Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas (original) (raw)
Abstract
EZH2, the catalytic subunit of the PRC2 complex, catalyzes the mono- through trimethylation of lysine 27 on histone H3 (H3K27). Histone H3K27 trimethylation is a mechanism for suppressing transcription of specific genes that are proximal to the site of histone modification. Point mutations of the EZH2 gene (Tyr641) have been reported to be linked to subsets of human B-cell lymphoma. The mutant allele is always found associated with a wild-type allele (heterozygous) in disease cells, and the mutations were reported to ablate the enzymatic activity of the PRC2 complex for methylating an unmodified peptide substrate. Here we demonstrate that the WT enzyme displays greatest catalytic efficiency (_k_cat/K) for the zero to monomethylation reaction of H3K27 and diminished efficiency for subsequent (mono- to di- and di- to trimethylation) reactions. In stark contrast, the disease-associated Y641 mutations display very limited ability to perform the first methylation reaction, but have enhanced catalytic efficiency for the subsequent reactions, relative to the WT enzyme. These results imply that the malignant phenotype of disease requires the combined activities of a H3K27 monomethylating enzyme (PRC2 containing WT EZH2 or EZH1) together with the mutant PRC2s for augmented conversion of H3K27 to the trimethylated form. To our knowledge, this is the first example of a human disease that is dependent on the coordinated activities of normal and disease-associated mutant enzymatic function.
Keywords: cancer, enzymology, epigenetics
Polycomb repressive complex 2 (PRC2) is a 4 or 5 protein complex that functionally represses gene transcription via posttranslational modification of the chromatin core protein, histone H3. More specifically, PRC2 functions as a histone methyltransferase, catalyzing the selective methylation of lysine residue 27 on histone H3 (H3K27). EZH2 is the catalytic subunit of the PRC2 complex, catalyzing the mono- through trimethylation of H3K27. Trimethylation of H3K27 is a mechanism for suppressing transcription of specific genes that are proximal to the site of histone modification.
Recently, somatic mutations of Tyr641 (Y641F, Y641N, Y641S, and Y641H) of EZH2 were reported to be associated with follicular lymphoma (FL) and the germinal-center B-cell like (GCB) subtype of diffuse large B-cell lymphoma (DLBCL) (1). In all cases, occurrence of the mutant EZH2 gene was heterozygous, and expression of both WT and mutant alleles was detected in the mutant samples profiled by transcriptome sequencing. It was also demonstrated that all of the mutant forms of EZH2 could be incorporated into the multiprotein PRC2 complex, but that the resulting complexes lacked the ability to catalyze methylation of the H3K27 equivalent residue of a peptidic substrate. Hence, it was concluded that the disease-associated changes at Tyr641 resulted in loss of function with respect to EZH2-catalyzed H3K27 methylation.
A presumptive reduction in the rate of H3K27 methylation due to enzyme heterozygosity seemed to us to be difficult to rationalize as the basis for a malignant phenotype (1, 2), especially in light of previous data indicating that overexpression of EZH2 (3–6), loss-of-function mutations in the corresponding H3K27 demethylase UTX (7), or overexpression of components of the PRC2, such as PHF19/PCL3 involved in increased H3K27 trimethylation (8–10), all result in malignant phenotypes in specific human cancers. Several recent reports indicate that an important component of early lymphomagenesis is the acquisition of stem cell-like characteristics (11, 12). Among these characteristics is enriched DNA methylation of PRC2 target genes, resulting in diminished transcription of these genes (13). Trimethylation of H3K27, catalyzed by the enzymatic activity of EZH2, similarly leads to diminished transcription of these same genes and has thus been reported as an additional, potential progenitor of lymphomagenesis (11–14). EZH2 levels have also directly been implicated in lymphogenesis. For example, Velichutina et al. (14) found that EZH2 mRNA level was directly correlated with cellular proliferation in primary germinal center diffuse large B-cell lymphoma tumors, whereas levels of many EZH2 target genes were negatively correlated with proliferation in these same tumors. Likewise, expression levels of EZH2 and the PRC1 component BMI1 have both been linked to lymphogenesis and the degree of malignancy of B-cell non-Hodgkins lymphomas (15). We were thus motivated to explore the enzymology of the EZH2 mutants in greater detail.
