Differentiation-specific histone modifications reveal dynamic chromatin interactions and alternative partners for the intestinal transcription factor CDX2 (original) (raw)
. Author manuscript; available in PMC: 2011 Nov 16.
SUMMARY
Cell differentiation requires remodeling of tissue-specific gene loci and activities of key transcriptional regulators, which are recognized for their dominant control over cellular programs. Using epigenomic methods, we characterized enhancer elements specifically modified in differentiating intestinal epithelial cells and found enrichment of transcription factor-binding motifs corresponding to CDX2, a critical regulator of the intestine. Directed investigation revealed surprising lability in CDX2 occupancy of the genome, with redistribution from hundreds of sites occupied only in proliferating cells to thousands of new sites in differentiated cells. Knockout mice confirmed distinct Cdx2 requirements in dividing and mature adult intestinal cells, including responsibility for the active enhancer configuration associated with maturity. Dynamic CDX2 occupancy corresponds with condition-specific gene expression and, importantly, to differential co-occupancy with other tissue-restricted transcription factors such as GATA6 and HNF4A. These results reveal dynamic, context-specific functions and mechanisms of a prominent transcriptional regulator within a cell lineage.
INTRODUCTION
Thousands of transcripts are coordinately regulated in differentiating cells, in part as a result of stable, heritable, cell type-specific changes in chromatin structure and in part through the actions of selected transcription factors (TFs) at many gene loci (Struhl, 1999). TFs that are highly restricted in their expression, regulate large numbers of genes within a cell lineage, and may hence confer a cell’s distinctive properties, are often considered “master regulators.” Examples of such TFs include PHA4 in the worm pharynx (Gaudet and Mango, 2002) and mammalian myogenic basic helix-loop-helix proteins (Molkentin and Olson, 1996). Although these TFs may function in both progenitor and terminally differentiated cells of a lineage, their interactions with alternative chromatin states are insufficiently characterized. Nor is it clear if they occupy _cis_-regulatory DNA elements stably throughout differentiation and whether distinct TF combinations control genes in different cell states within a lineage. We addressed these questions on a genome scale in the context of intestinal epithelial cells.
The adult intestine is a site of continual cell differentiation (Potten, 1998). Stem and dividing progenitor cells in the small bowel mucosa are confined to the crypts of Lieberkühn, whereas the differentiated post-mitotic cells that serve essential absorptive and secretory functions reside along villous projections. Gene expression differs substantially in crypts and villi (Tremblay et al., 2006) and, as with other self-renewing tissues, constitutive proliferation and arrested differentiation underlie intestinal tumorigenesis (van de Wetering et al., 2002). Gut epithelial cells hence serve as an excellent model to investigate gene regulation during differentiation.
Recent delineation of chromatin modifications at activated gene loci provides new tools to investigate the epigenomic basis of cell differentiation (Barski et al., 2007; Bernstein et al., 2006; Pokholok et al., 2005). Mono- and di-methylation of Lys4 (K4Me2) and acetylation of Lys27 (K27Ac) residues on Histone 3 (H3) are particularly associated with distant transcriptional enhancers and gene activation in cultured mammalian cells (Bernstein et al., 2005; He et al., 2010; Heintzman et al., 2009). Monitoring these chromatin modifications as cells transition from one state to another could identify the _cis_-elements responsible for differentiation in individual tissues, and characterization of such elements should help elucidate underlying transcriptional mechanisms. To identify _cis_-regulatory elements that are differentially active in proliferating and terminally mature intestinal epithelial cells, we mapped genome-wide histone H3 modifications. Our subsequent detailed analysis uncovered a prominent role and mechanisms for the homeodomain protein CDX2.
CDX2 expression is restricted at different developmental stages and its ability to specify cell fates or axial position in each context makes it a strong candidate “master regulator.” Expression of Cdx2 in mouse embryonic stem cells induces trophoblasts (Niwa et al., 2005) and expression in stomach or esophageal cells confers intestinal properties (Liu et al., 2007; Silberg et al., 2002). Conversely, Cdx2 loss in mouse embryos disrupts trophoblast, axial skeleton, and intestine development (Chawengsaksophak et al., 1997; Gao et al., 2009; Grainger et al., 2010). In adult mammals, Cdx2 expression is confined to the intestinal epithelium, where it is expressed in both progenitor and differentiated cells (James et al., 1994; Silberg et al., 2000). To examine if CDX2 behaves as a key regulator and to investigate its potentially diverse functions in dividing and mature cells, we assessed CDX2 binding properties during differentiation. We find that CDX2 interactions with the genome and with other sequence-specific TFs are surprisingly fluid; it interacts dynamically with active chromatin and co-occupies DNA differentially with other intestinal TFs, GATA6 or HNF4A, to regulate distinct genes in dividing and differentiated intestinal cells, respectively. A new conditional knockout mouse line revealed distinct Cdx2 functions in progenitor and differentiated cells in vivo and its requirement for differentiation-specific chromatin modifications. These results illustrate the dynamic functions and mechanisms of a critical TF in distinct cellular contexts within a continually differentiating tissue.
RESULTS
Epigenomic analysis of intestinal epithelial cells implicates CDX2 in cell maturation
Caco-2 human intestinal cells are widely used to investigate epithelial functions, intestinal gene expression, and transcriptional mechanisms of differentiation (Fleet et al., 2003; Halbleib et al., 2007; Soutoglou and Talianidis, 2002). These cells proliferate rapidly in sparse cultures but stop dividing at confluence (Fig. 1A) and develop morphologic features of mature enterocytes; the transcriptional changes that accompany cell differentiation mirror differences in gene expression between intestinal crypts and villi (Saaf et al., 2007; Tremblay et al., 2006). Accordingly, they serve as an ideal model to study pivotal cellular transitions. To identify _cis_-regulatory sequences associated with Caco-2 cell differentiation, we evaluated a distinct pattern of histone modifications attributed to active enhancer elements (He et al., 2010; Heintzman et al., 2009): abundant H3K4Me2 and H3K27Ac marks, with depletion of labile nucleosomes within an altered chromatin configuration (Fig. 1B). We digested chromatin from proliferating and post-confluent Caco-2 cells with micrococcal nuclease (MNase), precipitated the mononucleosome fraction with H3K4Me2- or H3K27Ac antibodies (Ab), and characterized the chromatin immunoprecipitates (ChIP) using Illumina high-throughput sequencing (ChIP-seq). Subsequent analysis of the two H3 modifications gave similar results (Pearson correlation coefficient 0.4 (_P_-value <10e−22), Supplemental Fig. S1); for simplicity, here we show the analysis of H3K4Me2 ChIP.
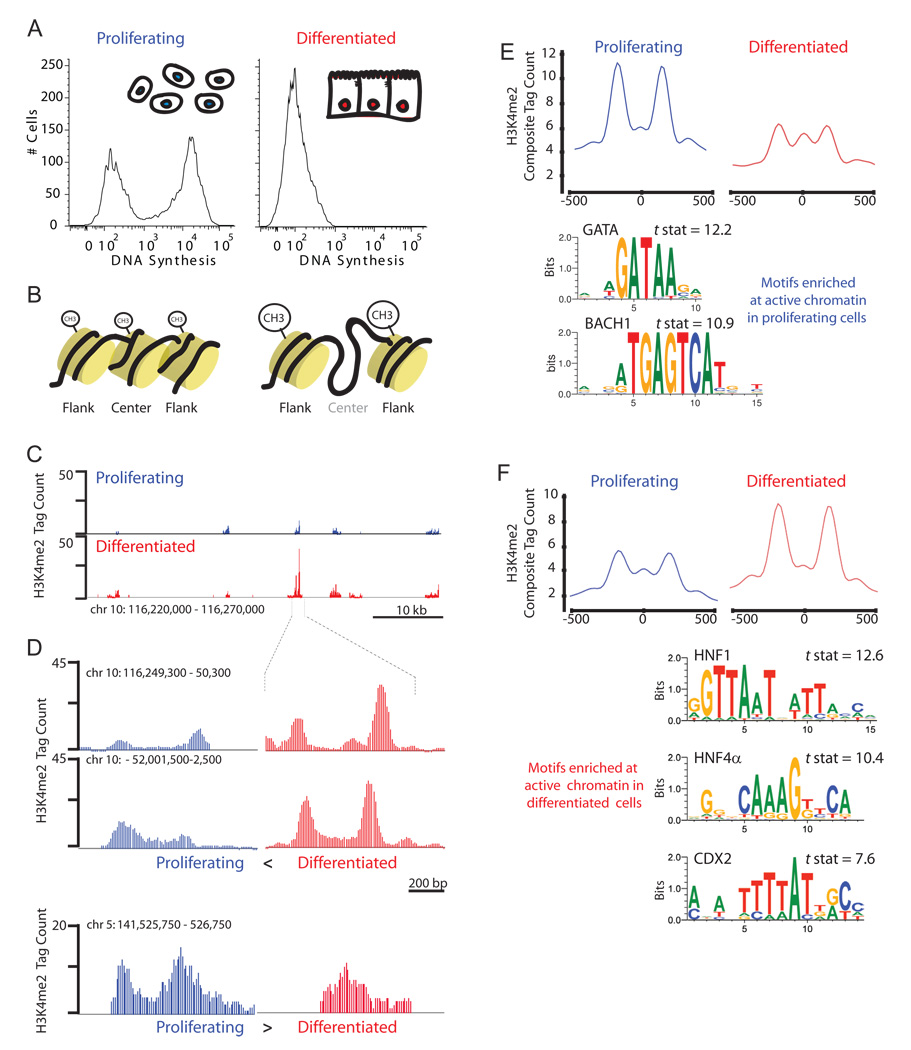
Figure 1. An epigenomic screen identifies CDX2 as a candidate regulator of intestinal epithelial maturation.
