CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex (original) (raw)
A conserved γ-tubulin complex–binding domain in CDK5RAP2 stimulates the microtubule-nucleating activity of γ-TuRC.
Abstract
CDK5RAP2 is a human microcephaly protein that contains a γ-tubulin complex (γ-TuC)–binding domain conserved in Drosophila melanogaster centrosomin and Schizosaccharomyces pombe Mto1p and Pcp1p, which are γ-TuC–tethering proteins. In this study, we show that this domain within CDK5RAP2 associates with the γ-tubulin ring complex (γ-TuRC) to stimulate its microtubule-nucleating activity and is therefore referred to as the γ-TuRC–mediated nucleation activator (γ-TuNA). γ-TuNA but not its γ-TuC–binding-deficient mutant stimulates microtubule nucleation by purified γ-TuRC in vitro and induces extensive, γ-TuRC-dependent nucleation of microtubules in a microtubule regrowth assay. γ-TuRC bound to γ-TuNA contains NME7, FAM128A/B, and actin in addition to γ-tubulin and GCP2–6. RNA interference–mediated depletion of CDK5RAP2 impairs both centrosomal and acentrosomal microtubule nucleation, although γ-TuRC assembly is unaffected. Collectively, these results suggest that the γ-TuNA found in CDK5RAP2 has regulatory functions in γ-TuRC–mediated microtubule nucleation.
Introduction
γ-Tubulin plays a critical role in microtubule nucleation occurring at least at centrosomes, chromatins, and spindle microtubules. There are two differently sized γ-tubulin complexes (γ-TuCs): the γ-tubulin small complex (γ-TuSC) and the γ-tubulin ring complex (γ-TuRC; Wiese and Zheng, 2006; Lüders and Stearns, 2007; Raynaud-Messina and Merdes, 2007). γ-TuSC is a tetramer consisting of two γ-tubulin and two other γ-complex proteins (GCPs), GCP2 and GCP3. In γ-TuRC, several γ-TuSCs are assembled into a distinct ring-shaped structure with additional γ-TuRC–specific proteins such as GCP4, GCP5, and GCP6 (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000; Kollman et al., 2010). However, the molecular assembly of γ-TuRC has not been fully understood.
The microtubule-nucleating activities of γ-TuCs are well controlled in cells. At centrosomes, γ-tubulin mediates microtubule nucleation and anchoring of the radial microtubule network. Structural studies of the Saccharomyces cerevisiae γ-TuCs have revealed that in both γ-TuSC and a γ-TuRC–like ring structure assembled by γ-TuSC, γ-tubulins are kept in distances incompatible with microtubule nucleation (Kollman et al., 2008, 2010). These observations have implied the activation of the nucleating activity by mechanisms in addition to γ-TuRC assembly. Indeed, salt-stripped centrosomes require not only γ-TuRC but also additional cytoplasmic factors to restore their microtubule-nucleating function (Moritz et al., 1998).
CDK5RAP2 is a human microcephaly protein that binds to the γ-TuCs and is involved in the centrosomal attachment of γ-tubulin (Bond et al., 2005; Fong et al., 2008). The γ-TuC–binding domain found in CDK5RAP2 is conserved in Drosophila melanogaster centrosomin and Schizosaccharomyces pombe Mto1p and Pcp1p, which are γ-TuC–tethering proteins in the respective organisms (Sawin et al., 2004; Fong et al., 2008). In this study, we demonstrate that this CDK5RAP2 domain associates with γ-TuRC to act as a γ-TuRC–mediated nucleation activator (γ-TuNA).