Results and Discussion
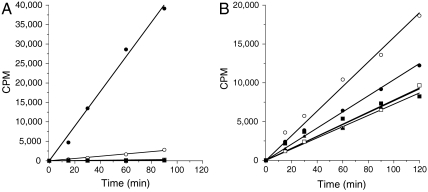
Recombinant PRC2 complexes (16) were prepared with WT and Tyr641 mutant versions of human EZH2 (see Materials and Methods). Equal concentrations (nominally 8 nM, based on protein determinations) of each complex were initially tested for the ability to catalyze 3H-methyl transfer from labeled SAM (_S_-adenosylmethionine) to an unmodified peptide representing the amino acid sequence surrounding H3K27 (H3:21-44) or to native avian erythrocyte oligonucleosomes. As previously reported (1), we found that the WT enzyme displayed robust activity for methyl transfer to this peptidic substrate, but that none of the mutant enzymes displayed significant methyltransferase activity (Fig. 1A). In contrast to the previously reported data and that in Fig. 1A, we found that all of the mutant EZH2 constructs were active methyltransferases against the avian nucleosome substrate (Fig. 1B).
Fig. 1.
B-cell lymphoma-associated mutants of EZH2 are active histone methyltransferases. In vitro methyltransferase activity of PRC2 complexes containing wild-type and various Y641 mutants of EZH2 was measured as (A) methyl transfer reactions using a peptide (H3 21-44) as substrate and (B) methyl transfer reactions using avian nucleosomes as substrate. Symbols: wild type (•), Y641F (○), Y641H (□), Y641N (▪), and Y641S (▴). CPM is counts per minute, referring to scintillation counting as a result of 3H radiation. See Materials and Methods for further information.
There are several potential explanations for the discordant activity of the mutant PRC2 complexes on peptide and nucleosome substrates. One possibility is that substrate recognition sites distal to the enzyme active site (i.e., exosites) are important determinants of substrate binding and turnover; these sites would engage complementary recognition elements on the nucelosome that are not available on small peptidic substrates. However, when _Escherichia coli_-expressed, recombinant human histone H3 was tested as a substrate for the WT and mutant PRC2 complexes, the resulting pattern of activity was identical to that seen for the peptide substrate (see Supporting Information); that is, the WT enzyme demonstrated robust methyltransferase activity against the H3 substrate, the Y641F mutant showed 7% the activity of WT complex, and all other mutants displayed ≤ 1% the activity of WT complex. Hence, exosite engagement seems an unlikely explanation for the current results. The nucleosome presents many lysine residues beyond H3K27 as potential sites of methylation that would not be present in the small peptidic substrate. Thus, another possibility is that mutation of Y641 alters the substrate specificity of EZH2 to result in methylation of lysine residues other than H3K27. This possibility is unlikely given the excellent agreement between mutant activity on small peptide and recombinant H3 protein substrates. To evaluate this possibility further, we tested the enzymatic activity of the WT and mutant PRC2 complexes against a panel of peptidic substrates that represent all possible lysine residues of histone H3 and histone H4 (see Materials and Methods). All of the enzyme forms showed significant activity only against peptides containing the equivalent of residue H3K27. The specific activity of the mutants, however, was greatly reduced relative to WT in the order WT≫Y641F > Y641S ∼ Y641H > Y641N, again consistent with previous reported findings. The nucleosomes isolated from the avian natural source represent an admixture of states of histone modification, including various states of H3K27 methylations as judged by Western blotting with H3K27 methylation specific antibodies. Hence, another possibility is that the Y641 mutants of EZH2 display differential activity against different methylation states of H3K27.
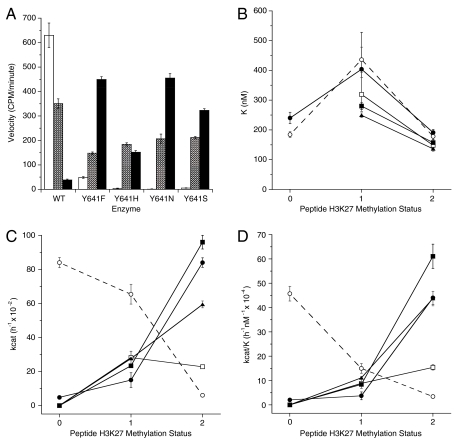
To understand further the enzymatic activity of these mutants, and to reconcile the apparent discrepancy between activity against peptidic and nucleosome substrates, we studied the ability of the enzyme forms to catalyze further methylation of various H3K27 methylation states in the context of the H3:21-44 peptide. As stated above, we found that all of the mutant enzymes were deficient catalysts of unmodified H3K27 peptide methylation, relative to the WT enzyme. To our surprise, however, all of the mutant enzymes were superior to the WT enzyme in catalyzing further methylation of the mono- and especially the dimethylated H3K27 peptides (Fig. 2). Thus, the data suggest that the WT enzyme is most efficient in catalyzing the zero to monomethylation reaction. The mutant enzymes are defective in catalyzing this initial step, but are more efficient than the WT enzyme in catalyzing the subsequent steps leading from monomethyl to di- and trimethyl H3K27.