A, Caco-2 cells show progenitor properties while proliferating (left); after reaching confluence, the cells differentiate to resemble mature intestinal villus enterocytes (right). Histograms depict EdU incorporation, a marker of DNA replication, 1 h after exposure, as measured by fluorescence intensity. Diagrams in the upper right represent cell morphologies. B, Chromatin modifications associated with nascent (left) and active (right) enhancers are depicted, the latter characterized by relative increase in H3K4Me2 on flanking nucleosomes and depletion of signal in the nucleosome in between. C, H3K4Me2 ChIP-seq results in proliferating and differentiated Caco-2 cells at a representative region on chromosome 10. D, H3K4Me2 ChIP-seq tag counts across indicated 1-kb intervals in proliferating (left, blue) and differentiated (right, red) cells, including high- resolution image of the data shown in C. The top 2 panels show gain of active chromatin structure upon differentiation; the lower panel shows a region selectively active earlier in proliferating cells. E, Composite plots of H3K4Me2 ChIP-seq tag counts from the 1,000 regions with the greatest differential chromatin structure in proliferating cells (left, blue) and the same 1,000 regions in differentiated cells (right, red), aligned at center nucleosomes. TF motifs most significantly over-represented at the valleys of the composite plots are shown (details in Experimental Procedures). A complete list of identified motifs is found in Table S1. F, The same analysis as in E, but on the 1,000 regions with the greatest differential chromatin structure in differentiated cells.
We analyzed 14.5 million and 13 million mappable sequence tags for H3K4Me2 ChIP from proliferating and mature cells, respectively, using published methods (He et al., 2010), and identified 219,266 and 208,762 positioned nucleosomes under the two conditions (Supplemental Fig. S2). H3K4Me2 marks were restricted either to transcriptional start sites (TSSs) or to dispersed short intergenic and intronic regions, with little background signal across the genome (examples in Fig. 1C). Approximately 60% of nucleosomes bearing the H3K4Me2 mark in each condition were present more than 5 kb from a TSS and hence represent putative distant _cis_-regulatory elements. We focused on these distal elements because previous work indicates that enhanced H3K4Me2 signals at flanking nucleosomes and a corresponding diminution of intervening signal typifies sites where nucleosomes are depleted or destabilized to allow TF binding (He et al., 2010; Heintzman et al., 2007; Jin et al., 2009). A computational method to infer increased H3K4Me2 signals at flanking nucleosomes relative to the intervening region uncovered thousands of individual sites that acquire or lose this signature during Caco-2 cell differentiation (sample tracings in Fig. 1D). We used the magnitude of difference in signal between replicating and mature cells to assign Nucleosome Stability-Destability (NSD) scores (He et al., 2010) and to rank genomic regions. The resulting patterns, displayed in composite plots of 1,000 sites with the highest NSD scores in proliferating (Fig. 1E) and differentiated (Fig. 1F) cells, underscore the considerable genome-wide modulation of chromatin during cell differentiation.
To identify proteins that might associate with labile regions, we searched for TF-binding sequence motifs that are over-represented at the centers of the most differentially modified chromatin domains (Supplemental Table S1). In regions active exclusively in proliferating cells, the most enriched motif matched the GATA protein family (Fig. 1E). Motifs enriched in chromatin that is active mainly in differentiated cells corresponded to the transcription factors HNF1, HNF4A and CDX2 (Fig. 1F, Supplemental Table S1), all of which are tissue-restricted proteins previously implicated in controlling genes expressed in mature enterocytes (Beck, 2004; D'Angelo et al., 2010; Mouchel et al., 2004; Stegmann et al., 2006). The selective over-representation of these sequences within differentially active enhancers reinforces the idea that NSD scores signify a regulatory function.
In vitro studies suggest that CDX2 controls intestinal genes like SI, LCT and CDH17 through their proximal promoters (Fang et al., 2000; Hinoi et al., 2002; Traber and Silberg, 1996), contributing to the idea of a “master” function. Indeed, Cdx2 is required to specify embryonic intestinal epithelium in vivo (Gao et al., 2009; Grainger et al., 2010) but in the adult intestine it is abundantly and equally expressed in crypt progenitors and differentiated villus cells (Silberg et al., 2000) and its function is unknown. The enrichment of CDX2 motifs at differentiation-associated enhancers suggested broad and distinct functions in controlling adult intestinal differentiation through distant _cis_-elements and provided fresh impetus to study its mechanisms.
CDX2 interacts dynamically with the genome of differentiating cells
CDX2 is expressed in both crypt and villus cells (Silberg et al., 2000) and could in principle regulate different loci in these two contexts. To verify CDX2 binding at enhancer regions, as the H3K4Me2 and H3K27Ac ChIP results suggested, and to determine if CDX2 interacts differently with the genome over the course of differentiation, we used ChIP-seq to identify CDX2 binding sites in Caco-2 cells. We detected CDX2 occupancy with high confidence (p ≤10−10) at 3,122 regions in sub-confluent, proliferating cells and at 16,198 sites in terminally differentiated cells (Fig. 2A). 679 sites were unique to proliferating cells and 13,755 were unique to differentiated cells (examples in Fig. 2B and Supplemental Fig. S3). CDX2-bound regions in both states were strongly enriched for the consensus CDX2 recognition motif and showed high, centered evolutionary conservation; most sites were far from TSSs (Supplemental Fig. S4), similar to findings with other TFs (Carroll et al., 2006). Thus, although CDX2 binds many regions common to the two states, its occupancy across the genome is surprisingly fluid, with hundreds of distinct “early” binding sites specific to proliferating cells thousands of “late” sites specific to mature cells. As CDX2 protein levels increase no more than 2- to 3-fold in differentiated cells (data not shown), changes in CDX2 binding are unlikely to reflect only the protein concentration; furthermore, occupancy at many early sites is selectively diminished in mature cells.
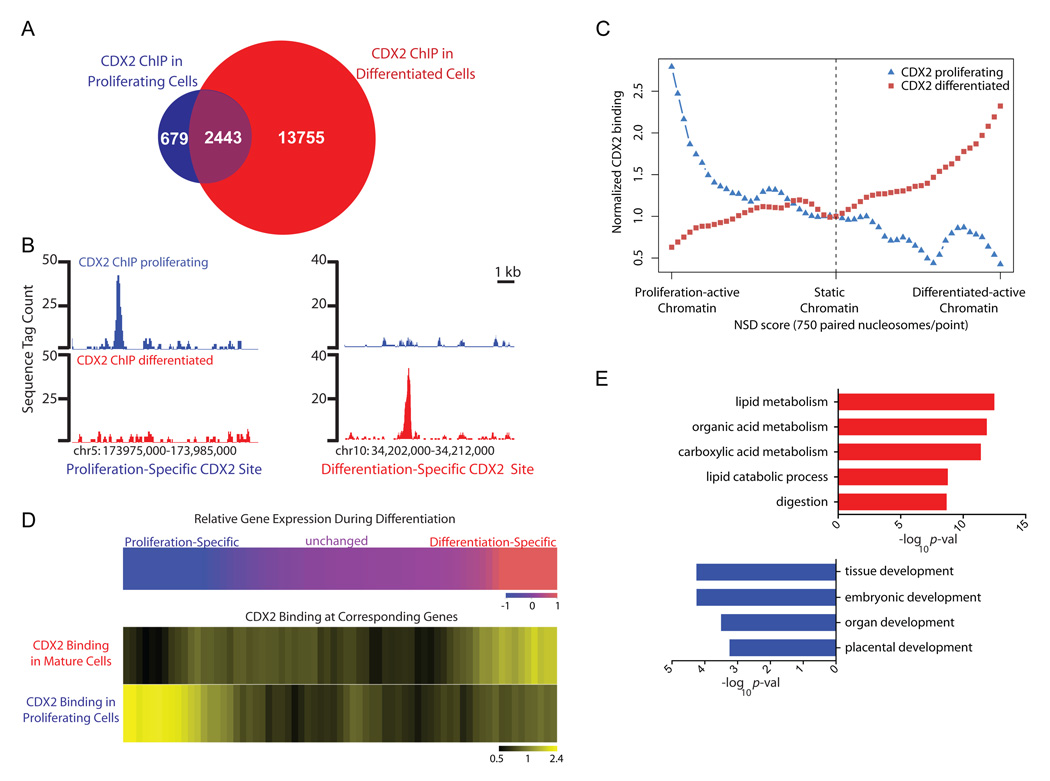
Figure 2. CDX2 interacts dynamically with the genome during intestinal cell differentiation.
