Hox cluster duplications and the opportunity for evolutionary novelties (original) (raw)
Abstract
Hox genes play a key role in animal body plan development. These genes tend to occur in tightly linked clusters in the genome. Vertebrates and invertebrates differ in their Hox cluster number, with vertebrates having multiple clusters and invertebrates usually having only one. Recent evidence shows that vertebrate Hox clusters are structurally more constrained than invertebrate Hox clusters; they exclude transposable elements, do not undergo tandem duplications, and conserve their intergenic distances and gene order. These constraints are only relaxed after a cluster duplication. In contrast, invertebrate Hox clusters are structurally more plastic; tandem duplications are common, the linkage of Hox genes can change quickly, or they can lose their structural integrity completely. We propose that the constraints on vertebrate Hox cluster structure lead to an association between the retention of duplicated Hox clusters and adaptive radiations. After a duplication the constraints on Hox cluster structure are temporarily lifted, which opens a window of evolvability for the Hox clusters. If this window of evolvability coincides with an adaptive radiation, chances are that a modified Hox cluster becomes recruited in an evolutionary novelty and then both copies of duplicated Hox clusters are retained.
Since their discovery, Hox genes, a family of linked transcription-factor genes sharing a DNA-binding domain (the homeobox) (1), have confronted biologists with surprising riddles. The first so-called Hox paradox was the discovery that homologous genes “code” for fundamentally different body plans. It is now widely accepted that the divergent body plans are based more, but not exclusively, on differences in the regulation of a conserved set of genes rather than different gene complements (2–4). This commentary discusses a second Hox paradox: Why is it that in the evolution of chordates (vertebrates) the number of Hox gene clusters has increased several times (Fig. 1), often in association with major radiations (5, 6), whereas no evidence exists for such a trend in invertebrates (7)? It is hard to believe that this difference should be due to differences in the frequency of genome and chromosome duplications between vertebrates and invertebrates. In this commentary, we argue that vertebrate Hox clusters, in the absence of duplication, are structurally less evolvable than their invertebrate counterparts. The constraint on Hox cluster structure may be temporarily lifted after cluster duplication, which may make an association between Hox cluster duplications and adaptive radiations more likely in vertebrates than in invertebrates.
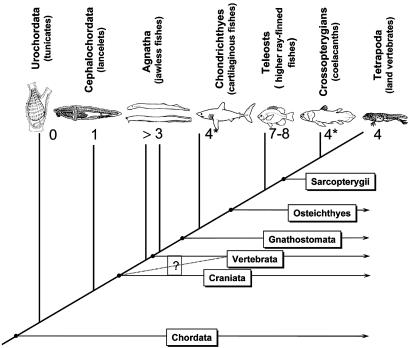
Fig. 1.
Number of Hox clusters in chordate phylogeny. The inferred numbers of Hox clusters are superimposed on the phylogeny. Asterisks indicate taxa under investigation whose Hox clusters have been isolated but not yet completely characterized. In the Agnatha, only lampreys have been investigated to date, and their Hox clusters appear to have originated by an independent duplication event(s); i.e., their Hox cluster duplicates are not orthologous with those in mammals (11–13). It should be noted that, although the increasing number of Hox clusters through phylogeny is consistent with Ohno's prediction (45) of genome duplications and vertebrate complexity, it is not entirely consistent with his proposed stepwise (2_R_) model.