Fig. 2.
PRC2 complexes containing mutant EZH2 preferentially catalyze di- and trimethylation of histone H3K27. (A) Methyltransferase activity of mutant and WT complexes on unmethylated peptide (open bars), monomethylated peptide (hatched bars), and dimethylated peptide (closed bars). (B) Affinity for peptide substrates as defined by _K_1/2 is similar across all peptide methylation states for PRC2 complexes containing wild-type (○),Y641F (•), Y641H (□), Y641N (▪), and Y641S (▴) EZH2. Note that the _K_1/2 values across all substrates and all enzyme forms varies less than 3.5-fold. For any particular methylation state of substrate, the variation in _K_1/2 value is less than 2-fold. (C) Enzyme turnover number (_k_cat) varies with substrate methylation status in opposing ways for WT and Y641 mutants of EZH2. The _k_cat decreases with increasing K27 methylation states for wild-type (○), but increases for Y641F (•), Y641H (□), Y641N (▪), and Y641S (▴) mutants of EZH2. (D) Catalytic efficiency (_k_cat/_K_1/2) decreases with increasing K27 methylation states for wild-type (○) but increases for Y641F (•), Y641H (□), Y641N (▪), and Y641S (▴) mutants of EZH2. (B_–_D) The lines drawn to connect the data points are not intended to imply any mathematical relationship; rather, they are intended merely to serve as a visual aid to guide the eye of the reader.
The origins of the differential substrate specificities of WT and mutant EZH2 were explored through steady-state enzyme kinetics. As summarized in Table 1, the mutations have minimal effects on ground-state substrate recognition, as demonstrated by the similar values of K m for nucleosome and of _K_1/2 for peptide substrates. In all cases the peptidic substrates displayed sigmoidal binding behavior; hence, the concentration of peptide resulting in half-maximal velocity is reported here as _K_1/2 instead of the more common Michaelis constant, K m (17). This sigmoidal behavior was seen only with peptidic substrates (i.e., nucleosome and recombinant histone substrates displayed classical Michaelis–Menten kinetics), and the origin of this effect is unclear at present. The SAM K m likewise displayed minimal variation among the enzyme forms, ranging from 208 ± 50 to 304 ± 64 nM. Instead, the differences in substrate utilization appear to have their origin in transition state recognition, as demonstrated by differences in _k_cat values among the enzymes for various substrates (Table 1); as a result, the catalytic efficiency, quantified as the ratio _k_cat/K (where K is either K m or _K_1/2, depending on substrate identity; see above), varies between the WT and mutant enzymes for different states of H3K27 methylation (Table 1).
Table 1.