A, Venn diagram representation of binding sites identified by CDX2 ChIP-seq in proliferating and mature Caco-2 cells. B, Histograms of normalized CDX2 ChIP-seq tags at representative sites bound exclusively early in proliferating (left) or late in differentiated cells (right). Additional examples appear in Figure S3. C, Condition-specific CDX2 binding corresponds to condition-specific active chromatin. H3K4Me2-marked putative _cis_-elements were ranked from most differentially active chromatin signatures in proliferating (left) to differentiated cells (right) and plotted along the x-axis in bins of 750. CDX2 occupancy was then plotted relative to occupancy in areas where active chromatin did not vary (center). CDX2 binding sites specific to proliferating (blue triangles) or differentiated cells (red squares) correlated with proliferating and differentiated cell-specific active chromatin, respectively. D, Heat maps demonstrating correlation of stage-specific CDX2 occupancy with stage-specific gene expression. Top: Early (left, blue) versus late (right, red) genes were ranked and binned in groups of 100; genes with unaltered expression are represented in the center. The color map (blue - red) presents the scale of log2 fold change (−1 to +1) in gene expression levels in the two stages. Bottom: The frequency of condition-specific CDX2 binding within 100 kb of each gene expression bin is indicated by the intensity of yellow shading. Numbers below the color map correspond to the minimum, mean, and maximum ratios (0.5, 1, and 2.4, respectively) between the average number of CDX2 binding sites within 100kb from the genes in the same bin over the average number of binding sites near all genes. E, Gene Ontology term analysis reveals that expressed genes with condition-specific CDX2 binding match functions classically attributed to differentiated (red) or proliferating (blue) cells. Complete analysis is found in Table S2.
To test the significance of condition-specific CDX2 occupancy, first we defined condition-specific binding rigorously, considering only high-stringency sites from one condition (p ≤10−10) that were unoccupied in the other, even at lower stringency (_p_≤10−3, Supplemental Fig. S3). To determine how these high-confidence, condition-specific CDX2 sites relate to H3K4Me2 modifications, we mapped binding sites onto the genome-wide distribution of nucleosomes that are differentially marked in replicating and mature Caco-2 cells. Early CDX2 binding occurred more frequently at enhancers that carry the active chromatin signature in proliferating cells, whereas late CDX2 binding predominated in regions of active chromatin in mature cells (Fig. 2C). CDX2 occupancy at early sites was also strongly associated with transcripts whose expression is enriched selectively in dividing Caco-2 cells (p<7.3e−6, Fisher’s exact test), whereas late CDX2 binding correlated better, and strongly, with transcripts that increase in differentiated cells (p<1.6e−23, Fig. 2D). Moreover, loci selectively occupied and expressed in dividing cells were enriched for Gene Ontology (GO) functions related to progenitors, including Embryonic, Organ and Tissue Development, whereas GO terms associated with differentiated enterocytes, such as Lipid Metabolism and Digestion, predominated among genes selectively occupied by CDX2 and expressed in terminally mature cells (Fig. 2E, Supplemental Table S2). Thus, labile CDX2 interactions with distant _cis_-elements in dividing and mature intestinal cells have observable counterparts in differential chromatin structure and gene expression. These results provided strong rationale to investigate the dynamic actions of a transcriptional regulator in the course of cell differentiation.
Cdx2 is required for both intestinal epithelial differentiation and proliferation in vivo
Whereas knockout mice have revealed Cdx2 functions in specifying fetal intestinal epithelium (Gao et al., 2009), its roles in the adult organ remain untested in vivo. To circumvent neonatal lethality associated with embryonic Cdx2 deficiency, we generated a new conditional null allele, Cdx2fV, with LoxP sites flanking exon 2 (Supplemental Fig. S5A–F), and crossed targeted mice with the intestine-specific, tamoxifen-inducible Cre recombinase-expressing transgenic strain Villin-CreER(T2) (el Marjou et al., 2004). Tamoxifen-treated adult Cdx2fV/fV;Villin-CreER(T2) mice showed loss of Cdx2 protein in >99% of small intestine villi and absence of Cdx2 mRNA (Fig. 3A,B; Supplemental Fig. S5G–H); expression of putative Cdx2 target genes, Sucrase Isomaltase and Intestine Specific Homeobox (Choi et al., 2006; Traber and Silberg, 1996), was profoundly reduced (Fig. 3B). These mice developed chronic diarrhea, rapidly lost weight (Fig. 3C) and became moribund, requiring humane euthanasia within 21 days. At necropsy they uniformly showed small intestine dilation and a dilated, fluid-filled cecum (Fig. 3D). Tissue architecture was overtly preserved in Cdx2fV/fV;Villin-CreER(T2) intestines (Fig. 3A,E). However, alkaline phosphatase activity was notably reduced along a proximal-distal gradient in mutant small bowel compared to tamoxifen-treated sibling controls (Fig. 3F), indicating enterocyte immaturity; enzyme activity was lowest in the ileum, where Cdx2 levels are the highest in wild-type mice (Silberg et al., 2000). The numbers of periodic acid Schiff-stained goblet (increased 2.1%, Fig. 3G), lysozyme+ Paneth (unchanged, Fig. 3H) and ChromograninA+ endocrine cells (decreased by 40%, _P_=0.0895, Fig. 3I) were altered subtly or not at all.
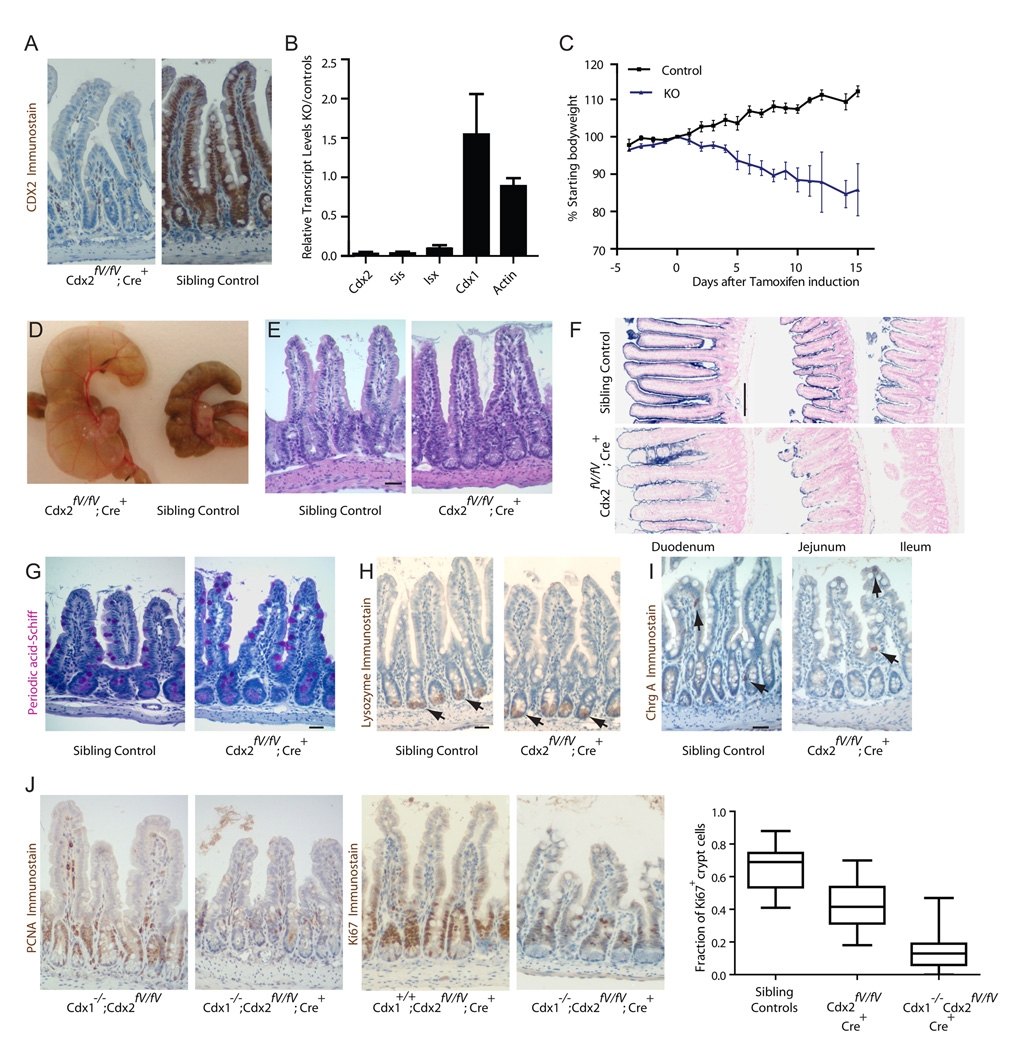
Figure 3. Cdx2 is required for proliferation and differentiation of the adult mouse intestinal epithelium.