Hox genes were first discovered through their effects on Drosophila development. Mutations cause dramatic transformations of the identity of specific body segments to those of different body segments, called homeotic transformations. Eventually these genes were characterized as coding for transcription factors from the family of homeobox-containing genes. Furthermore, it was found that the Hox genes tend to occur in tightly linked clusters that exhibit spatiotemporally coordinated expression along the anterior–posterior axis. In all bilaterian animals these Hox genes are responsible for patterning the main body axis (1). In addition, Hox genes have been recruited into secondary areas of expression, most notable the cranial neural crest in vertebrates, fins, and limbs and other organs. All invertebrate taxa extensively examined so far have only a single Hox cluster (reviewed in ref. 7). In sharp contrast, it was found that every major taxon of vertebrates has at least three if not up to eight such clusters (Fig. 1). First, it was shown that mammals have four Hox clusters, called A, B, C, and D (8), a condition which seems to be true for all tetrapods. The closest relative of vertebrates, Amphioxus, has a single Hox cluster with 14 genes, although this number might not be the ancestral condition for the vertebrate Hox gene number (9, 10). Recently, it was shown that the jawless vertebrate, Petromyzon marinus, the sea lamprey, has at least three clusters (11) (12). These clusters are, however, not orthologous to those in the mammals and have thus originated by an independent duplication event (13). From the horn shark, Heterodontus francisci, two Hox clusters have been described based on complete Hox cluster sequences (14), but more are likely to exist (unpublished data). Teleost fishes are the pinnacle of Hox cluster evolution, with at least seven Hox clusters in zebrafish (15) and Takifugu and Spheroides (16). Incomplete data from other teleosts are consistent with the hypothesis that all teleosts may have more Hox clusters than the mammals [killifish (17), tilapia (6), and striped bass (18)]. The expansion of Hox cluster number in higher ray-finned fishes is intriguing because it is associated with the teleost radiation, which gave rise to the largest taxon of extant vertebrates of about 24,000 species. It is currently not possible to draw a close association between these two events, cluster duplication and teleost radiation, however, because the Hox cluster situation among basal ray-finned fishes is not known. Currently, available data suggest that the cluster duplication happened before the most recent common ancestor of euteleosts and after the most recent common ancestor of the sturgeons and teleosts (K. Takahashi, J. Yoder, C.-h. Chiu, C.A., D. Nonaka, and G.P.W., unpublished work). Although the data for jawless vertebrates and cartilagenous fishes (sharks and relatives) are still incomplete, the most parsimonious scenario also associates the earlier Hox cluster duplications, leading to the four clusters found in humans, with major adaptive radiations (5, 19). One duplication might have occurred before the radiation of jawless vertebrates and one probably occurred before the radiation of the jawed vertebrates.
This pattern of Hox cluster number expansion in vertebrates is in striking contrast to the stasis of Hox cluster number in invertebrates. Yet, invertebrates experienced even more dramatic episodes of adaptive radiation and innovations of body design. More than 20 major clades of invertebrates differ so radically in body organization that they were formerly known as “phyla.” The largest metazoan radiation of all is that of insects, which gave rise to ≈1 Mio of described species with wildly different adaptations. If one includes crustaceans, spiders, and some minor taxa, any other phylogenetic event in animal phylogeny pales in comparison with the radiation of arthropods. Based on that evidence, Sean Carroll has argued persuasively that gene duplications play only a minor role in evolutionary innovations and adaptive radiations of invertebrates (20). We agree with that inference, but we maintain that we still must explain why, in vertebrate phylogeny, Hox cluster duplication and retention plays such a prominent role and what exactly this role is. Below we briefly review recent work on the structural and functional consequences of Hox cluster duplication, which lays the foundation for our proposal that Hox clusters in vertebrates are structurally less evolvable, in the absence of cluster duplications, than their invertebrate counterparts.
The most obvious evolutionary trend after Hox cluster duplication is gene loss, which can be differential among teleost lineages (6). In that respect, Hox genes are like all other genes, where the retention rate of duplicated genes is between 20% and 50% (21). But the retention rate is highly variable after different duplication events (Table 1). Another trend in the structural evolution of Hox clusters after duplication is the total size of the cluster. Invertebrate Hox clusters are huge, >1,000 kb (22). The closest relative of vertebrates, Amphioxus also has a comparatively large Hox cluster (≈450 kb; C.A., unpublished work). The Hox A cluster of shark and mammals is very similar and much smaller than the Amphioxus cluster, ≈100–120 kb. Most of the reduction is due to shortening of the intergenic regions, even in segments where neighboring genes are retained. The same phenomenon is seen in the zebrafish HoxA clusters, which are ≈58 kb for the HoxAa and 33 kb for the HoxAb cluster (23), and in Fugu (24) and Tilapia HoxA clusters (25). It has been argued that Hox cluster size is roughly correlated with genome size (25), but this correlation cannot fully account for the pattern. For instance, the shark and humans have different genome sizes, but their HoxA clusters are of the same size (23); and, within the teleosts, no correlation exists between genome size and cluster size (see figure 3 in ref. 25). The systematic size differences between vertebrates could reflect two strategies of evolving Hox gene regulation: (i) large intergenic distances might be involved with an elaboration of cis-regulatory elements of a few Hox genes. This strategy could be typical of invertebrates. (ii) Short intergenic distances could be indicative of simpler cis-regulatory elements but applied to a larger Hox gene number. This strategy could be what we see in vertebrates. The loss of noncoding sequence conservation after cluster duplication reviewed below is consistent with this model (see next paragraph).