Steady-state kinetic parameters for methylation reactions catalyzed by PRC2 containing wild-type or Y641 mutants of EZH2
| Enzyme | Substrate H3K27 methylation status | K, nM* | _k_cat, h-1 × 10-2 | _k_cat/K, h-1·nM-1 × 10-4 |
|---|---|---|---|---|
| WT | 0 | 184 ± 10 | 84.0 ± 3.0 | 45.7 ± 3.0 |
| 1 | 436 ± 42 | 65.4 ± 5.8 | 15.0 ± 2.0 | |
| 2 | 178 ± 16 | 6.0 ± 0.3 | 3.4 ± 0.3 | |
| Nucleosome | 141 ± 31 | 42.6 ± 2.6 | 30.2 ± 6.9 | |
| Y641F | 0 | 240 ± 19 | 4.8 ± 0.3 | 2.0 ± 0.2 |
| 1 | 404 ± 124 | 15.0 ± 4.3 | 3.7 ± 1.6 | |
| 2 | 191 ± 10 | 84.0 ± 2.8 | 44.0 ± 2.7 | |
| Nucleosome | 176 ± 19 | 65.4 ± 2.0 | 37.2 ± 4.2 | |
| Y641H | 0 | —† | — | — |
| 1 | 319 ± 57 | 28.2 ± 3.7 | 8.8 ± 2.0 | |
| 2 | 148 ± 9 | 22.8 ± 0.9 | 15.4 ± 1.1 | |
| Nucleosome | 140 ± 22 | 23.4 ± 1.0 | 16.7 ± 2.7 | |
| Y641N | 0 | — | — | — |
| 1 | 280 ± 11 | 23.4 ± 0.8 | 8.4 ± 0.4 | |
| 2 | 157 ± 11 | 96.0 ± 4.0 | 61.1 ± 5.0 | |
| Nucleosome | 191 ± 34 | 23.4 ± 1.3 | 12.3 ± 2.3 | |
| Y641S | 0 | — | — | — |
| 1 | 249 ± 8 | 27.6 ± 0.8 | 11.1 ± 0.5 | |
| 2 | 136 ± 8 | 59.4 ± 2.0 | 43.7 ± 3.0 | |
| Nucleosome | 137 ± 28 | 23.4 ± 1.4 | 17.1 ± 3.6 |
The mutation of Y641 to F, N, H, or S in EZH2 may facilitate multiple rounds of H3K27 methylation by impacting the H-bonding pattern and/or steric crowding in the active site of the enzyme–bisubstrate ternary complex, affecting the formation of a proper water channel for deprotonation of the reacting lysine (18). This inference is drawn by analogy to the crystallographic and molecular dynamic simulation results seen for tyrosine mutation in the related protein lysine methyltransferases LSMT, Dim-5, and SET7/9. For example, when tyrosine 245 of recombinant SET7/9 was mutated to alanine, a change in substrate specificity, similar to what is reported here for EZH2 Y641 mutations, was observed (19). The ability of the Y245A mutant SET7/9 to methylate an unmodified 20-residue peptide, representing the sequence surrounding H3K4, was reduced to ca. 20% of the WT enzyme (20). At the same time, the ability of the Y245A mutant to further methylate H3K4me1 and H3K4me2 peptides was greatly augmented (ca. 7-fold and 5-fold, respectively) relative to the WT enzyme. In contrast to the present results, however, mutation of SET7/9 Y245 to phenylalanine did not enhance mono- to dimethylation nor di- to trimethylation of the peptide; rather, the Y245F mutant displayed minimal catalytic activity for all peptidic substrates. Similarly, the wild-type enzyme G9a can dimethylate H3K9 but is unable to perform the di- to trimethylation reaction. Yet, when tyrosine 1067 of G9a (analogous to Y641 of EZH2) is mutated to phenylalanine, the enzyme now gains the ability to trimethylate H3K9 (21). The tolerance for multiple Y641 mutations in EZH2 suggests that a release of steric crowding may allow greater access for proper alignment of the larger dimethyl lysine as the substrate for the di- to trimethylation reaction. Crystallographic analysis of the protein methyltransferases SET7/9 and G9a reveals that the side chain hydroxyls of the active site tyrosine residues are involved in H-bonding interactions directly with the amine of the methyl-accepting lysine, or indirectly through an intervening water molecule. Although the larger active site of the Y641 mutants is favorable for di- and trimethylation, the loss of the tyrosine hydroxyl hydrogen bond acceptor may result in an unfavorable orientation of the active site for initial methyl transfer to the lysine amine. A complete understanding of the molecular basis for the dramatic reduction in the ability of these mutant enzymes to perform the initial methylation reaction will require additional study.
The implications of the present results for human disease are made clear by the data summarized in Table 1. Cells heterozygous for EZH2 would be expected to display a malignant phenotype, due to the efficient formation of H3K27me1 by the WT enzyme and the efficient, subsequent transition of this progenitor species to abnormally high levels of H3K27me3 by the mutant enzyme form(s). We note that H3K27me1 formation is not exclusively dependent on WT-EZH2 catalysis. Knockout studies of EZH2 and of another PRC2 subunit, EED, have demonstrated H3K27me1 formation can be catalyzed by PRC2 complexes containing either EZH2 or the related protein EZH1, as the catalytic subunit (22). Hence, catalytic coupling between the mutant EZH2 species and PRC2 complexes containing either WT-EZH2 or WT-EZH1 would suffice to augment H3K27me3 formation and thus produce the attendant malignant phenotype. Our data therefore suggest that the malignant phenotype of FL and DLBCL of the GCB subtype, associated with expression of mutant forms of EZH2 (1), is the result of an overall gain of function with respect to formation of the trimethylated form of H3K27.