Results and discussion
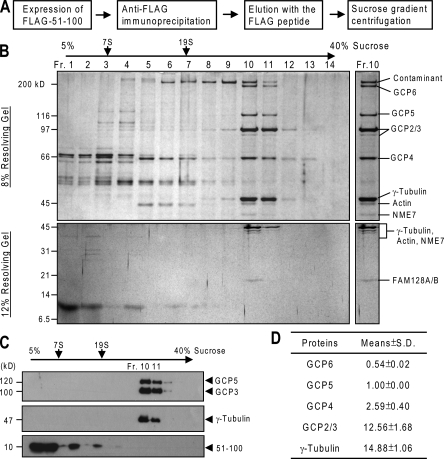
We set out to isolate γ-TuCs bound to γ-TuNA (i.e., 58–90) and to define the composition of the complexes. To this end, the γ-TuNA–containing construct 51–100 was used for immunoprecipitation through its ectopic tag (i.e., Flag). After elution using the tag peptide, the eluate was further separated by sedimentation through a sucrose gradient (Fig. 1 A). Each gradient fraction was analyzed by SDS-PAGE and immunoblotting. Proteins visualized in the peak fraction of γ-tubulin were identified by mass spectrometry. All γ-tubulin and GCP2–6 appeared exclusively in the γ-TuRC fractions (Fig. 1, B and C), revealing that γ-TuNA associates with γ-TuRC. In addition, mass spectrometry revealed the presence of NME7 (also known as NM23-H7 and NDPK7, a putative member of the NM23 family of nucleoside diphosphate kinases), FAM128A/B, and β/γ-actin from the γ-TuRC fraction (Fig. 1 B). A recent study also identified NME7 and FAM128A/B as components of γ-TuRC (Hutchins et al., 2010). The coisolation of actin is consistent with an observation of the Drosophila γ-TuRC (Oegema et al., 1999). Therefore, we obtained highly purified γ-TuRC from such an isolation procedure. It should be noted that during the isolation, the 51–100 protein was dissociated from γ-TuRC by the inclusion of the Flag peptide for elution and was then resolved into gradient fractions different from those of γ-TuRC (Fig. 1 C and Fig. S1 A).
Figure 1.
Isolation of γ-TuCs bound to γ-TuNA. (A) Schematic outline of the isolation procedure. (B) After gradient centrifugation, an aliquot of each fraction was resolved by SDS-PAGE followed by silver staining. Proteins resolved from the peak fraction of γ-TuRC (Fr. 10) were identified by mass spectrometry. The contaminant protein above GCP6 also appeared in the precipitates of blank beads. (C) The gradient fractions were analyzed by immunoblotting. (D) In a replicate gel stained with Sypro ruby, the relative amounts of γ-tubulin and GCPs were determined from the isolated γ-TuRC (Fr. 10) to derive the stoichiometry. The ratios of proteins to GCP5 are presented as mean ± SD from three independent experiments.
To determine the composition stoichiometry of the isolated γ-TuRC, we measured the intensity of fluorescent dye–stained proteins from the γ-TuRC peak fraction. After background subtraction, the stoichiometry was calculated as values relative to that of GCP5. Each γ-TuRC contains ∼14 copies of γ-tubulin, ∼12 copies of GCP2/3, which is equivalent to about six heterodimers of GCP2 and GCP3, two to three copies of GCP4, and one copy of GCP5 (Fig. 1 D), which is consistent with estimates made in the proposed γ-TuRC structure and in previous studies (Zheng et al., 1995; Fava et al., 1999; Oegema et al., 1999; Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000; Murphy et al., 2001). In addition, our data reveal that GCP6 is absent in a large fraction of the isolated γ-TuRC (Fig. 1 D).
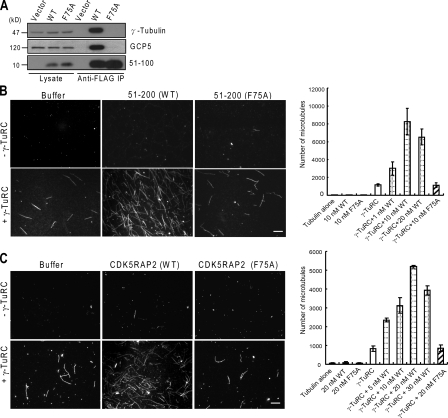
We subsequently explored whether γ-TuNA as well as the entire CDK5RAP2 protein can regulate the nucleating activity of γ-TuRC. To conduct microtubule-nucleating assays in vitro, γ-TuRC was isolated by coimmunoprecipitation with the γ-TuNA construct 51–100 and subsequently by gradient centrifugation. The purified γ-TuRC was devoid of the CDK5RAP2 fragment (Fig. 1 C). We also generated a γ-TuC–binding-deficient mutant of γ-TuNA to be used as a control in the nucleating assays. γ-TuNA contains predicted haptad repeats packed into an α-helical coiled coil. To generate such a mutant, we substituted Ala for Phe75, a conserved hydrophobic residue within the domain. This mutation abolished the binding of a γ-TuNA–containing construct, 51–100, as well as the entire CDK5RAP2 protein to γ-TuCs (Fig. 2 A and Fig. S2).
Figure 2.
