Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding (original) (raw)
Abstract
Hepatitis C virus infects an estimated 180 million people worldwide, prompting enormous efforts to develop inhibitors targeting the essential NS3/4A protease. Resistance against the most promising protease inhibitors, telaprevir, boceprevir, and ITMN-191, has emerged in clinical trials. In this study, crystal structures of the NS3/4A protease domain reveal that viral substrates bind to the protease active site in a conserved manner defining a consensus volume, or substrate envelope. Mutations that confer the most severe resistance in the clinic occur where the inhibitors protrude from the substrate envelope, as these changes selectively weaken inhibitor binding without compromising the binding of substrates. These findings suggest a general model for predicting the susceptibility of protease inhibitors to resistance: drugs designed to fit within the substrate envelope will be less susceptible to resistance, as mutations affecting inhibitor binding would simultaneously interfere with the recognition of viral substrates.
Keywords: drug design, hepatitis C, substrate envelope
Drug resistance is a major obstacle in the treatment of quickly evolving diseases. Hepatitis C virus (HCV) is a genetically diverse Hepacivirus of the Flaviviridae family infecting an estimated 180 million people worldwide (1). The viral RNA genome is translated as a single polyprotein and subsequently processed by host-cell and viral proteases into structural (C, E1, E2, and p7) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (2). The viral RNA-dependent RNA polymerase, NS5B, is inherently inaccurate and misincorporation of bases accounts for a very high mutation rate (3). While some mutations are neutral, others will alter the viability of the virus and propagate with varying efficiencies in each patient. Thus HCV infected individuals will develop a heterogeneous population of virus variants known as quasispecies (4). As patients begin treatment, the selective pressures of antiviral drugs will favor drug resistant variants (5). Therefore, an inhibitor must not only recognize one protein variant, but an ensemble of related enzymes. A detailed understanding of the atomic mechanisms of resistance is essential to effectively combat drug resistance against HCV antivirals.
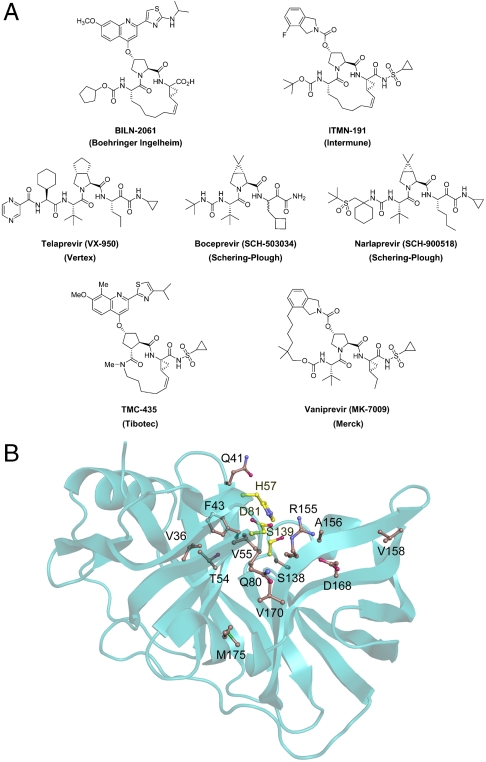
The essential HCV NS3/4A protease is an attractive therapeutic target responsible for cleaving at least four sites along the viral polyprotein. These sites share little sequence homology except for an acid at position P6, Cys or Thr at P1, and Ser or Ala at P1′ (Table S1). The first cleavage event at the 3-4A junction occurs in cis as a unimolecular process, while the remaining substrates are processed bimolecularly in trans. The NS3/4A protease also cleaves the human cellular targets TRIF and MAVS, which confounds the innate immune response to viral infection (6–8). Early drug design efforts were hampered by the relatively shallow, featureless architecture of the protease active site. The eventual observation of N-terminal product inhibition served as a stepping stone for the discovery of more potent peptidomimetic inhibitors (9, 10). Over the past decade, pharmaceutical companies have further developed these lead compounds. Many structure-activity-relationship (SAR) studies have been performed to evaluate the effect of different functional moieties on protease inhibition at positions P4-P1′ (11–17). Crystal structures have been determined of the NS3/4A protease domain bound to a variety of inhibitors as well as of several drug resistant protease variants, such as R155K and V36M (18, 19). These data elucidate the molecular interactions of NS3/4A with inhibitors and the effect of specific drug resistance mutations on binding. These efforts, conducted in parallel by several pharmaceutical companies, led to the discovery of many protease inhibitors. Proof-of-concept for the successful clinical activity of this drug class was first demonstrated by the macrocyclic inhibitor BILN-2061 (Boehringer Ingelheim) (20, 21), which was later dropped from clinical trials in 2006 due to cardiotoxicity (22). Many other NS3/4A protease inhibitors are currently in development, and telaprevir (Vertex), boceprevir (Schering-Plough), and ITMN-191 (Intermune) lead the way in advanced phases of human clinical trials (Fig. 1A).