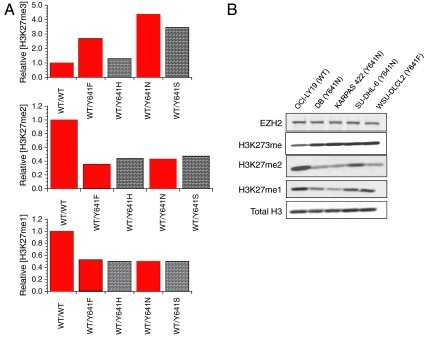
The steady-state kinetic parameters listed in Table 1 allow us to calculate the expected levels of different H3K27 methylation states for cells heterozygous for the various mutant EZH2 forms, relative to cells homozygous for the WT enzyme. To perform these simulations we made a number of simplifying assumptions: (i) that steady-state enzyme kinetics are relevant to PRC2-catalyzed H3K27 methylation in the cellular context and that all measurements are made at the same time point in cell growth; (ii) that the mutant and the WT enzyme are expressed at equal levels in heterozygous cells and that the total EZH2 level is equal in all cells; (iii) that the cellular concentration of SAM, relative to its K m, is saturating and does not change among the cells; (iv) that the cellular concentration of nucleosome is similar to its K m and likewise does not change among cells; (v) that EZH1 catalyzed methylation of H3K27 was insignificant and constant among the cells; and (vi) that any H3K27 demethylase activity was also constant among the cells. With these assumptions in place, we obtained the predictions illustrated in Fig. 3A for relative levels of H3K27me3 (Top), H3K27me2 (Middle), and H3K27me1 (Bottom). A clear pattern emerges from these simulations. The level of H3K27me3 increases relative to WT cells for all mutant-harboring cells, ranging from a 30% increase for the Y641H mutant to > 400% for the Y641N mutant. At the same time, the levels of H3K27me2 decrease to < 50% of WT for all of the mutants, and the levels of H3K27me1 are reduced by approximately half for all mutants, relative to WT. We then measured the relative levels of the H3K27 methylation states in B-cell lymphoma cell lines that are known to be homozygous for WT EZH2 (OCI-LY19) or heterozygous for EZH2 Y641N (DB, KARPAS 422, and SU-DHL-6) or EZH2 Y641F (WSU-DLCL2) by Western blotting (Fig. 3B). The pattern of relative H3K27 methylation states seen in Fig. 3B is in excellent agreement with the results of the simulations based on in vitro steady-state kinetic parameters, despite the crude assumptions used in the simulations and the use of a nonphysiological peptide surrogate as substrate. Thus, we observe increased H3K27me3 for all mutant-harboring cells relative to WT, decreased H3K27me2 for all mutant-harboring cells relative to WT, and decreased H3K27me1 for at least two of the four mutant cell lines. The near-comparable levels of H3K27me1 in WT and KARPAS 422 and SU-DHL-6 cells may reflect different expression levels of WT and mutant EZH2, different contributions of EZH1, or other factors not accounted for in our simulations. Nevertheless, the concordance between the predicted and experimental patterns of H3K27 methylation status is remarkable and supports our view that enzymatic coupling between WT and mutant EZH2 leads to increased H3K27me3, thus resulting in the malignant phenotype of cells that are heterozygous for these mutants.
Fig. 3.
Steady-state enzyme kinetics predicts the relative patterns of H3K27 methylation status in EZH2 WT and heterozygous mutant lymphoma cell lines. (A) Predicted relative levels of H3K27me3 (Top), H3K27me2 (Middle), and H3K27me1 (Bottom) for cells containing different EZH2 mutants. The simulations were performed using a coupled enzyme steady-state velocity equation (30) and the steady-state kinetic parameters reported in Table 1. All values are relative to the homozygous WT EZH2-containing cells and assume saturating concentrations of intracellular SAM, relative to K m and intracellular nucleosome concentrations similar to K m. Bars shown in red are meant to highlight WT and mutants for which experimental data are available for lymphoma cell lines in B. See text for further details. (B) Western blot analysis of relative patterns of H3K27 methylation status for lymphoma cell lines containing homozygous WT EZH2 or heterozygous for the indicated EZH2 Y641 mutation. (Top to Bottom) Results for probing with specific antibodies for the following: total EZH2 levels, H3K27me3, H3K27me2, H3K27me1, and total histone H3 as a loading control.