Stimulation of γ-TuRC for microtubule nucleation. (A) After immunoprecipitation of ectopically expressed proteins through the tag moiety (i.e., Flag), the immunoprecipitates and cell lysates were examined for γ-tubulin and GCP5. WT, 51–100 wild type; F75A, 51–100 (F75A). (B and C) Microtubule polymerization was performed with or without the isolated γ-TuRC and 51–200 (B) or the entire CDK5RAP2 protein (C). Representative microscopic fields of polymerized microtubules are shown. Microtubules were counted from 20 random fields to derive the mean numbers of microtubules. Data are shown as mean ± SD of three independent experiments. Bar, 10 µm.
In microtubule nucleation assays, spontaneous tubulin nucleation was negligible under the assay conditions (Fig. 2 B). The inclusion of the isolated γ-TuRC resulted in nucleation at a low level (Fig. 2 B). The γ-TuNA construct 51–200 or its F75A mutant did not induce nucleation in the absence of γ-TuRC (Fig. 2 B). When γ-TuRC was added with 51–200, the nucleating activity was increased in a manner dependent on the concentration of 51–200 with a maximal enhancement of ∼7.1-fold (Fig. 2 B). Such promoting effects were not observed when the wild type was replaced with the F75A mutant (Fig. 2 B). Therefore, the association with γ-TuNA significantly enhances the microtubule-nucleating activity of γ-TuRC.
The entire CDK5RAP2 protein and its F75A mutant were also tested in the nucleation assay. The CDK5RAP2 proteins were prepared by ectopic expression in HEK293T cells and subsequent immunoprecipitation through their tag moiety. Before elution, most coprecipitated proteins, including γ-tubulin and GCPs, were removed by washing with a high-salt and detergent-containing buffer (Fig. S1 B). In the nucleation assay, full-length CDK5RAP2 displayed a similar effect as γ-TuNA to stimulate γ-TuRC–dependent microtubule nucleation but not the F75A mutant (Fig. 2 C). These results indicate that the CDK5RAP2 protein has the capacity to stimulate the nucleating activity of γ-TuRC.
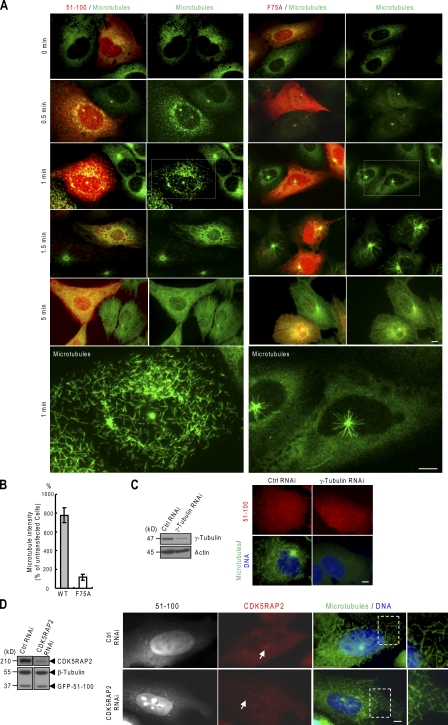
In a previous study, we conducted microtubule regrowth assays and terminated the assays by fixation with methanol at −20°C or 4% paraformaldehyde at room temperature (Fong et al., 2008). Under these conditions, we observed microtubule regrowth from centrosomes but were unable to monitor cytoplasmic microtubule nucleation, especially at the early stages of regrowth, mainly because of the high background staining of the cytoplasm. To reduce the cytoplasmic background, we modified the methods and performed fixation with 4% paraformaldehyde in the presence of 0.5% Triton X-100. Using this method, numerous microtubule speckles were found as early as 30 s of regrowth in the cytoplasm expressing 51–100, but such speckles did not appear in untransfected cells or those transfected with the F75A mutant (Fig. 3, A and B). Short microtubule segments were formed at 1 min in cells transfected with wild-type 51–100 (Fig. 3, A and B). When the regrowth proceeded, wild-type–transfected cells were quickly saturated with microtubules, whereas untransfected and 51–100 (F75A)-transfected cells contained scarce cytoplasmic microtubules beside a prominent centrosomal aster of microtubules (1.5 min; Fig. 3, A and B). After regrowth for ≥5 min, cells transfected with the wild type displayed a similar microtubule density in comparison with others except that microtubules were disorganized and without focus (Fig. 3, A and B).
Figure 3.
