Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway (original) (raw)
Abstract
Although activating mutations in RAS oncogenes are known to result in aberrant signaling through multiple pathways, the role of microRNAs (miRNAs) in the Ras oncogenic program remains poorly characterized. Here we demonstrate that Ras activation leads to repression of the miR-143/145 cluster in cells of human, murine, and zebrafish origin. Loss of miR-143/145 expression is observed frequently in KRAS mutant pancreatic cancers, and restoration of these miRNAs abrogates tumorigenesis. miR-143/145 down-regulation requires the Ras-responsive element-binding protein (RREB1), which represses the miR-143/145 promoter. Additionally, KRAS and RREB1 are targets of miR-143/miR-145, revealing a feed-forward mechanism that potentiates Ras signaling.
Keywords: Pancreatic cancer, Ras, miR-143/145, microRNA, primary transcript
Activating mutations in the RAS family of proto-oncogenes are among the most common in human malignancies and are particularly prominent in pancreatic ductal adenocarcinoma (PDAC), where mutations in KRAS2 (hereafter referred to as KRAS) are found in 90%–95% of cases (Maitra and Hruban 2008). Ras activation stimulates a number of downstream effectors, including the mitogen-activated protein kinase (MAPK) and phosphoinositide-3-kinase (PI3K) pathways, ultimately leading to enhanced cellular proliferation, survival, and motility (Hingorani and Tuveson 2003). The KRAS gene mutation principally encountered in pancreatic cancer and other malignancies is an alteration of codon 12 that prevents GTP hydrolysis, trapping Kras in the constitutively active configuration (Caldas and Kern 1995).
MicroRNAs (miRNAs) are 18- to 24-nucleotide (nt) ssRNAs that cause accelerated turnover and reduced translation of imperfectly complementary target messenger RNAs. Over the last decade, >700 human miRNAs have been identified and implicated in the regulation of a wide range of cellular processes, including differentiation, proliferation, and apoptosis (Ambros 2004). Accordingly, a large body of evidence has established an important role for miRNAs in cancer pathogenesis. Globally abnormal miRNA expression patterns are a ubiquitous feature of human cancers, including PDAC, and specific miRNAs have been shown to act as critical components of key oncogenic and tumor suppressor pathways (Bloomston et al. 2007; Lee et al. 2007; Szafranska et al. 2007; Lotterman et al. 2008; Kent et al. 2009). Moreover, delivery of anti-neoplastic miRNAs represents a highly effective therapeutic strategy in experimental cancer models (Esquela-Kerscher et al. 2008; Kumar et al. 2008; Kota et al. 2009).
Although both Ras signaling and miRNA activities can profoundly influence cancer cell behavior, the role of miRNAs in Ras-mediated phenotypes is poorly defined. To investigate a possible link between miRNA regulation and Ras-induced transformation, we characterized miRNA expression following expression of a constitutively active _KRAS_G12D allele in multiple model systems. We show that activated Kras signaling consistently leads to repression of the miR-143/145 cluster, and reduced expression of these miRNAs is necessary to maintain the tumorigenic potential of pancreatic cancer cells. Moreover, we elucidate an evolutionarily conserved regulatory pathway whereby Kras signaling activates Ras-responsive element-binding protein 1 (RREB1), which directly represses the miR-143/145 promoter. Both KRAS and RREB1 transcripts are direct targets of these miRNAs, demonstrating the existence of a feed-forward pathway that potentiates Kras-mediated tumorigenesis.