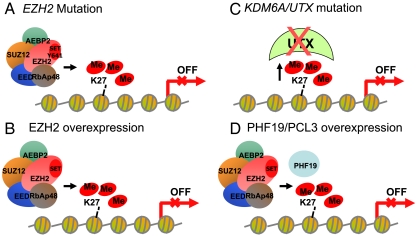
The foregoing interpretation of our data also helps to reconcile the existence of cancer-associated overexpression of EZH2 (3–6) or PRC2-associated proteins (e.g., PHF19/PCL3) (8–10) and also loss-of-function genotypes for the histone H3K27 demethylase UTX (7). Loss of UTX activity would be enzymatically equivalent to a gain of function for EZH2, in either situation resulting in greater steady-state levels of trimethylated H3K27 in cancer cells (Fig. 4). The mono-, di-, and trimethylation states of histone H3K27 are associated with different functions in transcriptional control. Histone H3K27 monomethylation is associated with active transcription of genes that are poised for transcription (23, 24). In contrast, trimethylation of histone H3K27 is associated with either transcriptionally repressed genes (23, 25) or genes that are poised for transcription when histone H3K4 trimethylation is in cis (26). Taken together, alterations in the PRC2 complex activity reported in cancer, including the Y641 mutation of EZH2, are predicted to result in an increase in the trimethylated state of histone H3K27 and to thus result in transcriptional repression.
Fig. 4.
Proposed mechanisms leading to aberrantly high levels of trimethylation on histone H3K27 in cancer include (A) mutation of Y641 in EZH2 resulting in a change in substrate preference from the nonmethylated to the mono- and dimethylated histone H3K27, (B) overexpression of EZH2, (C) mutations in UTX that inactivate enzyme function, causing a decrease in demethylation of H3K27me3, and (D) overexpression of the PRC2 complex subunit PHF19/PCL3 that leads to increases in recruitment of the PRC2 complex to specific genes and an increase in histone H3K27 trimethylation. In all four models the alteration leads to aberrant histone H3K27 trimethylation in the proximal promoter regions of genes resulting in transcriptional repression of key genes in cancer.
EZH2 and other protein methyltransferases have been suggested to be attractive targets for drug discovery (27–29). The present data also suggest an experimental strategy for development of FL and GCB lymphoma-specific drugs. As the differences in substrate recognition between the WT and disease-associated mutants derive from transition state interactions, small molecule inhibitors that selectively mimic the transition state of the mutant EZH2 over that of the WT enzyme should prove to be effective in blocking H3K27 methylation in mutation-bearing cells. Inhibitors of this type would be expected to display a large therapeutic index, as target-mediated toxicity would be minimal for any cells bearing only the WT enzyme. Transition state mimicry has proved to be an effective strategy for drug design in many disease areas (17, 30). Whether or not sufficient differentiation between the transition states of the WT and mutant EZH2 enzymes can be designed into a small molecule drug remains to be determined.
In summary, the present results point to a critical dependency on enzymatic coupling between enzymes that perform H3K27 monomethylation and the mutant forms of EZH2, for pathogenesis in FL and the GCB subtype of DLBCL. This is an example of a human disease that is dependent on such coupling of catalytic activity between normal (H3K27me1) and disease-associated mutant (H3K27me2/3) enzymes. We are unaware of other examples of this mechanism of pathogenesis in human disease.
Materials and Methods
Recombinant 5-Component PRC2.
Wild-type EZH2 (NM_004456) or Tyr641 mutants were coexpressed with wild-type AEBP2 (NM_153207), EED (NM_003797), SUZ12 (NM_015355), and RbAp48 (NM_005610) in Spodoptera frugiperda (Sf9) cells using a baculovirus expression system. An N-terminal FLAG tag on the EED was used to purify active PRC2 complex from cell lysates (BPS Bioscience, catalog number 51004). The purity of the final PRC2 preparations was assessed by SDS-PAGE with Coomassie blue staining.
Recombinant Human Histone H3.
Recombinant human histone H3.1, expressed in and purified from E. coli (catalog number M250S) was purchased from New England BioLabs, Inc.