Microtubule nucleation induced by γ-TuNA in transfected cells. (A and B) Microtubule regrowth was performed on U2OS cells expressing 51–100 or its F75A mutant. (bottom) Enlarged views of the boxed areas are shown. The microtubule intensities of regrowth for 1 min were quantified from cells expressing the proteins at similar levels and were expressed relative to untransfected cells (B). At least 20 cells were measured in each of three independent experiments. (B) Error bars indicate mean ± SD. (C) Microtubule regrowth (1 min) was performed after γ-tubulin was depleted by RNAi. (left) The expressions of γ-tubulin and actin were examined by immunoblotting. (right) Cells transfected with 51–100 were subjected to the regrowth assay. The depletion of γ-tubulin reduced microtubule regrowth to 9.20 ± 2.89% of control cells (n = 60 cells for each quantification). (D) 51–100 (GFP tagged) was expressed in the background of endogenous CDK5RAP2 depleted by RNAi or in the control. (left) Cell extracts were immunoblotted for GFP-51–100 (anti-GFP), endogenous CDK5RAP2, and β-tubulin. (right) The cells were stained for microtubules (anti–α-tubulin) and endogenous CDK5RAP2 after microtubule regrowth for 1 min. Arrows denote centrosomes. (right) Enlarged views of the boxed areas are shown. Bars, 10 µm.
Microtubule regrowth induced by γ-TuNA was tested when the expression of γ-tubulin or endogenous CDK5RAP2 was suppressed. The transfection of a _γ-tubulin_–targeting siRNA effectively reduced γ-tubulin level (by ∼85%) and almost completely inhibited microtubule formation even in the presence of 51–100 (Fig. 3 C). Next, the RNAi-mediated depletion of CDK5RAP2 had no impact on 51–100-induced regrowth of microtubules (Fig. 3 D). Thus, γ-TuNA induces microtubule growth in a manner dependent on γ-tubulin but independent on endogenous CDK5RAP2. Similarly, the transient expression of the entire CDK5RAP2 protein induced γ-tubulin–dependent growth of cytoplasmic microtubules, but the expression of its F75A mutant did not (Fig. S3, A and B).
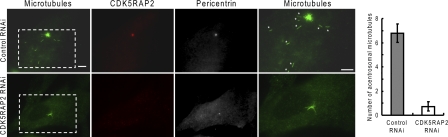
In our previous assays, the depletion of CDK5RAP2 was found to inhibit microtubule nucleation at centrosomes (Fong et al., 2008). In the following assays, we assessed its role in cytoplasmic nucleation with the modified fixation method. The depletion of CDK5RAP2 does not affect γ-TuRC assembly (Fong et al., 2008). In the assays, microtubules were observed to nucleate at dispersed sites in the cytoplasm in addition to the centrosomes (Figs. 3 and 4). In control cells, acentrosomal nucleation was most evident at 1.5 min of regrowth. The quantification of acentrosomal microtubules revealed that the disruption of CDK5RAP2 expression significantly reduced cytoplasmic nucleation (Fig. 4). These results combined with our previous study (Fong et al., 2008) indicate that CDK5RAP2 is required for efficient nucleation of microtubules both on centrosomes and in the cytoplasm.
Figure 4.
Cytoplasmic microtubule nucleation is impaired by the depletion of CDK5RAP2. (left) Microtubules regrew for 1.5 min after depolymerization in MRC-5 cells. Asterisks mark acentrosomal microtubules. Enlarged views of the boxed areas are shown. Bar, 10 µm. (right) The bar graph shows the number of acentrosomal microtubules. Data are shown as mean ± SD of three independent experiments; at least 20 cells were analyzed in each experiment.
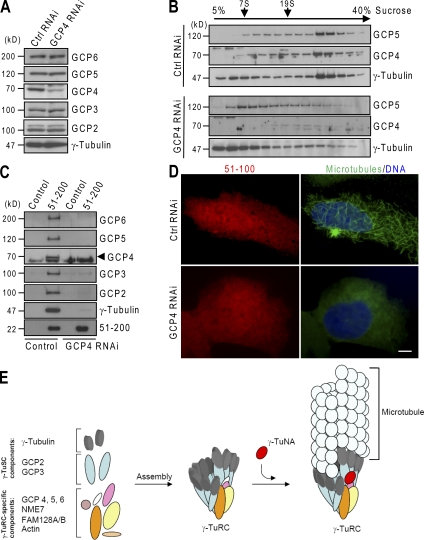
In Drosophila, Dgrip75 (a GCP4 orthologue) is required for the formation of γ-TuRC from γ-TuSCs (Vérollet et al., 2006). We attempted to disassemble γ-TuRC without affecting γ-TuSC by knocking down GCP4. The transfection of a _gcp4_-targeting siRNA effectively suppressed GCP4 expression without affecting the levels of γ-tubulin and other GCPs (Fig. 5 A). The expression suppression shifted most γ-tubulin and GCP5 as tested on the immunoblots to low molecular mass species (Fig. 5 B), indicating the disassembly of γ-TuRC. We then probed the binding of γ-TuRC proteins to γ-TuNA. In a pull-down assay, 51–200 did not bind to γ-tubulin and GCP2/3/5/6 in the extract depleted of GCP4, whereas the binding of γ-tubulin and GCPs was readily detected in the control extract (Fig. 5 C). Concomitantly, silencing GCP4 expression inhibited microtubule nucleation induced by 51–100 in the regrowth assay (Fig. 5 D). Therefore, γ-TuNA promotes microtubule growth via association with γ-TuRC. Using in vitro binding assays, we did not detect the direct binding of γ-TuNA to GCP4 or the newly identified γ-TuRC proteins NME7, FAM128A/B, and actin. These data revealed that γ-TuNA associates with γ-tubulin and GCPs only in the assembled γ-TuRC.