Results and Discussion
Expression of activated Kras leads to down-regulation of the miR-143/145 cluster in diverse model systems
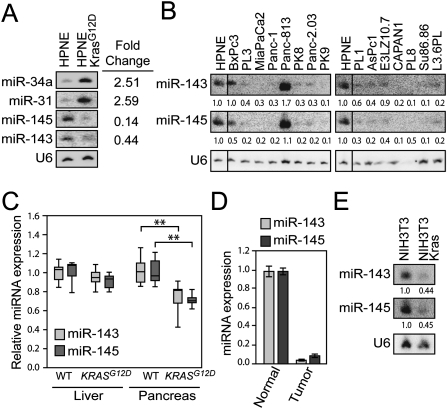
In order to identify miRNAs regulated by Kras signaling, we used a custom microarray to profile miRNA expression in a nontransformed pancreatic ductal epithelial cell line (HPNE) (KM Lee et al. 2003) stably overexpressing KrasG12D (Feldmann et al. 2008). Six miRNAs exhibited twofold or greater up-regulation (miR-34a, miR-199b, and miR-31) or down-regulation (miR-143, miR-145, and miR-27b) upon expression of KrasG12D (Supplemental Table S1). Of these, four miRNAs (miR-34a, miR-31, miR-143, and miR-145) showed similar expression changes when assessed by Northern blotting (Fig. 1A; data not shown). miR-34a has been demonstrated previously to be a direct transcriptional target of p53 and is up-regulated under conditions that trigger oncogene-induced senescence, including expression of activated Ras (He et al. 2007), likely explaining its increased expression in HPNE-KrasG12D cells. miR-31 has been shown previously to be overexpressed in tumor types where activating mutations of KRAS are common (Liu et al. 2010). Of particular interest, and the focus of this study, was the Kras-mediated repression of miR-143 and miR-145, two cotranscribed miRNAs located on human chromosome 5q. Significant evidence indicates that these miRNAs possess tumor suppressor activity. Reduced miR-143/145 expression is a common attribute of several tumor types, most notably colorectal carcinoma (Michael et al. 2003; Akao et al. 2007). Moreover, expression of these miRNAs inhibits proliferation and activates apoptosis of cancer cells in vitro and in vivo (Chen et al. 2009; Clape et al. 2009; Sachdeva et al. 2009).
Figure 1.
The miR-143/145 cluster is repressed by oncogenic Kras in multiple model systems. (A,B) Northern blot analysis of miRNA expression in HPNE cells (A) or PDAC cell lines (B). (C) Quantitative PCR (qPCR) analysis of relative miR-143/miR-145 expression in liver and pancreas of postnatal day 0 (P0) mice (n = 7 mice per genotype). Box plots show median (horizontal line), 25th and 75th percentiles (box), and full range of data (whiskers). (**) P < 0.01 (two-tailed _t_-test). (D) miR-143/miR-145 expression in normal zebrafish pancreas and pancreatic tumors induced by transgenic KrasG12V expression (Park et al. 2008). (E) miR-143/145 expression in NIH3T3 and _v-Kras_-transformed NIH3T3 cells.
Pancreatic cancer cell lines frequently exhibit low expression of miR-143/145 (Fig. 1B). Of note, one of two examined lines with retained miR-143/145 expression was BxPc3, the only cell line with wild-type KRAS in the panel (Sipos et al. 2003). Primary human pancreatic adenocarcinoma cells harboring activating KRAS mutations passaged as xenografts in mice (Rubio-Viqueira et al. 2006) also showed strong down-regulation of these miRNAs compared with normal pancreas tissue (Supplemental Fig. S1). Importantly, deletion of the miR-143/145 locus is an infrequent event in pancreatic cancer cells, as revealed by a previously published analysis of genomic copy number variation that included 10 of 15 cell lines in our panel (Calhoun et al. 2006).
To further demonstrate that oncogenic Kras represses miR-143 and miR-145 in vivo, we examined miRNA levels in pancreata from newborn PDX-1-Cre; LSL-KRASG12D mice, which harbor an activating mutation at the endogenous KRAS locus that is specifically expressed in pancreas and select other tissues (Hingorani et al. 2003). Repression of miR-143/145 was detectable in bulk pancreas in these animals relative to wild-type littermate controls (Fig. 1C). Moreover, tumors from a zebrafish model of pancreatic cancer induced by expression of oncogenic Kras (Park et al. 2008) exhibited nearly complete loss of expression of miR-143/145 relative to normal zebrafish pancreas (Fig. 1D). Finally, to determine if repression of miR-143/145 by Kras signaling is restricted to pancreas-derived cells, we examined their expression in the immortalized mouse fibroblast cell line NIH3T3 and its oncogenic _v-Kras_-transformed derivative, K:MoLV NIH3T3 (hereafter referred to as NIH3T3-Kras). miR-143 and miR-145 were strongly down-regulated by activated Kras in this setting as well (Fig. 1E). These experiments reveal the existence of an evolutionarily conserved pathway wherein activated Kras signaling leads to repression of the miR-143/145 cluster in diverse cell lineages.