A, Immunostaining verifies absence of intestinal Cdx2 expression in tamoxifen-treated Cdx2fV/fV;Villin-CreER(T2) mice (also see Supplemental Fig. S5). B, Reduced mRNA levels of Cdx2 and candidate target genes Sis and Isx in tamoxifen-treated Cdx2fV/fV;Villin-CreER(T2) intestines; Cdx1 transcript levels are unaffected. C, Cdx2fV/fV;CreER(T2) mice rapidly lose weight after tamoxifen treatment. D, Intestines of Cdx2fV/fV;Villin-CreER(T2) mice are dilated and the cecum is particularly engorged with fluid. E, H&E staining reveals intact tissue architecture in the Cdx2 mutant ileum. F, A profound deficiency is evident in Alkaline Phosphatase activity, a marker of enterocytes, especially in the distal small intestine. G–I, Periodic acid-Schiff (G), Lysozyme (H) and ChromograninA (I) were used as markers of goblet, Paneth, and enteroendocrine cells, respectively, in the ileum. J, In contrast to the respective single gene mutants, Cdx1−/−;Cdx2fV/fV;Villin-CreER(T2) intestinal crypts show markedly reduced expression of the proliferative markers Ki67 and Pcna. Quantitation of the Ki67+ fraction of crypt cells showed significant reduction in the double mutant (P<0.0001; two tailed t-test). Box plot indicate median Ki67+ counts per crypt, with lower and upper quartiles and whiskers indicating minimum and maximum counts. Error bars are SEM for B and C. Scale bars, 33.3 µm except for F, 200uM.
Although cell state-specific profiles in Caco-2 cells revealed distinct CDX2-occupied sites in proliferating and mature cells (Fig. 2A), the defects in adult Cdx2fV/fV;Villin-CreER(T2) intestines seemed confined to villus enterocytes. Crypt progenitors were also overtly normal and measures of cell proliferation - expression of Ki67 or Proliferating Cell Nuclear Antigen (Pcna) - were intact (Fig. 3J). Because Cdx1, a homologous factor normally expressed in crypt progenitors (Silberg et al., 2000; Subramanian et al., 1998) and present in tamoxifen-exposed mutant intestines (Fig. 3B), might compensate for Cdx2 loss in dividing cells, we crossed mice carrying the conditional Cdx2fV allele with Cdx1−/− mice, which have skeletal but no intestinal defects (Subramanian et al., 1995). Tamoxifen-treated Cdx1−/−;Cdx2fV/fV;Villin-CreER(T2) mice became markedly sicker than mice lacking only Cdx2 and survived fewer than 8 days after the first dose of hormone. Villi were stunted compared to single mutant littermates and the proliferation markers Ki67 and Pcna were expressed in significantly fewer crypt cells (Fig. 3J), revealing a redundant requirement for Cdx proteins in adult gut epithelial cell proliferation.
Consistent with the histologic analysis, mRNA profiling of isolated Cdx2fV/fV;Villin-CreER(T2) jejunal epithelium revealed lower levels of 991 transcripts (dChip analysis (Li and Wong, 2001), p<0.01) and Gene Ontology term analysis of affected genes pointed overwhelmingly to mature enterocyte functions, including solute transport and lipid and organic acid metabolism (Fig. 4A). Death from starvation and diarrhea in the face of overtly intact intestinal villus structure is best explained by malabsorption. Indeed, enterocyte products required for terminal digestion and nutrient absorption were prominent among transcripts that drop after tamoxifen treatment (Supplemental Table S3). Representative examples such as Dpp4, Lipe, and Abca1 showed lower expression throughout the small intestine (Fig. 4B). Thus, in agreement with the functions suggested by CDX2 occupancy in differentiated Caco-2 cells (Fig. 2E), Cdx2 is necessary for survival of adult mice and its absence profoundly affects gene expression in mature enterocytes. Moreover, genes affected by Cdx2 loss in mice are enriched in CDX2 occupancy near their human orthologs in differentiated Caco-2 cells, as shown in a representative example (Fig. 4C) and whole-genome analysis (Fig. 4D). These data further justify the use of Caco-2 cells to study gut epithelial differentiation and indicate conservation of Cdx2 function across species.
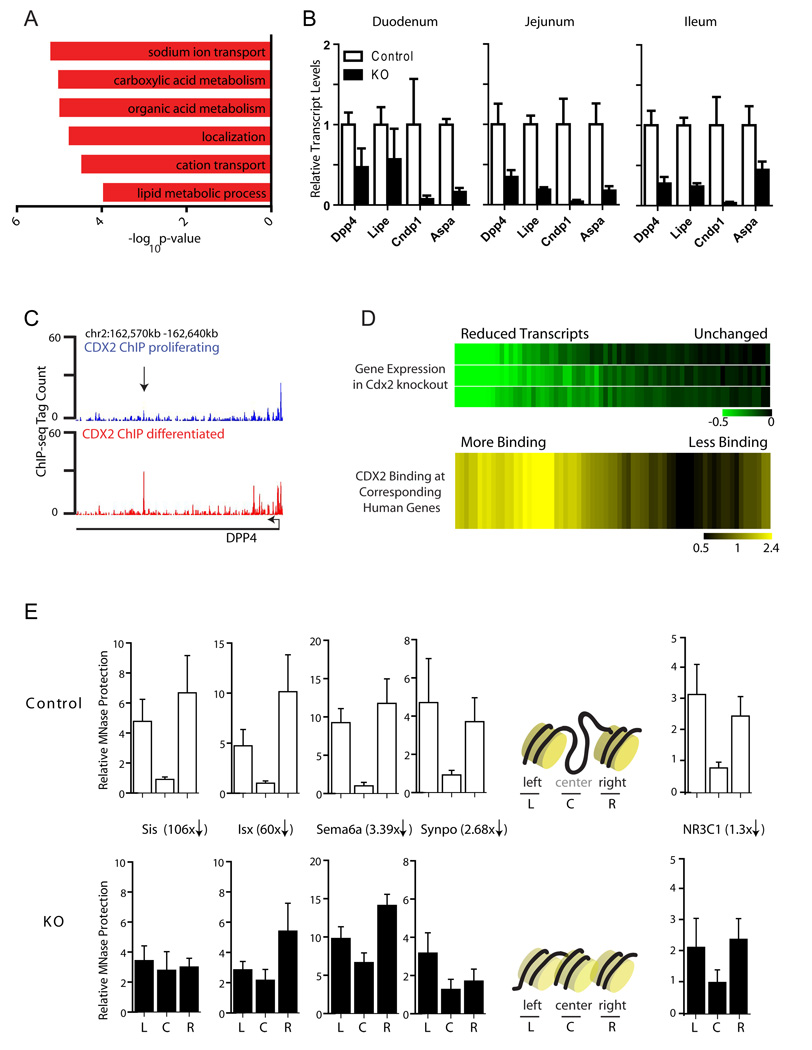
Figure 4. Cdx2 is required for active chromatin structure in vivo.
A, Gene Ontology terms associated with mature intestinal functions are notably enriched among transcripts reduced in Cdx2fV/fV;Villin-CreER(T2) intestinal mucosa. B, Examples of terminal digestive enzyme transcripts reduced in Cdx2-null duodenum, jejunum, and ileum and C, a representative example of binding data from Caco-2 cells showing differentiated cell-specific CDX2 occupancy (arrow). D, Top: Transcripts reduced or unchanged in Cdx2fV/fV;Villin-CreER(T2) mice were detected in triplicate samples and grouped into bins of 100 by a ratio relative to controls (−0.5 to 0). Bottom: Differentiated cell-specific CDX2 binding frequency within 100 kb of the corresponding orthologs in Caco-2 cells, indicated by intensity of yellow shading (a range of 0.5–2.4, similar to Fig. 2d). Binding is observed more frequently near loci with reduced expression, suggesting direct regulation of many targets and inter-species conservation. E, To determine the consequence of Cdx2 deletion on chromatin structure, oligonucleotides were designed to query the left, center, and right nucleosomes by PCR in MNase protection assays at nine CDX2-bound putative enhancers in control and tamoxifen-treated Cdx2fV/fV;Villin-CreER(T2) (KO) adult mice. MNase-protected DNA is shown relative to the central nucleosome at each region. In villi depleted of Cdx2, MNase protection was relatively enhanced at center nucleosomes, consistent with reappearance of nucleosomes and loss of active enhancer chromatin. Five of the 9 tested regions gave this pattern; 4 examples are shown. Results are depicted schematically on the right, together with 1 example where nucleosome protection was not significantly affected in KO mice. Reductions in transcript levels for each gene in the Cdx2-depleted tissue are indicated. All error bars are SEM.