Fig. 1.
NS3/4A protease inhibitors and reported sites of drug resistance. (A) The leading protease inhibitors in development mimic the N-terminal side of the viral substrates. (B) The majority of reported drug resistance mutations cluster around the protease active site with the catalytic triad depicted in yellow.
Despite these successes, the rapid acquisition of drug resistance has limited the efficacy of the most potent NS3/4A protease inhibitors in both replicon studies and human clinical trials (Fig. 1B and Table 1). In this study, we show that mutations conferring the most severe resistance occur where the protease extensively contacts the inhibitors but not the natural viral substrates. Four crystal structures of the NS3/4A protease domain in complex with the N-terminal products of viral substrates reveal a conserved mode of substrate binding, with the consensus volume defining the substrate envelope. The protease inhibitors ITMN-191 (3M5L), TMC435 (3KEE) (23), and boceprevir (2OC8) (24) protrude extensively from the substrate envelope in regions that correlate with known sites of resistance mutations. Most notably, the P2 moieties of all three drugs protrude to contact A156 and R155, which mutate to confer high-level resistance against nearly all drugs reported in the literature (25–30). These findings suggest that drug resistance results from a change in molecular recognition and imply that drugs designed to fit within the substrate envelope will be less susceptible to resistance, as mutations altering inhibitor binding will simultaneously interfere with the binding of substrates.
Table 1.
Drug resistance mutations reported in replicon studies and clinical trials*
| Residue | Mutation | Drug |
|---|---|---|
| V36 | A, M, L, G | Boceprevir, telaprevir |
| Q41 | R | Boceprevir, ITMN-191 |
| F43 | S, C, V, I | Boceprevir, telaprevir, ITMN-191, TMC435 |
| V55 | A | Boceprevir |
| T54 | A, S | Boceprevir, telaprevir |
| Q80 | K, R, H, G, L | TMC435 |
| S138 | T | ITMN-191, TMC435† |
| R155 | K, T, I, M, G, L, S, Q | Boceprevir, telaprevir, ITMN-191, BILN-2061, TMC435 |
| A156 | V, T, S, I, G | Boceprevir, telaprevir, ITMN-191, BILN-2061, TMC435 |
| V158 | I | Boceprevir |
| D168 | A, V, E, G, N, T, Y, H, I | ITMN-191, BILN-2061, TMC435 |
| V170 | A | Boceprevir, telaprevir |
| M175 | L | Boceprevir |
Results
Synthesis of ITMN-191.
We synthesized the macrocyclic inhibitor ITMN-191 using a convergent reaction sequence described in SI Text. Briefly, the P2 and P1-P1′ fragments were preassembled and the macrocyclic drug compound was generated by a four-step reaction sequence, including P2-P3 amide coupling, ester hydrolysis, coupling with the P1-P1′ fragment, and ring-closing metathesis. The P2-P3 fragment was assembled by coupling the commercially available Boc-protected amino acid (S)-2-(tert_-butoxycarbonylamino)non-8-enoic acid (Acme Biosciences, Inc) with the preassembled P2 fragment, (3_R, 5_S_)-5-(methoxycarbonyl)pyrrolidin-3-yl 4-fluoroisoindoline-2-carboxylate (31), using O-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU)/diisopropylethylamine (DIPEA). Hydrolysis of the P2-P3 methyl ester with LiOH.H2O in a mixture of THF-MeOH-H2O followed by coupling of the resulting acid under HATU/DIPEA conditions with the preassembled P1-P1′ fragment, (1_R_, 2_S_)-1-amino-N-(cyclopropylsulfonyl)-2-vinylcyclopropanecarboxamide (32), provided the bis-olefin precursor for ring-closing metathesis. Cyclization of the bis-olefin intermediate was accomplished using a highly efficient ring-closing metathesis catalyst Zhan 1B and provided the protease inhibitor ITMN-191.
Structure Determination of Inhibitor and Substrate Complexes.
Although NS3/4A cleaves the viral polyprotein of over 3,000 residues at four specific sites in vivo, we focused on the local interactions of the protease domain with short peptide sequences corresponding to the immediate cleavage sites. All structural studies were carried out with the highly soluble, single-chain construct of the NS3/4A protease domain described previously (33), which contains a fragment of the essential cofactor NS4A covalently linked at the N terminus by a flexible linker. A similar protease construct was shown to retain comparable catalytic activity to the authentic protein complex (34). Crystallization trials were initially carried out using the inactive (S139A) protease variant in complex with substrate peptides spanning P7-P5′. The 4A4B substrate complex revealed cleavage of the scissile bond and no ordered regions for the C-terminal fragment of the substrate. Similar observations were previously described for two other serine proteases where catalytic activity was observed, presumably facilitated by water, despite Ala substitutions of the catalytic Ser (35, 36). Thus all subsequent crystallization trials with the NS3/4A protease were performed using N-terminal cleavage products of the viral substrates spanning P7-P1.