Figure 5.
Depletion of GCP4 disrupts the association of γ-TuNA with γ-TuRC. (A) siRNA-transfected HEK293T extracts were immunoblotted for γ-tubulin and GCPs. (B) Cell extracts were subjected to sucrose gradient centrifugation. An aliquot of each gradient fraction was analyzed by immunoblotting. (C) His-Flag–CDK5RAP2 (51–200) was used in the pull down of γ-TuCs from siRNA-transfected extracts. The anti-Flag precipitates were analyzed by immunoblotting. Note that the protein band below GCP4 is a heat shock protein bound nonspecifically to anti-Flag beads. (D) Microtubule regrowth was performed on U2OS cells with the regrowth time of 1 min. GCP4 depletion reduced microtubule regrowth to 14.02 ± 3.84% of control cells (n = 60 cells for each quantification). Bar, 10 µm. (E) Model for the activation of γ-TuRC–mediated microtubule nucleation by γ-TuNA. After γ-TuRC assembly, γ-TuNA binds to the complex to stimulate its nucleation of microtubules.
As the principal nucleator of microtubules, γ-TuRC plays a pivotal role in microtubule organization. However, the molecular mechanisms responsible for the regulation of its microtubule-nucleating activity remain unknown. For the first time, we show that γ-TuNA identified from CDK5RAP2 is a stimulating factor of γ-TuRC–mediated microtubule nucleation. CDK5RAP2 is required for efficient microtubule nucleation by γ-TuRC, as the suppression of CDK5RAP2 expression causes remarkable defects of both centrosomal and acentrosomal microtubule nucleation. A mouse mutant with the deletion of the γ-TuNA region (exon 4) from CDK5RAP2 develops microcephaly and several other defects, providing additional evidence on the functional requirement of this domain (Lizarraga et al., 2010). In fission yeast, Mto1p, a CDK5RAP2-related protein, is indispensable for cytoplasmic microtubule nucleation (Sawin et al., 2004; Venkatram et al., 2004; Zimmerman and Chang, 2005). Furthermore, the mutations disrupting the function of its CM1 region, a sequence corresponding to γ-TuNA, lead to the inhibition of cytoplasmic nucleation (Samejima et al., 2008), implying that CM1 may have a similar function as γ-TuNA. Therefore, the γ-TuNA identified in this study from CDK5RAP2 may represent an evolutionarily conserved regulation of the microtubule-nucleating function of γ-TuCs.
We have found that the assembly of γ-tubulin and GCPs as γ-TuRC is a prerequisite for their association with γ-TuNA. The γ-TuNA–associated γ-TuRC was found to contain NME7, FAM128A/B, and actin in addition to γ-tubulin and GCP2–6. Moreover, the molecular assembly of γ-TuRC is more precisely defined with the stoichiometries determined for γ-tubulin and GCPs within the complex. These nine proteins may serve as the major constituents of γ-TuRC that is activated by CDK5RAP2 for microtubule nucleation (Fig. 5 E), as we did not detect any other protein from the purified complex. In particular, GCP-WD/NEDD1, a protein involved in the centrosomal attachment of γ-TuRC (Haren et al., 2006; Lüders et al., 2006), was not found in the γ-TuNA–bound complex. Within γ-TuRC, GCP4 and perhaps other γ-TuRC–specific proteins participate in tethering γ-TuSCs into the large ring structure (Vérollet et al., 2006; this study).