Table 1. Retention rate of duplicated Hox genes in vertebrate evolution.
| Ancestral genes, n | Derived genes, n | Retention rate, % | |
|---|---|---|---|
| Two-cluster ancestor | 14 | 23 | 64 |
| Four-cluster ancestor | 23 | 42 | 83 |
| Mammals | 23 | 39 | 70 |
| Zebrafish | 42 | 47 | 12 |
| Takifugu | 42 | 45 | 7 |
Conservation of noncoding sequences is another feature strongly affected by Hox cluster duplication. A detailed comparison of intergenic sequences between the HoxA clusters of shark and human shows extensive regions of strong sequence conservation, which is largely absent in the zebrafish (23) and fugu (24). It can be expected that the cis-regulatory elements of genes get modified when two copies are retained to resolve genetic redundancy (26). Surprisingly, however, conservation is also lost when only one paralog is retained in the zebrafish, which by inference is expected to be necessary to maintain the ancestral gene function. Apparently, this function was important enough to conserve those very same sequences since the most recent common ancestor of sharks and humans, at least for the HoxA cluster. But functional studies of zebrafish Hox genes show that even the notion of retained ancestral functions is misleading in the Hox genes. Prince and collaborators have shown that the zebrafish ortholog of the mammalian Hoxa-1, Hoxa-1a, is not expressed in the hindbrain, but its function has been taken over by Hoxb-1b. In contrast, in the mouse the Hoxb-1 gene is not essential for hindbrain development (27). These findings suggest that, after duplication, functions can be divided among paralogs or shifted to other members of the gene family. Furthermore, orthologous genes among teleosts do not retain the same functions. For instance, the Hoxb-2b gene from zebrafish is expressed in the neural crest, as is the Hoxb-2 ortholog in mouse; however, this does not hold true for the Hoxb-2b ortholog in the striped bass (28). The evolution of the Hox proteins among teleost paralogs is even more confusing. In all cases, the rate of nonsynonymous substitutions in Hox genes is increased after duplication (29, 30), and evidence shows that both directional selection and increased mutation rate contribute to this rate increase (K. Takahashi, J. Yoder, C.-h. Chiu, C.A., D. Nonaka, and G.P.W., unpublished data). In many cases, however, the biochemical functions of the proteins are not affected (31). Extensive sequence divergence is also found among paralogous genes in the four-cluster animals despite functional conservation (32), but, on the other hand, considerable conservation occurs among orthologous genes between species.
The picture that emerges from this admittedly somewhat spotty evidence is that, immediately after a Hox cluster duplication in vertebrates, the Hox clusters undergo rapid and extensive evolutionary change. Loss of genes, loss of coding, and noncoding sequence conservation and function are rampant. Some time after a duplication event, however, things tend to settle down as apparently has been the case in the most recent common ancestor of gnathostomes. Since that time the molecular evolution of vertebrate Hox clusters seems to be more conserved than in many invertebrates. The high level of structural and noncoding sequence conservation observed in vertebrate Hox clusters corroborate this tenet (14, 23). Vertebrate Hox clusters exclude repeated elements, most likely because of the deleterious effects of insertions in these regions (14, 33). No such phenomenon is yet documented for invertebrate Hox clusters, which are large and can also add genes by tandem duplication within the cluster. For instance, the zen-related gene z2 in Drosophila melanogaster is absent in the closely related Drosophila. pseudoobscura (34). Another example is bicoid, which arose from a Hox-3 paralog and acquired a function in the establishment of the A–P axis within the dipteran insects (35). Within the genus Drosophila, the Hox cluster “broke apart,” i.e., tight linkage is only retained among two subgroups of genes, the Antp and the Ubx complexes, such that two Hox clusters occur in insects that are not the result of duplication but of cluster splitting (36). This happened at least twice, because the insertion of non-Hox cluster sequences different between D. melanogaster and D. pseudoobscura, on the one hand, and D. virilis, on the other hand (36, 37), where the split in D. melanogaster is between Antp and Ubx and in D. virilis and D. repleta, between Ubx and AbdB (38, 39). In addition, inverted Hox gene orientation has been documented for dipterans. For instance, the Deformed gene (Hox4) is the same as the other Hox genes in vertebrates, the mosquito (22, 40), D. pseudoobscura (34), and D. virilis (38), but inverted in D. melanogaster. Similarly the orientation of ftz is also different between D. hydei and D. melanogaster (37). These facts suggest a highly dynamic and complex history of Hox cluster evolution in dipteran insects (38). In the nematode Caenorhabditis elegans seven Hox genes exist, but they are dispersed over >3 Mb of DNA with thousands of intervening genes, i.e., no Hox cluster is maintained (41) and, in the urochordate Ciona intestinalis, the Hox genes have also been dispersed with many intervening genes (42). No comparable structural changes have been reported for gnathostome Hox clusters, although the earliest history of vertebrate Hox clusters (jawless vertebrates) remains poorly documented. Only after cluster duplication do we observe structural changes of similar magnitude, but different in nature. For instance, the case of a tandem duplication of a Hox gene in a jawed vertebrate still has not been reported.