Cdx2 is required for the chromatin configuration associated with active enhancers
The in vivo requirement for Cdx2 in differentiated and proliferating cells provides a strong functional correlate for the distinctive site occupancy in different cell states. To investigate Cdx2 requirements in gene regulation, we examined its role in chromatin structure. We and others define active enhancers as distal chromatin elements carrying two strongly positioned, H3K4Me2-marked nucleosomes around a central region with destabilized or absent nucleosomes (Fig. 1B). This designation facilitated genome-wide mapping of condition-sensitive epigenomic marks and triggered elucidation of CDX2 requirements in vivo. To determine if Cdx2 is necessary for the active chromatin configuration in differentiated mouse intestinal epithelium, we studied the consequence of its absence in tamoxifen-treated Cdx2fV/fV;Villin-CreER(T2) mouse villi. We used MNase digestion followed by quantitative real-time PCR to interrogate CDX2-bound regions located near genes that are dysregulated in the mutant mice. We designed oligonucleotide primers to detect nucleosome-protected DNA at representative CDX2-binding sites near differentiation-associated genes and both flanking nucleosomes. Villus cells isolated from tamoxifen-treated sibling control mice gave the pattern predicted for active nucleosome positioning at all 9 tested regions (Fig. 4E and data not shown). This active nucleosome signature was not apparent at 5 of the 9 regions in Cdx2-depleted villus cells, which showed relatively higher MNase sensitivity at flanking nucleosomes and greater protection in the center. This labile, Cdx2-dependent pattern most likely reflects displacement of a center nucleosome when Cdx2 is available and its reappearance when Cdx2 is absent. Our results thus indicate that Cdx2 is necessary to produce or maintain key chromatin modifications at loci associated with mature intestinal epithelium, an activity that may account for its control over a large number of genes required for intestinal function.
CDX2 partners with different transcription factors in different cell states
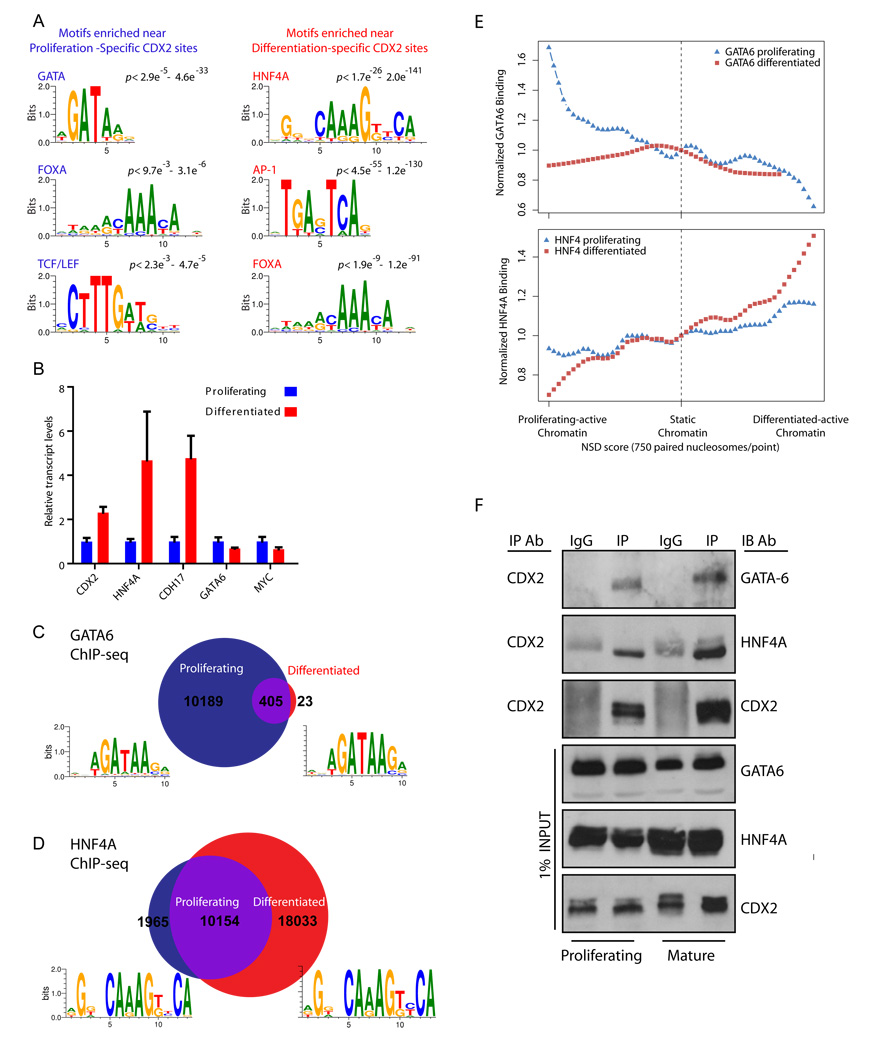
The demonstration of distinct CDX2 binding sites and functions in intestinal progenitors and mature cells suggested the possibility of distinct classes of regulatory _cis_-elements. To address the particular possibility of condition-specific partnership with other TFs, we searched CDX2-occupied regions in the two different cell states for other consensus sequence motifs. At early CDX2-occupied regions, the SeqPos motif analysis tool (Lupien et al., 2008) detected significant over-representation of GATA binding sites (Fig. 5A); thus, in dividing cells GATA sequences preferentially abound not only in chromatin with an active enhancer configuration (Fig. 1E) but also very close to CDX2-occupied sites. By contrast, CDX2-occupied regions specific to differentiated cells showed notable enrichment of a canonical HNF4A motif, the same sequence that is enriched among enhancers activated in mature Caco-2 cells (Fig. 1F) but not among early CDX2 sites (Fig. 5A). Adding credence to these results, the FOXA motif, matching a factor previously implicated in regulating endoderm-derived tissues (Sekiya et al., 2009; Zaret, 1999), was over-represented at CDX2 sites in both dividing and differentiated cells, whereas Wnt-responsive TCF/LEF motifs, associated with intestinal cell proliferation (Clevers, 2006), were enriched selectively at early CDX2 sites (Verzi et al., 2010). Mirroring the sequence motif enrichments, GATA6 mRNA levels decline with Caco-2 cell differentiation, as do those of the proliferation marker MYC; as noted previously (Soutoglou and Talianidis, 2002), HNF4A transcripts and the representative differentiation marker CDH17 increase markedly over the same period (Fig. 5B). Thus, CDX2 may partner with specific TFs in different cellular states; in particular, with a GATA protein early on and with HNF4A in differentiated cells.
Figure 5. Differential occupancy of proliferating and differentiated intestinal cell genomes by GATA6 and HNF4A.
A, DNA motif analysis at condition-specific CDX2-binding regions shows, in addition to CDX2 motifs, substantial enrichment of GATA motifs in proliferating (left) and of HNF4A motifs in differentiated cells (right). B, Over the course of Caco-2 differentiation, HNF4A transcript levels increase and GATA6 levels decrease; known markers of maturation (CDH17) or proliferation (MYC) increase or decrease, respectively, as expected. Error bars are SEM. C–D, ChIP-seq for GATA6 and HNF4A in proliferating (blue) and differentiated (red) cells reveals differences in preferred binding contexts. Venn diagrams indicate the number of sites bound uniquely in proliferating or differentiated cells (p<1e−10). HNF4A binding sites are more numerous later in differentiation, whereas GATA6 binds predominantly early in proliferating cells. For each occupancy dataset, the most enriched de novo binding motif is indicated (all within the statistical limit of p<1e−30,) and each is similar to empirically defined TRANSFAC motifs (as shown in Fig. 3A). Identified binding regions were highly conserved and mainly found far from promoters (Figure S6). E, As in figure 2C, TF occupancy at regions of differentially active chromatin is compared to binding frequency at static chromatin regions. Top, GATA6 binding is better associated with the active H3K4Me2 pattern in proliferating than in differentiated cells, whereas HNF4A occupancy (bottom) correlates better with active H3K4Me2 in differentiated cells. F, GATA6 and HNF4A co-immunoprecipitate with CDX2, but not with IgG controls, indicating their presence in CDX2 complexes.
To test this possibility, we conducted ChIP-seq analysis for GATA6 and HNF4A in mature and proliferating Caco-2 cells. In agreement with the motif search results, we identified 10,594 GATA6 binding sites (p ≤10−10) in dividing cells and fewer than 500 sites in differentiated cells (Fig. 5C). Conversely, HNF4A bound more than twice as many sites in mature cells than in the proliferative phase (Fig. 5D; 28,187 vs 12,119 sites, p ≤10−10). Regions occupied by GATA6 and HNF4A were highly conserved across species (Supplemental Fig. S6) and showed substantial enrichment of the expected sequence motifs (Fig 5C–D). Because CDX2 binding to DNA in replicating and mature Caco-2 cells correlates with active chromatin in the respective cell states (Fig. 2C), we asked if binding of these candidate partners is also associated with the chromatin state. Indeed, GATA6 preferentially occupies sites that carry the active-enhancer mark in proliferating but not in differentiated cells, whereas HNF4A binding shows a marked preference for chromatin that is selectively active in mature cells (Fig. 5E). Thus, although GATA6 occupancy is more state-selective than HNF4A binding, histone H3 modifications are tightly coupled to experimentally determined binding of all three proteins and our results identify putative co- regulatory complexes for CDX2 function. Corroborating the existence of such complexes, immunoprecipitation of CDX2 from Caco-2 nuclear extracts revealed association with GATA6 and HNF4A (Fig. 5F). These associations were not intrinsically cell state-specific, probably reflecting interactions at constitutively occupied regions (represented in the intersection set in Fig. 2A).