The structural analyses of the budding yeast γ-TuSC and a γ-TuRC–like ring structure assembled by γ-TuSC have provided insights into the control of microtubule nucleation by γ-TuCs. Within γ-TuSC, the two Tub4p molecules are separated in distances that are incompatible with that of adjacent protofilaments in the microtubule lattice (Kollman et al., 2008). Notably, such an incompatible spacing of Tub4p is not changed by γ-TuSC assembly into the γ-TuRC–like structure (Kollman et al., 2010). In mammalian cells, γ-TuSC assembles with γ-TuRC–specific proteins into the γ-TuRC that has the capacity of binding to γ-TuNA (Fig. 5 E). The binding of γ-TuNA plausibly induces a conformational change to bring the γ-tubulins into a position competent for nucleation, resulting in γ-TuRC activation. Future studies are awaited to analyze the activation mechanism used by γ-TuNA.
Materials and methods
Reagents
The CDK5RAP2 plasmids, antibodies, and siRNAs were used as reported previously (Fong et al., 2008, 2009). The following proteins were prepared in fusion with a His6 tag by bacterial expressions for assays or for immunizing rabbits: CDK5RAP2 (51–200), GCP2 (734–902), GCP4 (202–371), GCP5 (1–180), and GCP6 (805–1060). The antibodies generated against GCP2, GCP4, and GCP6 were purified using respective antigens prepared in fusion with GST and without His6. The GCP5 antibody was purified using immobilized GCP5 (121–150). The specificity of the purified antibodies was tested by immunoblotting cell extracts with or without expression silencing of respective proteins (Fig. S1 C). Alternatively, the specificity was tested by preblocking antibodies with respective antigens for immunoblotting (Fig. S1 D). The generation of an anti-GCP3 antibody was previously described (Fong et al., 2008). The following antibodies were purchased: rabbit anti-GFP (Santa Cruz Biotechnology, Inc.), anti-Flag (monoclonal M2 and rabbit polyclonal; Sigma-Aldrich), goat anti–pericentrin 2 (C-16; Santa Cruz Biotechnology, Inc.), anti–α-tubulin (B-5-1-2; Sigma-Aldrich), anti–β-tubulin (TUB2.1; Sigma-Aldrich), anti–γ-tubulin (GTU88 and rabbit polyclonal; Sigma-Aldrich), and anti–β-actin (AC-15; Sigma-Aldrich). The following secondary antibodies were used in immunostaining: Alexa Fluor dye (Alexa Fluor 488, 568, or 647)–conjugated antibodies (Invitrogen). siRNA duplexes were synthesized against the following human sequences: γ-tubulin (5′-AGGAGGACATGTTCAAGGA-3′), GCP2 (5′-GCATGAAATTAGATGGCGA-3′), GCP3 (5′-GGGCGAAGCGGATGGAATA-3′), GCP4 (5′-GCAATCAAGTGGCGCCTAA-3′), and GCP5 (5′-GGAACATCATGTGGTCCATC-3′). The Flag peptide (DYKDDDK) was synthesized at >90% purity from GenScript.
Cell culture and microscopy
HEK293T and U2OS were grown in DME (Invitrogen) containing 10% fetal bovine serum at 37°C in 5% CO2. Human fetal lung fibroblasts MRC-5 were grown in MEM (Invitrogen) supplemented with 10% fetal bovine serum. To perform microtubule regrowth, cells grown on fibronectin or poly-d-lysine–coated cover glasses were placed on ice water for 1 h to depolymerize microtubules. Microtubule regrowth was initiated at 37°C and proceeded for various time periods. Cells were fixed for 10 min at room temperature with 4% paraformaldehyde in a buffer of 60 mM Pipes, 25 mM Hepes, pH 6.9, 10 mM EGTA, 1 mM MgCl2, and 0.5% Triton X-100 before immunostaining. Nuclear DNA was stained with Hoechst 33258 (Sigma-Aldrich). Wide-field microscopy was performed with an inverted microscope (Eclipse TE2000E; Nikon) equipped with 100× 1.40 NA Plan Apo oil objective and a camera (SPOT-RT1200; Diagnostic Instruments, Inc.). Images were acquired and processed with MetaMorph software (Universal Imaging). To quantify the intensity of cellular microtubules, the fluorescence intensity was taken from an entire cell using MetaMorph software. After subtraction of background measured from a no-cell area of the same size, the data were presented as percentages of control cells.