NS3/4A crystal structures in complex with ITMN-191 and peptide products 4A4B, 4B5A, and 5A5B were determined and refined at 1.25 Å, 1.70 Å, 1.90 Å, and 1.60 Å resolution, respectively (Table S2). The complexes crystallized in the space groups _P_212121 and _P_21 with one, two, or four molecules in the asymmetric unit. The average B factors range from 16.8–29.7 _Å_2 and there are no outliers in the Ramachandran plots. These structures represent the highest resolution crystal structures of NS3/4A protease reported to date.
Overall Structure Analysis.
The NS3/4A protease domain adopts a tertiary fold characteristic of serine proteases of the chymotrypsin family (37, 38). A total of nine protease molecules were modeled in the four crystal structures solved in this study with an overall rms deviation (rmsd) of 0.28 Å. The rmsds reveal the five most variable regions of the protease to be (Fig. S1): (i) the linker connecting cofactor 4A at the N terminus, (ii) the loop containing residues 65–70, (iii) the zinc-binding site containing residues 95–105, (iv) the 310 helix region spanning residues 128–136, and (v) the active site antiparallel β-sheet containing residues 156–168. These structural differences likely indicate inherent flexibility in the protease and do not appear to correlate with ligand type or active site occupancy.
Analysis of Product Complexes.
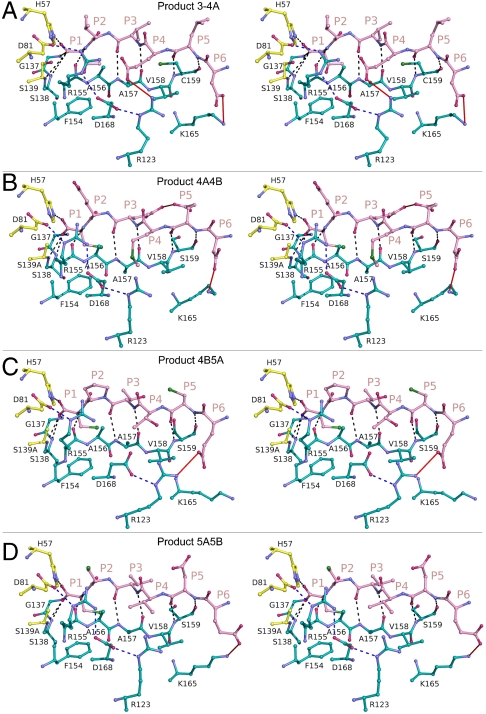
Product complexes 4A4B, 4B5A, and 5A5B were further analyzed with the C terminus of the full-length NS3/4A structure (1CU1), which contains the N-terminal cleavage product of viral substrate 3-4A (39). All four products bind to the protease active site in a conserved manner (Fig. 2), forming an antiparallel β-sheet with residues 154–160 and burying 500–600 _Å_2 of solvent accessible surface area as calculated by PISA (40). The peptide backbone torsions are very similar, being most conserved at position P1 and deviating slightly toward position P4. Eight hydrogen bonds between backbone amide and carbonyl groups are completely conserved, involving protease residues S159 (C159 in product 3-4A), A157, R155, S139A, S138, and G137. S159 (C159 in product 3-4A), and A157 each contribute two hydrogen bonds with the P5 and P3 peptide residues, respectively. All P1 terminal carboxyl groups sit in the oxyanion hole, hydrogen bonding with the Nϵ atom of H57 and the amide nitrogens of residues 137–139. Although only product 4B5A contains a proline at P2, the other substrate sequences still adopt constrained P2 φ torsion angles. Thus products bind similarly despite their high sequence diversity.
Fig. 2.
Stereo view of N-terminal cleavage product binding to NS3/4A protease. N-terminal protease cleavage products (A) 3-4A, (B) 4A4B, (C) 4B5A, and (D) 5A5B are depicted as they bind to the protease active site. All conserved interactions are indicated by black dashes, while red lines depict interactions that are not present in all structures. The electrostatic network involving residues R123, D168, R155, and D81 is indicated by blue dashes.
The P1 and P6 residues are most conserved among the substrate sequences, as are most of their interactions with the protease. The P1 side chains interact with the aromatic ring of F154. In all structures but product complex 4B5A, K165 forms salt-bridges with the P6 acids, while residues R123, D168, R155, and the catalytic D81 form an ionic network along one surface of the bound products (Fig. 2). In complex 4B5A, R123 interacts directly with the P6 acid, while D168 reorients and no longer contacts R155. Other molecular interactions in the product complexes are more diverse. Notably, K136 interacts differently with the cleavage products, forming: (i) a hydrogen bond with the P2 backbone carbonyl oxygen of 3-4A; (ii) a salt-bridge with the P3 glutamate of product 4A4B; and (iii) nonspecific van der Waals interactions with the P2 and P3 side chains of products 4B5A and 5A5B. Also, in product complex 4A4B, an intramolecular hydrogen bond forms between the P3 and P5 glutamate residues, while the unique P4 acid of product 3-4A forms salt-bridges with the guanidinium groups of R123 and R155. Thus distinct patterns of side chain interactions underlie the set of conserved features involved in NS3/4A cleavage product binding.