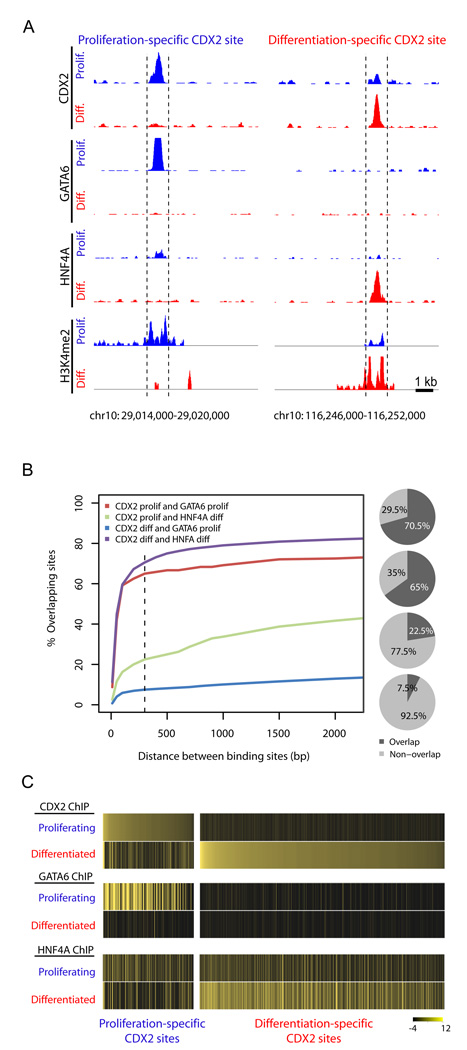
Importantly, in analysis of empirical binding, CDX2-binding sites specific to dividing cells were more likely to be identified as GATA6-binding sites in ChIP-seq on dividing cells, whereas CDX2-binding sites in differentiated cells were more likely to be identified as HNF4A binding sites in ChIP-seq on differentiated cells. Individual examples of this duality are illustrated in Fig. 6A and Supplemental Fig. S7. Furthermore, CDX2 and its condition-specific partners typically occupy DNA within at most a few hundred bp of each other (Fig. 6B), suggesting they target the same regulatory elements. These relationships are reflected well in heat maps for genome-wide binding of each factor in relation to CDX2 occupancy (Fig. 6C). These data collectively indicate that CDX2 interacts with intestinal DNA in state-specific pairings with GATA6 and HNF4A at regions of differentially modified chromatin.
Figure 6. Condition-specific CDX2 binding is accompanied by GATA6 in proliferating cells and HNF4A in differentiated cells.
A, Individual examples of ChIP-seq in proliferating (blue) and differentiated (red) cells show condition-specific GATA6 and CDX2 occupancy at the same region in proliferating but not in differentiated cells; HNF4A and CDX2 co-occupy a region bound selectively in differentiated cells. Condition-specific H3K4me2 signals are evident at the regions selectively occupied by the TFs and demarcated here by the dotted lines. Additional examples appear in Figure S7. B, Frequency of co-occupancy among TFs in proliferating and differentiated cells as a function of the distance between peak factor-binding sites. Condition-specific CDX2 binding sites more frequently lie near GATA6-bound regions in proliferating cells and near HNF4A-occupied regions in mature cells. The frequency of overlap in binding peaks within a 300bp distance (dotted line) is indicated in pie charts (right). C, Heat maps of all condition-specific CDX2-bound sites, showing the extent of GATA6 and HNF4A co-occupancy at these regions and confirming the relationship between these factors in binding across the genome. Sites are ordered by robustness of CDX2 occupancy in each state of differentiation. The measure of each TF’s occupancy was defined as the relative ChIP signal in relation to the input signal at binding sites (see Experimental Procedures for mathematical details) and distributed between −4 and 12 (color map scale).
DISCUSSION
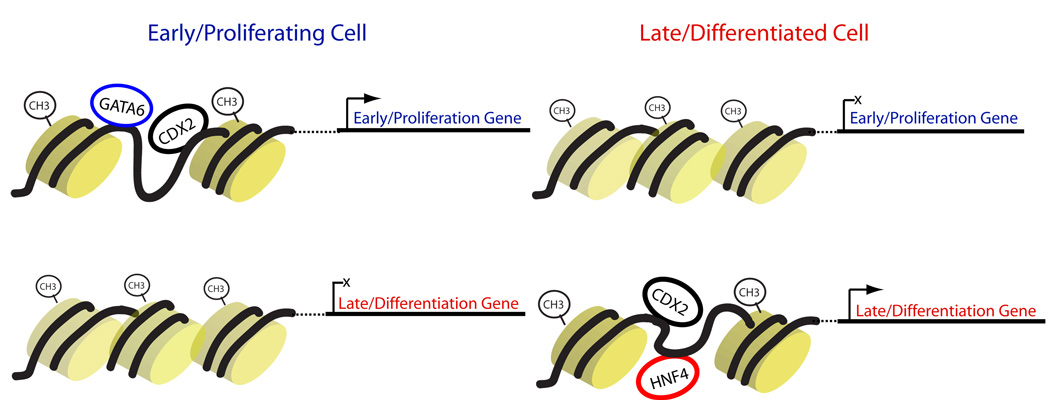
Progenitor cells and their mature progeny elicit substantially different properties from genomes that are usually identical. Analysis of gene knockouts in vivo and lineage-specific _cis_-elements in vitro support the idea that key regulatory TFs participate materially in differentiation, using mechanisms that are diverse and incompletely understood. Our identification of the CDX2 recognition motif at enhancers active in differentiated Caco-2 cells prompted detailed investigation of a TF whose highly restricted tissue expression, activity at intestine-specific gene promoters in vitro, and embryonic requirement encompass the properties of a “master regulator” (Beck, 2004; Gao et al., 2009). Findings in Cdx2-null mice support this designation in the adult intestine and reveal its requirement for cellular functions, gene expression, and active enhancer chromatin in distinct states. We also present evidence for dynamic CDX2 associations in a regenerative tissue, including labile interactions with chromatin and other TFs that correlate strongly with gene expression in dividing and mature cells (Fig. 7). The sum of these results provides a comprehensive view of the actions and mechanisms of a critical lineage-specific TF.
Figure 7. Model for dynamic CDX2 function during intestinal differentiation.
In proliferating cells, CDX2 binds with relative selectivity, frequently accompanied by GATA6, at H3K4Me2 regions with selectively open chromatin at loci active in progenitor cells. Genes associated with epithelial maturity are not expressed in these cells and their enhancers have a relatively closed structure with less H3K4Me2. As cells differentiate, CDX2 relocates from progenitor-specific target genes to those associated with maturity, now accompanied by HNF4A; the appearance of an active H3K4Me2 pattern at many loci depends on CDX2.
Dual activity in progenitor and differentiated cells is a feature of several lineage-restricted TFs. For example, Pu.1 functions in blood progenitor commitment and proliferation as well as terminal macrophage differentiation (DeKoter et al., 1998), much as MITF does in melanocyte proliferation, survival, and maturation (McGill et al., 2002). In development, the worm FoxA homolog PHA4 functions throughout pharynx formation, as do Pax6 and its homologs in the metazoan eye (Ashery-Padan and Gruss, 2001). Even in the face of such examples, it is unclear if these factors directly control tissue-specific genes throughout a cell’s ontogeny, if their genome associations are stable across cellular transitions, and if Cdx2’s distinct activities in dividing and differentiated cells reflect a general property of lineage-determining TFs. The muscle regulator MyoD, for example, functions in both myoblast progenitors and mature myotubes, and its binding to DNA was recently reported to vary little during differentiation (Cao et al., 2010). In contrast, redistribution of CDX2 during intestinal cell maturation exposes diversity among mechanisms of key TFs. Investigation of other regulators, ideally coupled with delineation of chromatin modifications, will resolve whether their associations with DNA are largely invariant, as reported for MyoD, or dynamic, as we observe with CDX2.