Are vertebrate and invertebrate Hox clusters different? Their patterns of evolution certainly corroborate such a difference. But, with our current knowledge, it is not possible to say exactly why. For Drosophila Hox clusters the presence of boundary elements that make the function of cis-regulatory elements more modular could be a factor contributing to their structural flexibility (43, 44). The nature of the differences, however, is obvious, with vertebrate Hox clusters being more constrained than invertebrate Hox clusters. It is plausible then that the evolvability of vertebrate Hox clusters is, for some structural or functional reason, lower than their invertebrate counterparts. Hox cluster duplications could temporarily open a window of evolvability in which these constraints are relaxed. A vertebrate lineage may take advantage of this window of opportunity, given the right ecological and developmental boundary conditions. This window of opportunity may explain why duplicated Hox clusters in vertebrates tend to be associated with major radiations, like those of the gnathostomes and the teleosts.
Acknowledgments
We thank Tom Powers, Jutta Roth, Geff Stopper, and Terri Williams and an anonymous reviewer for useful comments on an earlier draft of this article. This research is supported by National Science Foundation Grants IBN-9905403, IBN-0321470, and IBN-9905408.
References
- 1.Duboule, D. (1994) Guidebook to the Homeobox Genes (Oxford Univ. Press, Oxford).
- 2.Davidson, E. (2001) Genomic Regulatory Systems (Academic, San Diego).
- 3.Carroll, S. B., Grenier, J. K. & Weatherbee, S. D. (2001) From DNA to Diversity (Blackwell Science, Malden, MA).
- 4.Wray, G. A. (2001) Science 292**,** 2257. [Google Scholar]
- 5.Holland, P. W. & Garcia-Fernandez, J. (1996) Dev. Biol. 173**,** 382–395. [DOI] [PubMed] [Google Scholar]
- 6.Malaga-Trillo, E. & Meyer, A. (2001) Am. Zool. 41**,** 676–686. [Google Scholar]
- 7.Martinez, P. & Amemiya, C. T. (2002) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133**,** 571–580. [DOI] [PubMed] [Google Scholar]
- 8.Schughart, K., Kappen, C. & Ruddle, F. H. (1987) Br. J. Cancer 58**,** 9–13. [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Fernández, J. & Holland, P. W. H. (1994) Nature 370**,** 563–566. [DOI] [PubMed] [Google Scholar]
- 10.Ferrier, D. K., Minguillón, C., Holland, P. W. H. & Garcia-Fernández, J. (2000) Evol. Dev. 2**,** 284–293. [DOI] [PubMed] [Google Scholar]
- 11.Force, A., Amores, A. & Postlethwait, J. H. (2002) J. Exp. Zool. 294**,** 30–46. [DOI] [PubMed] [Google Scholar]
- 12.Irvine, S. Q., Carr, J. L., Bailey, W. J., Kawasaki, K., Shimizu, N., Amemiya, C. T. & Ruddle, F. H. (2002) J. Exp. Zool. 294**,** 47–62. [DOI] [PubMed] [Google Scholar]
- 13.Fried, C., Prohaska, S. J. & Stadler, P. F. (2003) J. Exp. Zool. 299**,** 18–25. [DOI] [PubMed] [Google Scholar]
- 14.Kim, C.-B., Amemiya, C., Bailey, W., Kawasaki, K., Mezey, J., Miller, W., Minoshima, S., Shimizu, N., Wagner, G. & Ruddle, F. (2000) Proc. Natl. Acad. Sci. USA 97**,** 1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amores, A., Force, A., Yan, Y.-L., Joly, L., Amemiya, C., Fritz, A., Ho, R. K., Langeland, J., Prince, V., Wang, Y.-L., et al. (1998) Science 282**,** 1711–1714. [DOI] [PubMed] [Google Scholar]
- 16.Amores, A., Suzuki, T., Yan, Y.-L., Pomroy, J., Singer, A., Amemiya, C. & Postlethwait, J. (2003) Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 17.Misof, B. Y. & Wagner, G. P. (1996) Mol. Phylogenet. Evol. 5**,** 309–322. [DOI] [PubMed] [Google Scholar]