γ-TuRC isolation
γ-TuRC was isolated by immunoprecipitation from a mixture of Flag-51–100-transfected and untransfected HEK293T cells at the ratio of 1:2. After the preparation of cell extracts in lysis buffer (50 mM Hepes, pH 7.2, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 0.5% NP-40, and Complete protease inhibitors [Roche]) containing 0.1 mM GTP, immunoprecipitation was performed by incubating the extracts with anti-Flag M2-coupled beads. After washing with the lysis buffer and subsequently with the buffer but containing 0.01% NP-40, Flag-51–100 and bound proteins were eluted by incubating the beads for 30 min with 0.2 mg/ml Flag peptide in the washing buffer. The eluate was subjected to sucrose gradient (5–40%) centrifugation. After centrifugation, each fraction was resolved by SDS-PAGE for Sypro ruby (Bio-Rad Laboratories) or silver staining and for immunoblotting. Protein bands were excised from γ-TuRC–containing gel lanes for tryptic digestion followed by mass spectrometry. Mass spectra were acquired on a mass spectrometer (LTQ FT Ultra; Thermo Fisher Scientific) for searching gene databases to reveal protein identity. To determine the stoichiometry of isolated γ-TuRC proteins, images of SDS-PAGE gels acquired on an imager (Typhoon Trio; GE Healthcare) were analyzed using ImageQuant TL software (GE Healthcare).
Preparation of CDK5RAP2 proteins
HEK293T cells transfected with Flag-CDK5RAP2 or its F75A mutant were extracted in lysis buffer. The extracts were clarified by centrifugation at 100,000 g (30 min at 4°C) before anti-Flag immunoprecipitations were performed. The immunoprecipitates were extensively washed with the lysis buffer containing 1 M NaCl, 1% NP-40, and 12 mM deoxycholate and were subsequently washed with the lysis buffer containing 0.01% NP-40. After elution in the buffer supplemented with 0.2 mg/ml Flag peptide, the CDK5RAP2 proteins were dialyzed against HB buffer (50 mM Hepes, pH 7.2, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, and 20 µM GTP). The CDK5RAP2 proteins were verified by immunoblotting.
Microtubule nucleation in vitro
Purification of porcine brain α/β-tubulin was described previously (Hou et al., 2007). The purified γ-TuRC was dialyzed against HB buffer. Before use in assays, α/β-tubulin was clarified by centrifugation at 287,000 g (TLA100.1 rotor; Beckman Coulter), and other proteins were centrifuged at 100,000 g for 10 min at 4°C. Microtubule nucleation was performed as described previously with modifications (Oegema et al., 1999; Vinh et al., 2002). In brief, microtubule polymerization was conducted with 1 µg/µl α/β-tubulin (a mixture of rhodamine-labeled [Cytoskeleton, Inc.] and unlabeled tubulin at the ratio of 1:12) at 37°C for 3 min in 5 µl BRB80 (80 mM K-Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) supplemented with 1 mM GTP. γ-TuRC (∼2 nM GCP5) was included in the assay with or without CDK5RAP2 proteins (preincubation on ice for 30 min). Microtubule polymerization was terminated by the addition of 50 µl prewarmed 1% glutaraldehyde/BRB80 and further incubation at room temperature for 3 min. The samples were then diluted in 1 ml ice-cold BRB80. An aliquot of the samples was centrifuged at 173,000 g (8 min at 4°C) in tubes (TLS55; Beckman Coulter) equipped with a Teflon platform to sediment microtubules through a 15% glycerol/BRB80 cushion onto cover glasses. For each sample, microtubule numbers were taken from 20 random fields under a fluorescence microscope.
Online supplemental material
Fig. S1 shows the characterization of CDK5RAP2 proteins and reagents. Fig. S2 shows that the mutation of Phe75 abolishes the association of CDK5RAP2 with γ-TuCs. Fig. S3 shows that the overexpression of CDK5RAP2 induces γ-tubulin–dependent nucleation of microtubules. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007030/DC1.
Acknowledgments
This work was supported by the Research Grants Council (General Research Fund and Collaborative Research Fund) and the University Grants Committee (Area of Excellence Scheme and Special Equipment Grant) of Hong Kong and the Nanoscience and Nanotechnology Program of The Hong Kong University of Science and Technology.