In C. elegans pharynx development, PHA4 controls early and late genes by binding unique target sites with different affinities. Early genes contain high-affinity sites and bind first; late targets carry sites with distinct sequences and lower affinity, and engage only after PHA4 levels increase late in development (Gaudet and Mango, 2002). CDX2 levels change little during intestinal differentiation and the consensus CDX2 motifs we identified in different conditions are almost identical (Supplemental Fig. S4), indicating a limited role for primary DNA sequence or TF concentrations in directing occupancy. Hence, post-translational modifications, alternative partners, chromatin structure, or a combination of these factors, likely underlie CDX2’s cell state-specific complexes. Known post-translational alterations of CDX2 do not affect DNA affinity in vitro (Gross et al., 2005; Rings et al., 2001) but could in principle affect alternative complexes. Biochemical analysis of modifications on CDX2 and its partner proteins might advance understanding of the mechanisms underlying nucleosome lability at target enhancers. In identifying GATA6 and HNF4A as state-specific CDX2 partners at differentially modified regions of chromatin, we take an important step toward characterizing tissue-restricted TFs that produce or interact with altered chromatin states to enable intestinal cell differentiation. Future work might determine if the factors are co-dependent or restructure chromatin in an ordered hierarchy; understanding these mechanisms will help define the networks that control intestinal cell transitions (Davidson and Levine, 2008). Although findings in mutant mice corroborate our conclusions, the bulk of the present analysis was conducted in a tumor cell line that replicates the distinction between progenitor and differentiated states only partially. Primary intestinal crypt and villus cells would provide a physiologic model to test these ideas.
Studies in cancer cell lines paradoxically implicate CDX2 as both an oncogene and a tumor suppressor (Aoki et al., 2003; Guo et al., 2004); context-dependent CDX2 occupancy and function may account for distinct roles in cell proliferation and differentiation and help explain the discrepancy. Contextual genome occupancy and cooperation with other TFs might also underlie CDX2 requirements in embryonic axial patterning and trophectoderm formation. Indeed, CDX2 binding in the corresponding tissues suggests remarkable diversity in target loci (our unpublished data), underscoring the idea that TF activities are strongly influenced by cellular context. Another TF with many target genes, PPARG, was recently shown to utilize different binding sites and partners in two cell types, adipocytes and macrophages (Lefterova et al., 2010).
The continually renewing gut epithelium, a frequent target of malignant transformation, is an ideal model system to study transcriptional mechanisms of differentiation. Our integrated approach toward genome-wide chromatin analysis, TF binding, and mRNA expression can also be extended to uncover mechanisms of lineage-specific gene regulation in other tissues.
EXPERIMENTAL PROCEDURES
Nucleosome-resolution ChIP and identification of mononucleosomes
Cells were collected from Caco-2 cultures or mouse intestinal villi and resuspended in a digestion buffer (50 mM Tris-HCl, pH7.6, 1 mM CaCl2, 0.2% Triton X-100, 5 mM Na butyrate, complete protease inhibitors) and treated with 0.2 U micrococcal nuclease (MNase, Sigma Aldrich) for 8 min at 37°C. The reaction was terminated by adding 5 mM EDTA in 10 mM Tris, pH7.6, and samples were dialyzed in chromatin RIPA buffer (10 mM Tris, pH7.6, 1 mM EDTA, 0.1% SDS, 0.1% Na deoxycholate, 1% Triton X-100) before overnight IP at 4°C with Histone H3K27Ac (Abcam ab4729) or H3K4Me2 (Millipore 07–030) antibodies. IP material was subsequently handled as described below for conventional ChIP of sonicated genomic DNA. Mononucleosomes carrying H3K4Me2 or H3K27Ac marks were identified using Nucleosome Positioning from Sequencing (NPS) with default parameters (Zhang et al., 2008b), as described (Extended Experimental Procedures).
Detection of state-specific open chromatin structures and identification of transcription factor motifs
NPS was used to identify mononucleosomes with H3K4Me2. Two adjacent nucleosomes were declared a pair if the distance between their center locations was between 250 and 400 nucleotides. For each nucleosome pair, relative depletion of a nucleosome in the inter-nucleosomal region in one state against the other was estimated using the Nucleosome Stability-Destability (NSD) scoring scheme defined previously (He et al., 2010). In this context, the NSD score of a nucleosome pair indicates the degree of differential chromatin structure between proliferating and mature intestinal cells by quantifying the relative change in nucleosome signal between cell states. Thus, nucleosome pairs with larger NSD scores can be interpreted to represent genomic regions more open to TF binding in one cell state compared to static regions (i.e., regions with NSD scores ~0). Conversely, negative NSD scores correspond to open chromatin structure in the other state (Supplemental Table S1). A hierarchical mixture model was used to search for DNA motifs enriched at the center of nucleosome pairs (the region of depleted signal corresponding to the middle nucleosome). Enrichment of each motif represented in the TRANSFAC database was weighed by the NSD score distribution of other regions with similar enrichment levels of the same motif (He et al., 2010) (and Meyer CA, unpublished) and ranked using t-statistics.
Chromatin Immunoprecipitation (ChIP) and co-immunoprecipitation for transcription factors, ChIP-seq, and data analysis
Standard procedures were used for ChIP (Extended Experimental Procedures) on sub-confluent or 26-day post-confluent Caco-2 cells. For ChIP-seq, 10 ng each of ChIP and input DNA from up to 3 pooled experiments were processed for deep sequencing according to manufacturer’s instructions (Illumina). Prior to sequencing, qPCR was used to verify that positive and negative control ChIP regions amplified in the linear range.
Binding peaks for CDX2, GATA6, and HNF4A were identified using Model-Based Analysis of ChIP-Seq (MACS) (Zhang et al., 2008a) with a p-value cutoff of 10−10 and default values for other parameters. Sequences were mapped to reference genomes using ELAND tools (Illumina software suite). All sequence information is relative to human genome build 18. CDX2 binding sites specific to proliferating or differentiated cells were delineated by eliminating sites that might potentially be occupied in the other condition had the much less stringent _p_-value cut-off of 10−3 been applied. Binding sites determined by ChIP-Seq were scanned for sequence motifs using the SeqPos algorithm (He et al., 2010) as described (Extended Experimental Procedures). Co-immunoprecipitation was done using standard techniques and anti-CDX2 (BD Pharmingen 560171; Extended Experimental Procedures). All ChIP-seq data have been deposited in GEO (Series GSE23436).
Association assessment of empirical transcription factor binding with paired nucleosomes and gene expression
To calculate the relative frequency of nucleosome pairs occupied by a TF (Figs. 2C, 5E), paired nucleosomes were first sorted according to their NSD scores and grouped into bins of 750. For each bin, the number of paired nucleosomes bound by the factor was counted and scaled with respect to binding in bins representing static chromatin (i.e,. paired nucleosomes with NSD score ~0). To determine association of protein binding with gene expression (Fig. 2D), Caco-2 expression microarray data (Fleet et al., 2003) (GEO accession number GSE 1614) were enumerated based on log-fold changes in differentiated over proliferating cells using SAM analysis and genes were binned into groups of 100. Similar to Heintzman et al (Heintzman et al., 2009), the ratio of the average number of CDX2 binding sites within 100 kb of genes belonging in the same bin and the average number of binding sites near all genes was calculated and transformed to a logarithmic scale.
Association assessment of transcription factor partners
Associations between binding partners, such as CDX2 and GATA6 in proliferating or CDX2 and HNF4A in mature cells (Fig. 6B), were interrogated using their ChIP-Seq signals. First, the union of binding sites of CDX2, GATA6, and HNF4A in both conditions was obtained and the ChIP signals of each individual protein at the union sites were computed using the equation T=TTF×Sinput−Tinput×STFSinput. T and S stand for the maximum tag count for a binding site and the sequence depth, respectively; subscripts TF and input indicate a transcription factor or input. This equation quantifies relative signal strength over the input on account of the sequence depth of TF and input data sets.
Knockout mice
LoxP sites were placed flanking exon 2 of mouse Cdx2 in a targeting construct containing an FRT-flanked Neomycin resistance (NeoR) cassette immediately downstream of the 3’ LoxP site. For ES cell screening, southern blots on HindIII-digested DNA were hybridized with 5’ (chr5:148120111+148120519, mm9) and 3’ (chr5:148107065-148107467, mm9) probes. Two properly targeted clones were injected into blastocysts. Chimeric progeny were crossed with R26R-FLP mice (Farley et al., 2000) to excise the NeoR gene and resulting progeny were crossed with _Villin-Cre_ER(T2) transgenic mice (el Marjou et al., 2004), yielding Mendelian transmission of all alleles. Experimental and littermate control mice were treated with 1 mg tamoxifen by intraperitoneal injection for 5 consecutive days. Control mice included Cre+ animals and no effects of Cre toxicity were observed. For RNA microarray analysis, mice were harvested on the 4th day after completion of the tamoxifen course. For MNase assays, mice were analyzed at least 1 week after conclusion of tamoxifen treatment. Histologic analysis was done 2 days after the last tamoxifen dose. Cdx1−/− mice were maintained and genotyped as described (Subramanian et al., 1998).