H3, H4 Peptide Panel.
A library consisting of 44 peptides of 15 amino acids each was synthesized by 21st Century Biochemicals. This peptide panel encompassed all of the amino acids of human histones H3 and H4 with 10 residue overlaps between consecutive peptide sequences. The N terminus of each peptide was appended with biotin, and the C termini were represented as the amide. Purity (> 95%) and identity were confirmed by liquid chromatography/mass spectral analysis.
For study of the H3K27 methylation status dependence of enzyme activity, peptides were synthesized representing the amino acid sequence of human H3 from residues 21–44 (H3:21–44) with lysine 27 represented as the unmodified, monomethylated, dimethylated, or trimethylated side chain amine. These peptides were purchased from New England Peptide with biotin appended to the C terminus of each peptide.
In Vitro Assays of PRC2 Methyltransferase Activity.
Flashplate assay with peptide substrate.
For initial comparison of WT and Y641 mutants of EZH2, biotinylated histone H3:21-44 peptide containing unmethylated K27 (New England Peptide), monomethylated K27 (Millipore), or dimethylated K27 (Millipore) at a concentration of 800 nM was combined with a mixture of SAM at 1,700 nM, and 300 nM tritiated SAM (PerkinElmer, Inc.). This substrate combination was then added to the PRC2 in assay buffer [20 mM N,_N_-bis(2-hydroxyethyl)glycine (Bicine), 1 mM DTT, 0.002% Tween20, 0.005% bovine skin gelatin (BSG), pH 7.6]. Reactions were allowed to proceed for the indicated time interval and then quenched by addition of excess cold SAM (600-μM final concentration). Quenched reaction mixtures were transferred to a streptavidin-coated Flashplate (PerkinElmer, catalog number SMP410), allowed to bind for 1 h, and then detected on a TopCount NXT HTS (PerkinElmer). Each time point represents the average of six individual reactions. Steady-state kinetic parameters were determined under identical reaction conditions except that the concentration of peptide or SAM was varied, while at saturating conditions of the other substrate. Velocity was plotted as a function of varied substrate concentration and the data were fitted to the untransformed version of the Michaelis–Menten equation or the untransformed version of a sigmoidal kinetic equation (30) to calculate values of K and _k_cat (30). Standard errors of fitted parameters are listed in Table 1 and were used to construct the error bars illustrated in Fig. 2 B and C. Errors associated with _k_cat/K (Table 1) were calculated according to standard methods of error propogation; the fractional error of _k_cat/K was determined as
 |
[1] |
|---|
where _μk_cat is the standard error of _k_cat and μK is the standard error of K.
Filterplate assay with histone and oligonucleosome.
Chicken erythrocyte oligonucleosomes were purified as previously described (31). Nucleosomes or histones were combined with a mixture of SAM and tritiated SAM and added to PRC2 in assay buffer (20 mM Bicine, 100 mM KCl, 1 mM DTT, 0.002% Tween20, 0.005% BSG, pH 7.6). Reactions were run and quenched as above. Quenched reaction mixture was transferred to a glass fiber filterplate (Millipore, catalog number MSFBN6B) and washed three times with 10% trichloroacetic acid and allowed to dry. Microscint Zero (30 μL) was added and tritium incorporation was detected on a TopCount. Steady-state parameters for reactions with nucelosomes were determined under identical reaction conditions except that the concentration of nucleosome or SAM was varied while at saturating conditions of the other substrate. Velocity was plotted as a function of varied substrate concentration and fitted to the untransformed version of the Michaelis–Menten equation to derive the values of K m and _k_cat as described above.
Evaluation of H3K27 methylation status in cells.