The Substrate Envelope.
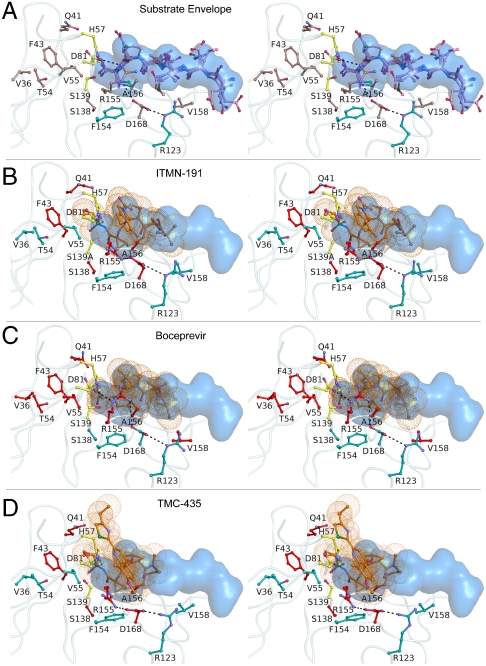
To further analyze the structural similarities of the four NS3/4A product complexes, the active sites were superposed on the Cα atoms of residues 137–139 and 154–160, revealing that both the active site residues and substrate products spanning P6-P1 align closely with an average Cα rmsd of 0.24 Å and 0.35 Å, respectively. The consensus van der Waals volume shared by any three of the four cleavage products was then calculated to generate the NS3/4A substrate envelope (Fig. 3A). This shape could not be predicted by the primary sequences alone and highlights the conserved mode of viral substrate recognition despite their high sequence diversity.
Fig. 3.
Stereo view of the NS3/4A substrate envelope and protease inhibitors. (A) After active site superpositions, the overlapping van der Waals volume shared by any three of the four cleavage products defines the substrate envelope, depicted in blue. NS3/4A protease residues which mutate to confer drug resistance are shown in brown. (B) ITMN-191, (C) boceprevir and (D) TMC435 protrude from the substrate envelope at several locations, which correlate with known sites of drug-resistant mutations to each inhibitor, shown in red.
Analysis of Inhibitor Complexes.
ITMN-191, TMC435, and boceprevir are all peptidomimetic NS3/4A protease inhibitors. Active site superpositions of these drug complexes reveal that the inhibitors interact with many of the same protease residues as the cleavage products. Despite the P3-P1 cyclization of ITMN-191 and TMC435, the functional groups are positioned similarly in all three inhibitor complexes. The P1 cysteine surrogates interact with the aromatic ring of F154, while the P2 and P3 moieties overlap closely. Although TMC435 does not contain a P4 substituent, the P4 _tert_-butyl groups of ITMN-191 and boceprevir also align closely. In addition, the P1 and P3 backbone atoms of all inhibitors hydrogen bond with the carbonyl oxygens of R155 and A157, respectively. These observations verify the peptidomimetic nature of these drugs and support their observed mechanism as competitive active site inhibitors.
The largest variation between these three protease inhibitors occurs at P2 where the aromatic rings of ITMN-191 and TMC435 stack against the guanidinium group of R155 (Fig. 3). This molecular interaction alters the electrostatic network involving R123, D168, R155, and D81. R155 rotates nearly 180° around Cδ relative to its conformation observed in product complexes, losing its hydrogen bond with D81 but maintaining interaction with D168. Mutations at R155 or D168 would disrupt the electrostatic network and destabilize this packing thereby lowering the affinity of these macrocyclic drugs. This observation provides a structural rationale for the drug resistance mutations R155K, as previously proposed (19), and D168A/V, which both confer a selective advantage in vitro in the presence of ITMN-191 or TMC435 (26, 30). In addition, the TMC435 complex reveals that R155 is stabilized by a hydrogen bond with Q80, which also mutates to confer resistance to TMC435 (30). Thus many of the primary drug resistance mutations can be explained by the disruption of atomic interactions involving the P2 functional groups of the drugs.
Insights into Drug Resistance.
To determine the locations where the inhibitors protrude from the substrate envelope, the inhibitor and product complexes were also superposed using residues 137–139 and 154–160. The van der Waals volumes of inhibitor protrusion from the substrate envelope (_V_out) (41, 42) were calculated for each drug and compared with published EC50 fold-change data for drug resistance variants (30). The magnitudes of the EC50 fold-change data determined for each NS3/4A mutant generally trend with the _V_out values for the three drugs. The P2 moieties of boceprevir, ITMN-191 and TMC435 protrude most extensively from the substrate envelope with _V_out values of 105, 294, and 496 _Å_3, respectively (Table 2). However, the precise level of drug resistance observed is also determined by the particular change in molecular interaction occurring for a given mutation. For example, A156 and R155 pack with the P2 moieties of these three inhibitors where they protrude beyond the substrate envelope. Mutations of A156 to bulkier side chains would result in a steric clash with the P2 drug moieties. Indeed, the rigid dimethylcyclopropane group of boceprevir protrudes from the substrate envelope at the P2 subsite, and A156V or A156T confer 65 and 75 fold-changes in EC50, respectively (Table 2). Similarly, molecular changes at R155 and D168 would result in a substantial loss of interactions with P2. The most extensive protrusions of ITMN-191 and TMC435 at P2 trend with their greatest fold-change in potency of nearly 450 and 600, respectively, from mutations in this subsite. Thus the extent by which an inhibitor protrudes from the substrate envelope in a given subsite is indicative of its vulnerability to resistance.