Expression analyses
Cell and tissue RNAs were harvested using Trizol reagent (Invitrogen) and reverse transcribed using SuperScript enzyme (Invitrogen) or labeled for microarray (Affymetrix). cDNA was assessed by qPCR using gene-specific primers (Extended Experimental Procedures) and SYBR green master mix (Applied Biosystems). Additional details are found in Extended Experimental Procedures. For immunostaining, routine histological techniques were applied (Extended Experimental Procedures) using the following antibodies: CDX2 (Biogenex, 1:50), Ki67 (Novocastra Laboratories, 1:1000), Chromogranin A (Immunostar, 1:500), Lysozyme (Invitrogen, 1:50), and PCNA (NeoMarkers, 1:1000). All expression data have been deposited to GEO (Series GSE23436).
Gene association and GO analyses
were performed using MetaCore and DAVID Gene Functional Analysis tools (http://david.abcc.ncifcrf.gov/) as described (Extended Experimental Procedures).
Association of CDX2 binding sites in Caco-2 cells with genes dysregulated in Cdx2-deficient intestine
To determine associations between transcripts that change upon Cdx2 loss in mouse intestine and CDX2 binding near their human orthologs in Caco-2 (Fig. 4D), we first used Ensembl BioMart (http://uswest.ensembl.org/index.html) to match mouse genes with human orthologs, which were then arranged according to expression levels of the corresponding mouse gene and grouped into bins of 100. For each such bin, the ratio between the average number of CDX2 binding sites within 100 kb of genes in the same bin and the average number of binding sites near all genes was estimated and displayed (Fig. 4D).
MNase protection
Cells were harvested from knockout or control mouse intestinal villi and treated with MNase as described above. Instead of incubation with antibody, DNA was purified after Proteinase K digestion and compared to control samples (no MNase treatment) by qPCR using the indicated oligonucleotides (Extended Experimental Procedures).
Cell proliferation
Proliferating (50% confluent) and 26-day post-confluent Caco-2 cells were treated for 1 h with EdU. DNA synthesis was measured by fluorescent detection of EdU according to manufacturer’s protocols (Invitrogen).
HIGHLIGHTS
- A histone modification screen flagged CDX2 as a regulator of gut differentiation
- CDX2 interactions with the genome change materially as cells differentiate
- Adult knockout mice reveal distinct Cdx2 functions in progenitor and mature cells
- Context-specific CDX2 binding occurs with specific partner proteins GATA6 or HNF4A
Supplementary Material
01
02
03
ACKNOWLEDGMENTS
Supported by National Institutes of Health (NIH) grants RC2CA148222 and R01DK082889 (RAS), R01HG004069 (XSL), R01DK054111 (JCF), and P50CA127003 (Dana-Farber/Harvard Cancer Center), and a gift from the Caring for Carcinoid Foundation (RAS). MPV was supported by NIH training grant T32DK07477 and Fellowship No. 1987 from the Crohn’s & Colitis Foundation of America. We thank Steve Krasinski for critical appraisal of the work; Madeline Lemieux and Andrew Kung for helpful discussions; Sylvie Robine for permission to use, and Eric Martin for providing, Villin-CreER(T2) transgenic mice; and Peter Gruss for permission to use, and Gary Gilliland and Sumin Koo for providing, Cdx1−/− mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
MPV and RAS conceived the study, analyzed data, and wrote the manuscript; MPV, RS and RKM performed experiments; MPV, HS, HHH, CAM, JCF and XSL developed the algorithms and performed computational analyses; HS, JCF, MB and XSL interpreted data and edited the report.
REFERENCES
- Aoki K, Tamai Y, Horiike S, Oshima M, Taketo MM. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/− compound mutant mice. Nat Genet. 2003;35:323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Gruss P. Pax6 lights-up the way for eye development. Curr Opin Cell Biol. 2001;13:706–714. doi: 10.1016/s0955-0674(00)00274-x. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beck F. The role of Cdx genes in the mammalian gut. Gut. 2004;53:1394–1396. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- Choi MY, Romer AI, Hu M, Lepourcelet M, Mechoor A, Yesilaltay A, Krieger M, Gray PA, Shivdasani RA. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development. 2006;133:4119–4129. doi: 10.1242/dev.02537. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- D'Angelo A, Bluteau O, Garcia-Gonzalez MA, Gresh L, Doyen A, Garbay S, Robine S, Pontoglio M. Hepatocyte nuclear factor 1alpha and beta control terminal differentiation and cell fate commitment in the gut epithelium. Development. 2010;137:1573–1582. doi: 10.1242/dev.044420. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci USA. 2008;105:20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter RP, Walsh JC, Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 1998;17:4456–4468. doi: 10.1093/emboj/17.15.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Fang R, Santiago NA, Olds LC, Sibley E. The homeodomain protein Cdx2 regulates lactase gene promoter activity during enterocyte differentiation. Gastroenterology. 2000;118:115–127. doi: 10.1016/s0016-5085(00)70420-3. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Fleet JC, Wang L, Vitek O, Craig BA, Edenberg HJ. Gene expression profiling of Caco-2 BBe cells suggests a role for specific signaling pathways during intestinal differentiation. Physiol Genomics. 2003;13:57–68. doi: 10.1152/physiolgenomics.00152.2002. [DOI] [PubMed] [Google Scholar]
- Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Grainger S, Savory JG, Lohnes D. Cdx2 regulates patterning of the intestinal epithelium. Dev Biol. 2010 doi: 10.1016/j.ydbio.2009.12.025. In Press. [DOI] [PubMed] [Google Scholar]
- Gross I, Lhermitte B, Domon-Dell C, Duluc I, Martin E, Gaiddon C, Kedinger M, Freund JN. Phosphorylation of the homeotic tumor suppressor Cdx2 mediates its ubiquitin-dependent proteasome degradation. Oncogene. 2005;24:7955–7963. doi: 10.1038/sj.onc.1208945. [DOI] [PubMed] [Google Scholar]
- Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3:593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Saaf AM, Brown PO, Nelson WJ. Transcriptional modulation of genes encoding structural characteristics of differentiating enterocytes during development of a polarized epithelium in vitro. Mol Biol Cell. 2007;18:4261–4278. doi: 10.1091/mbc.E07-04-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey S, Wei G, Wang Q, Zhany Y, Xu K, Ni M, Lupien M, et al. Positioned nucleosomes flanking a labile nucleosome characterize transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hinoi T, Lucas PC, Kuick R, Hanash S, Cho KR, Fearon ER. CDX2 regulates liver intestine-cadherin expression in normal and malignant colon epithelium and intestinal metaplasia. Gastroenterology. 2002;123:1565–1577. doi: 10.1053/gast.2002.36598. [DOI] [PubMed] [Google Scholar]
- James R, Erler T, Kazenwadel J. Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J Biol Chem. 1994;269:15229–15237. [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang X, So CK, Wang S, Wang P, Yan L, Myers R, Chen Z, Patterson AP, Yang CS, Chen X. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis. 2007;28:488–496. doi: 10.1093/carcin/bgl176. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Defining the regulatory networks for muscle development. Curr Opin Genet Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- Mouchel N, Henstra SA, McCarthy VA, Williams SH, Phylactides M, Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Potten CS. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci. 1998;353:821–830. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rings EH, Boudreau F, Taylor JK, Moffett J, Suh ER, Traber PG. Phosphorylation of the serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- Saaf AM, Halbleib JM, Chen X, Yuen ST, Leung SY, Nelson WJ, Brown PO. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell. 2007;18:4245–4260. doi: 10.1091/mbc.E07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122:689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Stegmann A, Hansen M, Wang Y, Larsen JB, Lund LR, Ritie L, Nicholson JK, Quistorff B, Simon-Assmann P, Troelsen JT, Olsen J. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol Genomics. 2006;27:141–155. doi: 10.1152/physiolgenomics.00314.2005. [DOI] [PubMed] [Google Scholar]
- Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Meyer B, Evans GS. The murine Cdx1 gene product localises to the proliferative compartment in the developing and regenerating intestinal epithelium. Differentiation. 1998;64:11–18. doi: 10.1046/j.1432-0436.1998.6410011.x. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Meyer BI, Gruss P. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell. 1995;83:641–653. doi: 10.1016/0092-8674(95)90104-3. [DOI] [PubMed] [Google Scholar]
- Traber PG, Silberg DG. Intestine-specific gene transcription. Annu Rev Physiol. 1996;58:275–297. doi: 10.1146/annurev.ph.58.030196.001423. [DOI] [PubMed] [Google Scholar]
- Tremblay E, Auclair J, Delvin E, Levy E, Menard D, Pshezhetsky AV, Rivard N, Seidman EG, Sinnett D, Vachon PH, Beaulieu JF. Gene expression profiles of normal proliferating and differentiating human intestinal epithelial cells: a comparison with the Caco-2 cell model. J Cell Biochem. 2006;99:1175–1186. doi: 10.1002/jcb.21015. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, Clevers H, Liu XS, Shivdasani RA. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. PNAS. 2010;107(34):15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008a;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008b;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03