- 18.Snell, E. A., Scemama, J. L. & Stellwag, E. J. (1999) J. Exp. Zool. 285**,** 41–49. [PubMed] [Google Scholar]
- 19.Holland, P. W. H., Garcia-Fernández, J., Williams, N. A. & Sidow, A. (1994) Development (Cambridge, U.K.) Suppl., 125–133. [PubMed]
- 20.Carroll, S. B. (1995) Nature 376**,** 479–485. [DOI] [PubMed] [Google Scholar]
- 21.Lynch, M. & Force, A. (2000) Genetics 154**,** 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers, T. P., Hogan, J., Ke, X., Dymbrowski, K., Wang, X., Collins, F. H. & Kaufman, T. C. (2000) Evol. Dev. 2**,** 311–325. [DOI] [PubMed] [Google Scholar]
- 23.Chiu, C.-h., Amemiya, C., Dewar, K., Kim, C.-B., Ruddle, F. & Wagner, G. P. (2002) Proc. Natl. Acad. Sci. USA 99**,** 5492–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prohaska, S. J., Fried, C., Flamm, C., Wagner, G. P. & Stadler, P. F. (2003) Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 25.Santini, S., Boore, J. L. & Meyer, A. (2003) Genome Res. 13**,** 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151**,** 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClintock, J. M., Kheirbek, M. A. & Prince, V. E. (2002) Development (Cambridge, U.K.) 129**,** 2339–2354. [DOI] [PubMed] [Google Scholar]
- 28.Scemama, J.-L., Hunter, M., McCallum, J., Prince, V. & Stellwag, E. (2002) J. Exp. Zool. 294**,** 285–299. [DOI] [PubMed] [Google Scholar]
- 29.Chiu, C.-H., Nonaka, D., Xue, L., Amemiya, C. T. & Wagner, G. P. (2000) Mol. Phylogenet. Evol. 17**,** 305–316. [DOI] [PubMed] [Google Scholar]
- 30.Peer, Y. V. d., Taylor, J. S., Braasch, I. & Meyer, A. (2001) J. Mol. Evol. 53**,** 436–446. [DOI] [PubMed] [Google Scholar]
- 31.Bruce, A. E. E., Oates, A. C., Prince, V. E. & Ho, R. K. (2001) Evol. Dev. 3**,** 127–144. [DOI] [PubMed] [Google Scholar]
- 32.Greer, J. M., Puetz, J., Thomas, K. R. & Capecchi, M. R. (2000) Nature 403**,** 661–665. [DOI] [PubMed] [Google Scholar]
- 33.Hart, C. P., Bogarad, L. D., Fainsod, A. & Ruddle, F. H. (1987) Nucleic Acids Res. 15**,** 5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randazzo, F. M., Seeger, M. A., Huss, C. A., Sweeney, M. A., Cecil, J. K. & Kaufman, T. C. (1993) Genetics 133**,** 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stauber, M., Prell, A. & Schmidt-Ott, U. (2002) Proc. Natl. Acad. Sci. USA 99**,** 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.VonAllmen, G., Hogga, I., Spierer, A., Karch, F., Bender, W., Gyurkovics, H. & Lewis, E. (1996) Nature 380**,** 116. [DOI] [PubMed] [Google Scholar]
- 37.Maier, D., Preiss, A. & Powell, J. R. (1990) EMBO J. 9**,** 3957–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis, E. B., Barret, B. D., Mathog, D. R. & Celniker, S. E. (2003) Curr. Biol. 13**,** R587–R588. [DOI] [PubMed] [Google Scholar]
- 39.Ranz, J. M., Casals, F. & Ruiz, A. (2001) Genome Res. 11**,** 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devenport, M. P., Blass, C. & Eggelston, P. (2000) Evol. Dev. 2**,** 326–339. [DOI] [PubMed] [Google Scholar]
- 41.Ruvkun, G. & Hobert, O. (1998) Science 282**,** 2033–2041. [DOI] [PubMed] [Google Scholar]
- 42.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298**,** 2157–2167. [DOI] [PubMed] [Google Scholar]
- 43.Galloni, M., Gyurkovics, H., Schedl, P. & Karch, F. (1993) EMBO J. 12**,** 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drewell, R. A., Bae, E., Burr, J. & Lewis, E. B. (2002) Proc. Natl. Acad. Sci. USA 99**,** 16853–16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).