Table 2.
EC50 fold-change (FC) data * for several NS3/4A drug resistant variants tabulated with _V_out, the van der Waals volume of protrusion from the substrate envelope, at each subsite of the enzyme
| Boceprevir | ITMN-191 | TMC435 | |||||
|---|---|---|---|---|---|---|---|
| Subsite | Resistance mutation | _V_out (_Å_3) | EC50 FC | _V_out (_Å_3) | EC50 FC | _V_out (_Å_3) | EC50 FC |
| Total | 292 | 500 | 649 | ||||
| P1 | 76 | 67 | 64 | ||||
| P2 | 105 | 294 | 496 | ||||
| Q80R | 0.5 | 3.5 | 6.9 | ||||
| Q80K | 0.8 | 2.3 | 7.7 | ||||
| R155K | 4.7 | 447 | 30 | ||||
| A156V | 75 | 63 | 177 | ||||
| A156T | 65 | 41 | 44 | ||||
| D168A | 0.7 | 153 | 594 | ||||
| D168E | 0.8 | 75 | 40 | ||||
| P3 | 34 | 70 | 67 | ||||
| P4 | 76 | 69 | 0 | ||||
| V158I | 3.3† | ND | ND |
Further structural analyses with the substrate envelope provide insights into other NS3/4A drug resistance mutations. The P1′ sulfonamide groups of ITMN-191 and TMC435, as well as the P1′ ketoamide of boceprevir, protrude from the substrate envelope near residues Q41 and F43, which both mutate to confer low-level resistance to these drugs (25, 30, 43). The keto group of boceprevir also projects outside the substrate envelope near T54 and V55. T54A/S confers low-level resistance to boceprevir, while V55A was recently identified in patient isolates after treatment with boceprevir (44). The analogous carbonyl groups of ITMN-191 and TMC435, however, are orientated in the opposite direction and protrude toward S138. In fact, in vitro studies reveal reduced activity for ITMN-191 and TMC435 against S138T variants, while boceprevir remains fully active (30, 43). The bulky P4 _tert_-butyl group of boceprevir extends outside the substrate envelope contacting V158; the V158I variant has lower affinity for this drug, likely due to a steric clash (45). This variant may also impact the affinity of ITMN-191, as its P4 _tert_-butyl also protrudes at the same location. These findings demonstrate that in regions outside the P2 subsite, positions where ITMN-191, TMC435, and boceprevir protrude from the substrate envelope also correlate with many other known sites of drug resistance mutations.
Discussion
The emergence of drug resistance is a major obstacle in modern medicine that limits the long-term usefulness of the most promising therapeutics. By considering how HCV NS3/4A protease inhibitors bind relative to natural viral substrates, we discovered that primary sites of resistance occur in regions of the protease where drugs protrude from the substrate envelope. In particular, R155 and A156, which mutate to confer severe resistance against ITMN-191, TMC435, and boceprevir, interact closely with the P2 drug moieties where they protrude most extensively from the substrate envelope. Molecular changes at these residues confer resistance by selectively weakening inhibitor binding without compromising the binding of viral substrates. We further speculate that these mutations will not considerably affect the binding of the host cellular substrates TRIF and MAVS, which likely fit well within the substrate envelope as they share many features with the viral substrates. However, TRIF contains a track of eight proline residues instead of an acidic residue at position P6, which may modulate its binding. Further structural studies are warranted to better ascertain the molecular details of how these cellular substrates are recognized by the NS3/4A protease.
Although this study focuses on ITMN-191, TMC435, and boceprevir, other NS3/4A protease inhibitors in clinical trials, including telaprevir, narlaprevir, and vaniprevir (Fig. 1A), contain similar functional groups that likely protrude from the substrate envelope. Most notably, all these drug candidates contain bulky P2 moieties and are therefore susceptible to cross-resistance against mutations at R155 and A156. R155 and A156 mutations have been shown to confer telaprevir resistance in treated patients (46). Cross-resistance studies have also shown that narlaprevir displays similar fold losses in activity against most of the known drug resistance mutations for telaprevir and boceprevir (47). Ultimately, to slow the emergence of multidrug resistant viral strains, inhibitors should be confined within the substrate envelope, particularly at the P2 position. To compensate for the loss of binding affinity that will likely accompany these changes, additional interactions could potentially be optimized spanning the S4-S6 subsites of the protease and the catalytic triad.