The cell lines OCI-LY19 (ACC 528), KARPAS-422 (ACC 32), and WSU-DLCL2 (ACC 575) were obtained from DSMZ. The cell lines DB (CRL-2289) and SU-DHL2 (CRL-2959) were obtained from ATCC. OCI-LY19, WSU-DLCL2, and DB cell lines were grown in RPMI-1640 with 10% FBS, and KARPAS-422 and SU-DHL2 cell lines were grown in RPMI medium 1640 plus 20% FBS. Cells were grown to a density of 1.5–2 × 106 cells/mL and 1 × 107 cells were harvested by centrifugation at 264 × g, washed in ice-cold PBS, and lysed by resuspension in a 10× pellet volume of RIPA lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.25% deoxycholate, 1% NP-40, and 1mM EDTA (Millipore #20-188), plus 0.1% SDS and protease inhibitor tablets (Roche # 1836153). Lysates were sonicated by two rounds of ten 1-s bursts at setting 3 with a Misonix XL-2000 to ensure efficient histone extraction, and cleared by centrifugation at 4° using a bench top centrifuge at 14,000 rpm for 10 min. Protein concentration was determined by bicinchoninic acid assay (Pierce). Four micrograms of each lysate was fractionated on 4–20% Tris-Glycine gel (Invitrogen), transferred to PVDF, and probed with the following antibodies in Odyssey blocking buffer: mouse anti-EZH2 (CST 3147; 1∶2,000 dilution), rabbit anti-H3K27me3 (CST 9733; 1∶10,000 dilution), rabbit anti-H3K27me2 (CST 9755; 1∶5,000 dilution), rabbit anti-H3K27me1 (Active Motif 39377; 1∶5,000 dilution), and mouse antiTotal H3 (CST 3638; 1∶20,000 dilution). Following primary Ab incubation, membranes were probed with IRDye 800CW donkey-anti-mouse IgG (LiCOR #926-32212) or Alexa Fluor 680 goat-anti-rabbit IgG (Invitrogen #A-21076) secondary Ab and imaged using the LiCOR Odyssey system.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Dr. Robert Gould and Professor Christopher T. Walsh for helpful suggestions regarding this manuscript, and Professors Gregg Morin and Marco Marra for access to the recombinant wild-type and mutant PRC2 complexes and useful discussions.
Footnotes
Conflict of interest statement: All authors are employees and stockholders of Epizyme, Inc.
This article is a PNAS Direct Submission.
References
- 1.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Garcia E, Licht JD. Deregulation of H3K27 methylation in cancer. Nat Genet. 2010;42:100–101. doi: 10.1038/ng0210-100. [DOI] [PubMed] [Google Scholar]
- 3.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 6.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 7.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Robertson GP, Zhu J. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343:69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Cao R, et al. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaknovich R, et al. DNA methylation signatures define molecular subtypes of diffuse large B cell lymphoma. Blood. 2010 doi: 10.1182/blood-2010-05-285320. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss TJ, Wallrath LL. Connections between epigenetic gene silencing and human disease. Mutat Res. 2007;618:163–174. doi: 10.1016/j.mrfmmm.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Subero JI, et al. New insights into the biology and origin of mature aggressive B-cell lymphomas by combined epigenomic, genomic, and transcriptional profiling. Blood. 2009;113:2488–2497. doi: 10.1182/blood-2008-04-152900. [DOI] [PubMed] [Google Scholar]
- 14.Velichutina I, Shaknovich R, Geng H, Melnick A, Elemento O. EZH2 mediates DNA methylation-independent epigenetic silencing of a germinal center specific transcriptional program that contributes to cellular proliferation and lymphomagenesis; Proceedings of the 51st American Society of Hematology Annual Meeting and Exposition; Washington, DC: American Society of Hematology; 2009. A25356. [Google Scholar]
- 15.van Kemenade FJ, et al. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97:3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- 16.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery. A Guide for Medicinal Chemists and Pharmacologists. Hoboken, NJ: Wiley; 2005. [PubMed] [Google Scholar]
- 18.Zhang X, Bruice TC. Enzymatic mechanism and product specificity of SET-domain protein lysine methyltransferases. Proc Natl Acad Sci USA. 2008;105:5728–5732. doi: 10.1073/pnas.0801788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: Protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao B, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 21.Wu H, et al. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 27.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 28.Copeland RA, Olhava EJ, Porter Scott M. Targeting epigenetic enzymes for drug discovery. Curr Opin Chem Biol. 2010;14:505–510. doi: 10.1016/j.cbpa.2010.06.174. [DOI] [PubMed] [Google Scholar]
- 29.Pollock RM, Richon VM. Epigenetic approaches to cancer therapy. Drug Discov Today. 2010;6:71–79. [Google Scholar]
- 30.Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism and Data Analysis. 2nd Ed. Hoboken, NJ: Wiley; 2000. [Google Scholar]
- 31.Fang J, Wang H, Zhang Y. Purification of histone methyltransferases from HeLa cells. Methods Enzymol. 2004;377:213–226. doi: 10.1016/S0076-6879(03)77012-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information