Footnotes
Abbreviations used in this paper:
GCP
γ-complex protein
γ-TuC
γ-tubulin complex
γ-TuNA
γ-TuRC–mediated nucleation activator
γ-TuRC
γ-tubulin ring complex
γ-TuSC
γ-tubulin small complex
References
- Bond J., Roberts E., Springell K., Lizarraga S.B., Lizarraga S., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Fava F., Raynaud-Messina B., Leung-Tack J., Mazzolini L., Li M., Guillemot J.C., Cachot D., Tollon Y., Ferrara P., Wright M. 1999. Human 76p: a new member of the γ-tubulin–associated protein family. J. Cell Biol. 147:857–868 10.1083/jcb.147.4.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.W., Choi Y.K., Rattner J.B., Qi R.Z. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the γ-tubulin ring complex. Mol. Biol. Cell. 19:115–125 10.1091/mbc.E07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.W., Hau S.Y., Kho Y.S., Jia Y., He L., Qi R.Z. 2009. Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol. Biol. Cell. 20:3660–3670 10.1091/mbc.E09-01-0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Remy M.H., Bazin I., Callebaut I., Wright M., Merdes A. 2006. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172:505–515 10.1083/jcb.200510028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Li Q., He L., Lim H.Y., Fu X., Cheung N.S., Qi D.X., Qi R.Z. 2007. Microtubule association of the neuronal p35 activator of Cdk5. J. Biol. Chem. 282:18666–18670 10.1074/jbc.C700052200 [DOI] [PubMed] [Google Scholar]
- Hutchins J.R., Toyoda Y., Hegemann B., Poser I., Hériché J.K., Sykora M.M., Augsburg M., Hudecz O., Buschhorn B.A., Bulkescher J., et al. 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 328:593–599 10.1126/science.1181348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating T.J., Borisy G.G. 2000. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat. Cell Biol. 2:352–357 10.1038/35014045 [DOI] [PubMed] [Google Scholar]
- Kollman J.M., Zelter A., Muller E.G., Fox B., Rice L.M., Davis T.N., Agard D.A. 2008. The structure of the γ-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell. 19:207–215 10.1091/mbc.E07-09-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman J.M., Polka J.K., Zelter A., Davis T.N., Agard D.A. 2010. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 466:879–882 10.1038/nature09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga S.B., Margossian S.P., Harris M.H., Campagna D.R., Han A.P., Blevins S., Mudbhary R., Barker J.E., Walsh C.A., Fleming M.D. 2010. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 137:1907–1917 10.1242/dev.040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. 2007. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8:161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., Stearns T. 2006. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Moritz M., Zheng Y., Alberts B.M., Oegema K. 1998. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 142:775–786 10.1083/jcb.142.3.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M.B., Guénebaut V., Heuser J., Agard D.A. 2000. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2:365–370 10.1038/35014058 [DOI] [PubMed] [Google Scholar]
- Murphy S.M., Preble A.M., Patel U.K., O’Connell K.L., Dias D.P., Moritz M., Agard D., Stults J.T., Stearns T. 2001. GCP5 and GCP6: two new members of the human γ-tubulin complex. Mol. Biol. Cell. 12:3340–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Wiese C., Martin O.C., Milligan R.A., Iwamatsu A., Mitchison T.J., Zheng Y. 1999. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144:721–733 10.1083/jcb.144.4.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina B., Merdes A. 2007. γ-tubulin complexes and microtubule organization. Curr. Opin. Cell Biol. 19:24–30 10.1016/j.ceb.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Samejima I., Miller V.J., Groocock L.M., Sawin K.E. 2008. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J. Cell Sci. 121:3971–3980 10.1242/jcs.038414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., Lourenco P.C., Snaith H.A. 2004. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14:763–775 10.1016/j.cub.2004.03.042 [DOI] [PubMed] [Google Scholar]
- Venkatram S., Tasto J.J., Feoktistova A., Jennings J.L., Link A.J., Gould K.L. 2004. Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Mol. Biol. Cell. 15:2287–2301 10.1091/mbc.E03-10-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vérollet C., Colombié N., Daubon T., Bourbon H.M., Wright M., Raynaud-Messina B. 2006. Drosophila melanogaster γ-TuRC is dispensable for targeting γ-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172:517–528 10.1083/jcb.200511071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh D.B., Kern J.W., Hancock W.O., Howard J., Davis T.N. 2002. Reconstitution and characterization of budding yeast γ-tubulin complex. Mol. Biol. Cell. 13:1144–1157 10.1091/mbc.02-01-0607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. 2000. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2:358–364 10.1038/35014051 [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. 2006. Microtubule nucleation: γ-tubulin and beyond. J. Cell Sci. 119:4143–4153 10.1242/jcs.03226 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., Mitchison T. 1995. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 378:578–583 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
- Zimmerman S., Chang F. 2005. Effects of γ-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell. 16:2719–2733 10.1091/mbc.E04-08-0676 [DOI] [PMC free article] [PubMed] [Google Scholar]