Our findings further suggest a general model for using the substrate envelope to predict patterns of drug resistance in other quickly evolving diseases. For drug resistance to occur, mutations must selectively weaken a target’s affinity for an inhibitor without significantly altering its natural biological function. Mutations occurring outside the substrate envelope are better able to achieve this effect, as these molecular changes can selectively alter inhibitor binding without compromising the binding of natural substrates. Whenever the interaction of a drug target with its biological substrates can be structurally characterized, we predict that drugs designed to fit within the substrate envelope will be less susceptible to resistance. Structure-based design strategies can utilize this model as an added constraint to develop inhibitors that fit within the substrate envelope. In fact, previous work in our laboratory provides proof-of-concept for the successful incorporation of the substrate envelope in the design of unique HIV protease inhibitors, which maintain high affinities against a panel of multidrug resistant variants of HIV-1 protease (42, 48–53). As a general paradigm, design efforts incorporating the substrate envelope would facilitate a more rationale evaluation of drug candidates and lead to the development of more robust inhibitors that are less susceptible to resistance.
Materials and Methods
Protein Crystallization.
The NS3/4A protease construct was expressed and purified as reported previously (33, 54), detailed in SI Text. Purified protein was concentrated to ∼3 mg/mL and loaded on a HiLoad Superdex75 16/60 column equilibrated with 25 mM 2-(N-morpholino)ethanesulfonic acid (MES) at pH 6.5, 500 mM NaCl, 10% glycerol, 30 μM zinc chloride, and 2 mM DTT. The protease fractions were pooled and concentrated to 20–25 mg/mL with an Amicon Ultra-15 device (10 kD; Millipore). The concentrated samples were then incubated at 4 °C for 1 h with 2–20 M excess of viral substrate 4A4B, peptide products 4B5A or 5A5B, or ITMN-191. Information about the synthesis of viral peptides and ITMN-191 is provided in SI Text. Diffraction-quality crystals were obtained overnight for all ligands by mixing equal volume of concentrated protein solution with precipitant solution (20–26% PEG-3350, 0.1 M sodium MES buffer at pH 6.5, and 4% ammonium sulfate) in 24-well VDX hanging drop trays.
Data Collection and Structure Solution.
Crystals were flash-frozen in liquid nitrogen and mounted under constant cryostream. X-ray diffraction data were collected at Advanced Photon Source BioCARS 14-IDB, 14-BMC, and LS-CAT 21-ID-F. Diffraction intensities of the complexes were indexed, integrated, and scaled using the programs HKL2000 (55) and XDS (56). 5% of the data was used to calculate R-free (57). All structure solutions were generated using isomorphous molecular replacement with PHASER (58) or AMORE. The NS3/4A protease domain (PDB code 2A4G) (59) was used for molecular replacement in solving the product 4A4B structure, and this structure was subsequently used for solving the other complexes. In all cases, initial refinement was carried out in the absence of modeled ligand, which was subsequently built in. Phases were improved using ARP/wARP (60). Iterative rounds of translation, libration, and screw (TLS) and restrained refinement with CCP4 (61) and graphical model building with COOT (62) until convergence was achieved. The final structures were evaluated with MolProbity (63) prior to deposition in the protein data bank.
Structural Analysis.
Double-difference plots (64) were used to determine the structurally invariant regions of the protease, consisting of residues 32–36, 42–47, 50–54, 84–86, and 140–143. Structures were superposed with PyMOL (65) using the Cα atoms of these residues for all protease molecules from the solved structures (nine total). The B chain of product complex 4A4B was used as the reference structure in all alignments. Fit of individual inhibitors into the substrate envelope was quantified by mapping the substrate envelope and the van der Waals volume of each inhibitor on a three-dimensional grid with spacing of 0.5 Å. _V_out for each drug moiety was computed by counting the grid cells, which were occupied by any inhibitor atom of that site but not the substrate envelope, and multiplied by the grid cell size, 0.125 _Å_3 (41, 42).
Substrate Envelope and Inhibitor Analyses.
NS3/4A substrate envelope was computed using product complexes 4A4B (B chain), 4B5A (D chain), and 5A5B (A chain). In structures with multiple protease molecules in the asymmetric unit, the one containing the most ordered peptide product was used for the alignment. The protease domain of the full-length NS3/4A structure (A chain; PDB code 1CU1) (39), including the C-terminal six amino acids, was included as a product complex 3-4A. All active site alignments were performed in PyMOL using Cα atoms of protease residues 137–139 and 154–160. After superposition, Gaussian object maps were generated in PyMOL for each cleavage product. Four consensus Gaussian maps were then calculated, representing the intersecting volume of a group of three object maps. Finally, the summation of these four consensus maps was generated to construct the substrate envelope, depicting the van der Waals volume shared by any three of the four products. The previously determined boceprevir complex (PDB code 2OC8) (24) and TMC435 complex (PDB code 3KEE) (23) were used in this study (66).
