:1298–1306. doi: 10.1182/blood-2002-11-3541. [DOI] [PubMed] [Google Scholar]
- 44.Nemeth MJ, Kirby MR, Bodine DM. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci U S A. 2006;103:13783–13788. doi: 10.1073/pnas.0604006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan B, Neilson KM, Moody SA. Notch signaling downstream of foxD5 promotes neural ectodermal transcription factors that inhibit neural differentiation. Dev Dyn. 2009;238:1358–1365. doi: 10.1002/dvdy.21885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan B, Neilson KM, Moody SA. foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev Biol. 2009;329:80–95. doi: 10.1016/j.ydbio.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C, Tang Y, Li F, Turner S, Li K, et al. 17beta-estradiol (betaE2) protects human retinal Muller cell against oxidative stress in vitro: evaluation of its effects on gene expression by cDNA microarray. Glia. 2006;53:392–400. doi: 10.1002/glia.20291. [DOI] [PubMed] [Google Scholar]
- 48.Gustafsson K, Wang X, Severa D, Eriksson M, Kimby E, et al. Expression of cannabinoid receptors type 1 and type 2 in non-Hodgkin lymphoma: growth inhibition by receptor activation. Int J Cancer. 2008;123:1025–1033. doi: 10.1002/ijc.23584. [DOI] [PubMed] [Google Scholar]
- 49.Sander B, Flygare J, Porwit-Macdonald A, Smith CI, Emanuelsson E, et al. Mantle cell lymphomas with low levels of cyclin D1 long mRNA transcripts are highly proliferative and can be discriminated by elevated cyclin A2 and cyclin B1. Int J Cancer. 2005;117:418–430. doi: 10.1002/ijc.21166. [DOI] [PubMed] [Google Scholar]
- 50.Matthews J, Wihlen B, Tujague M, Wan J, Strom A, et al. Estrogen receptor (ER) beta modulates ERalpha-mediated transcriptional activation by altering the recruitment of c-Fos and c-Jun to estrogen-responsive promoters. Mol Endocrinol. 2006;20:534–543. doi: 10.1210/me.2005-0140. [DOI] [PubMed] [Google Scholar]