Supplementary Material
Supporting Information
Acknowledgments.
We thank H. Klei for helpful discussions. We also thank Z. Wawrzak, M. Bolbat, and K. Brister of the LS-CAT beamline at Argonne National Laboratory for data collection of the ITMN-191 complex; M. Nalam and R. Bandaranayake for assistance with structural refinement; A. Ozen for providing _V_out calculations; and S. Shandilya and Y. Cai for computational support. The National Institute of Health (NIH) Grants R01-GM65347 and R01-AI085051 supported this work. Use of Advanced Photon Source (APS) was supported by the Department of Energy (DOE), Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357. Use of the BioCARS Sector 14 was supported by NIH-NCRR RR007707. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology TriCorridor under Grant 085P1000817.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3M5L, 3M5M, 3M5N, and 3M5O).
References
- 1.World Health Organization. . Vaccine research: hepatitis C. In: Barnes E, editor. Hepatitis C Virus: Disease Burden. 2010. [March 15, 2010]. Available at http://www.who.int/vaccine_research/diseases/viral_cancers/en/index2.html. [Google Scholar]
- 2.Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi SA. Hepatitis C virus—biology, host evasion strategies, and promising new therapies on the horizon. Med Res Rev. 2007;27:353–373. doi: 10.1002/med.20063. [DOI] [PubMed] [Google Scholar]
- 4.Martell M, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paolucci S, et al. Analysis of HIV drug-resistant quasispecies in plasma, peripheral blood mononuclear cells and viral isolates from treatment-naive and HAART patients. J Med Virol. 2001;65:207–217. doi: 10.1002/jmv.2022. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, et al. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol. 2007;81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llinas-Brunet M, et al. Peptide-based inhibitors of the hepatitis C virus serine protease. Bioorg Med Chem Lett. 1998;8:1713–1718. doi: 10.1016/s0960-894x(98)00299-6. [DOI] [PubMed] [Google Scholar]
- 10.Steinkuhler C, et al. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry. 1998;37:8899–8905. doi: 10.1021/bi980313v. [DOI] [PubMed] [Google Scholar]
- 11.Arasappan A, et al. P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease. Bioorg Med Chem Lett. 2006;16:3960–3965. doi: 10.1016/j.bmcl.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Bogen S, et al. Hepatitis C virus NS3-4A serine protease inhibitors: SAR of new P1 derivatives of SCH 503034. Bioorg Med Chem Lett. 2008;18:4219–4223. doi: 10.1016/j.bmcl.2008.05.060. [DOI] [PubMed] [Google Scholar]
- 13.Malancona S, et al. SAR and pharmacokinetic studies on phenethylamide inhibitors of the hepatitis C virus NS3/NS4A serine protease. Bioorg Med Chem Lett. 2004;14:4575–4579. doi: 10.1016/j.bmcl.2004.05.093. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson M, et al. Synthesis and SAR of potent inhibitors of the hepatitis C virus NS3/4A protease: exploration of P2 quinazoline substituents. Bioorg Med Chem Lett. 2010;20:4004–4011. doi: 10.1016/j.bmcl.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 15.Venkatraman S, et al. Discovery and structure-activity relationship of P1-P3 ketoamide derived macrocyclic inhibitors of hepatitis C virus NS3 protease. J Med Chem. 2009;52:336–346. doi: 10.1021/jm800940u. [DOI] [PubMed] [Google Scholar]
- 16.Raboisson P, et al. Structure-activity relationship study on a novel series of cyclopentane-containing macrocyclic inhibitors of the hepatitis C virus NS3/4A protease leading to the discovery of TMC435350. Bioorg Med Chem Lett. 2008;18:4853–4858. doi: 10.1016/j.bmcl.2008.07.088. [DOI] [PubMed] [Google Scholar]
- 17.Perni RB, et al. Inhibitors of hepatitis C virus NS3.4A protease 2. Warhead SAR and optimization. Bioorg Med Chem Lett. 2004;14:1441–1446. doi: 10.1016/j.bmcl.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, et al. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother. 2008;52:110–120. doi: 10.1128/AAC.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, et al. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J Biol Chem. 2007;282:22619–22628. doi: 10.1074/jbc.M610207200. [DOI] [PubMed] [Google Scholar]
- 20.Lamarre D, et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 21.Hinrichsen H, et al. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology. 2004;127:1347–1355. doi: 10.1053/j.gastro.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Vanwolleghem T, et al. Ultra-rapid cardiotoxicity of the hepatitis C virus protease inhibitor BILN 2061 in the urokinase-type plasminogen activator mouse. Gastroenterology. 2007;133:1144–1155. doi: 10.1053/j.gastro.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Cummings MD, et al. Induced-fit binding of the macrocyclic noncovalent inhibitor TMC435 to its HCV NS3/NS4A protease target. Angew Chem Int Ed Engl. 2010;49:1652–1655. doi: 10.1002/anie.200906696. [DOI] [PubMed] [Google Scholar]
- 24.Prongay AJ, et al. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]–6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization. J Med Chem. 2007;50:2310–2318. doi: 10.1021/jm060173k. [DOI] [PubMed] [Google Scholar]
- 25.Tong X, et al. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 2008;77:177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 26.He Y, et al. Relative replication capacity and selective advantage profiles of protease inhibitor-resistant hepatitis C virus (HCV) NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob Agents Chemother. 2008;52:1101–1110. doi: 10.1128/AAC.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarrazin C, et al. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology. 2007;132:1270–1278. doi: 10.1053/j.gastro.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 28.Kieffer TL, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46:631–639. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 29.Yi M, Ma Y, Yates J, Lemon SM. Transcomplementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 2009;5:e1000403. doi: 10.1371/journal.ppat.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz O, et al. In vitro resistance profile of the HCV NS3/4A protease inhibitor TMC435. Antimicrob Agents Chemother. 2010;54:1878–1887. doi: 10.1128/AAC.01452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arasappan A, Njoroge FG, Girijavallabhan VM. PCT Int.Appl. WO 2005/113581. 2005. Patent Application.
- 32.Wang XA, et al. 6995174. US Patent. 2006
- 33.Wittekind M, Weinheirner S, Zhang Y, Goldfarb V. 6333186. US Patent. 2002
- 34.Taremi SS, et al. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 1998;7:2143–2149. doi: 10.1002/pro.5560071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 36.Krishnan R, Sadler JE, Tulinsky A. Structure of the Ser195Ala mutant of human alpha—thrombin complexed with fibrinopeptide A(7—16): evidence for residual catalytic activity. Acta Crystallogr D . 2000;56:406–410. doi: 10.1107/s0907444900001487. [DOI] [PubMed] [Google Scholar]
- 37.Kim JL, et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 38.Bode W, Huber R. Crystal structure analysis and refinement of two variants of trigonal trypsinogen: trigonal trypsin and PEG (polyethylene glycol) trypsinogen and their comparison with orthorhombic trypsin and trigonal trypsinogen. FEBS Lett. 1978;90:265–269. doi: 10.1016/0014-5793(78)80382-2. [DOI] [PubMed] [Google Scholar]
- 39.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Nalam MN, et al. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J Virol. 2010;84:5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chellappan S, Kairys V, Fernandes MX, Schiffer C, Gilson MK. Evaluation of the substrate envelope hypothesis for inhibitors of HIV-1 protease. Proteins. 2007;68:561–567. doi: 10.1002/prot.21431. [DOI] [PubMed] [Google Scholar]
- 43.Kuntzen T, et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 2008;48:1769–1778. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Susser S, et al. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology. 2009;50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 45.Qiu P, et al. Identification of HCV protease inhibitor resistance mutations by selection pressure-based method. Nucleic Acids Res. 2009;37:e74. doi: 10.1093/nar/gkp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarrazin C, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132:1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 47.Tong X, et al. Preclinical characterization of the antiviral activity of SCH 900518 (Narlaprevir), a novel mechanism-based inhibitor of hepatitis C virus NS3 protease. Antimicrob Agents Chemother. 2010;54:2365–2370. doi: 10.1128/AAC.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chellappan S, et al. Design of mutation-resistant HIV protease inhibitors with the substrate envelope hypothesis. Chem Biol Drug Des. 2007;69:298–313. doi: 10.1111/j.1747-0285.2007.00514.x. [DOI] [PubMed] [Google Scholar]
- 49.Ali A, et al. 238th ACS National Meeting, Washington, DC, United States. Washington, DC: American Chemical Society; 2009. Substrate envelope based design of new HIV-1 protease inhibitors active against drug-resistant HIV-1. MEDI-102. [Google Scholar]
- 50.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Prabu-Jeyabalan M, et al. Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 2006;50:1518–1521. doi: 10.1128/AAC.50.4.1518-1521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altman MD, Nalivaika EA, Prabu-Jeyabalan M, Schiffer CA, Tidor B. Computational design and experimental study of tighter binding peptides to an inactivated mutant of HIV-1 protease. Proteins. 2008;70:678–694. doi: 10.1002/prot.21514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altman MD, et al. HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J Am Chem Soc. 2008;130:6099–6113. doi: 10.1021/ja076558p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallinari P, et al. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 57.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 58.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arasappan A, et al. Hepatitis C virus NS3-4A serine protease inhibitors: SAR of P′2 moiety with improved potency. Bioorg Med Chem Lett. 2005;15:4180–4184. doi: 10.1016/j.bmcl.2005.06.091. [DOI] [PubMed] [Google Scholar]
- 60.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr D. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 61.Collaborative Computational Project Number 4. The CCP4 Suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prabu-Jeyabalan M, Nalivaika EA, Romano K, Schiffer CA. Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate. J Virol. 2006;80:3607–3616. doi: 10.1128/JVI.80.7.3607-3616.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific LLC; 2008. [Google Scholar]
- 66.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information