Reduced expression of miR-143 and miR-145 is necessary for Kras-mediated transformation
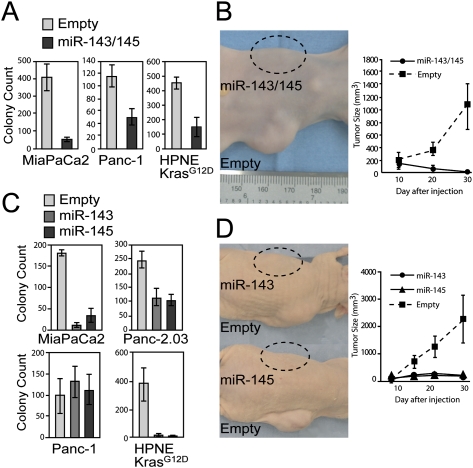
In order to assess whether down-regulation of miR-143/145 is necessary for cellular transformation induced by oncogenic Kras expression, we used a retroviral construct to express the miRNA cluster at physiologic levels in two PDAC cell lines harboring activating KRAS mutations (MiaPaCa2 and Panc-1) and in HPNE-KrasG12D cells (Supplemental Fig. S2A; Supplemental Table S2). Despite normal rates of proliferation in culture (Supplemental Fig. S2B), cells expressing miR-143/145 exhibited greatly reduced anchorage-independent growth (Fig. 2A; Supplemental Fig. S2C). MiaPaCa2 and Panc-1 cells expressing these miRNAs were also unable to form tumors in immunocompromised mice (Fig. 2B; data not shown). These data document that expression of miR-143/145 fully abrogates the transformed phenotype of PDAC cells. Future studies will be necessary to elucidate the underlying mechanisms through which the disparate effects on cellular proliferation versus transformation are elicited.
Figure 2.
Expression of miR-143/145 inhibits transformation and tumorigenesis in pancreatic cancer cells. (A) Anchorage-independent growth of retrovirally infected cell lines (average of three independent experiments shown). Error bars for this and subsequent panels represent standard deviations. (B) Tumorigenesis assays with retrovirally infected MiaPaCa2 cells. Images depict a representative animal injected in each flank with cells infected with empty virus or miR-143/145 virus. The graph shows the average tumor volume (n = 5 mice per cell line). (C) Anchorage-independent growth of retrovirally infected cell lines expressing either miR-143 or miR-145. (D) Tumorigenesis assays with MiaPaCa2 cells infected with empty virus or virus expressing miR-143 or miR-145 (n = 5 mice per cell line).
We next determined whether the observed anti-tumorigenic effects of miR-143/145 expression could be attributed to either of the individual miRNAs in the cluster. miR-143 or miR-145 were expressed individually in HPNE-KrasG12D, MiaPaCa2, Panc-1, or an additional PDAC cell line, Panc-2.03, at physiologically relevant levels (Supplemental Fig. S3A; Supplemental Table S2). Again, no effect on proliferation rates was observed (Supplemental Fig. S3B). Independent expression of miR-143 or miR-145 strongly inhibited soft agar colony formation in HPNE-KrasG12D, MiaPaCa2, and Panc-2.03 cells but not in Panc-1 cells (Fig. 2C; Supplemental Fig. S3C). Likewise, miR-143 or miR-145 were able to independently block tumor formation by MiaPaCa2 cells (Fig. 2D). These observations indicate that both miRNAs in the miR-143/145 cluster exhibit tumor suppressor activity in pancreatic cancer cells, although in some settings both miRNAs must be coexpressed for the cluster to exert its full anti-tumorigenic potential.
Kras signaling leads to transcriptional repression of the miR-143/145 cluster in a RREB1-dependent manner
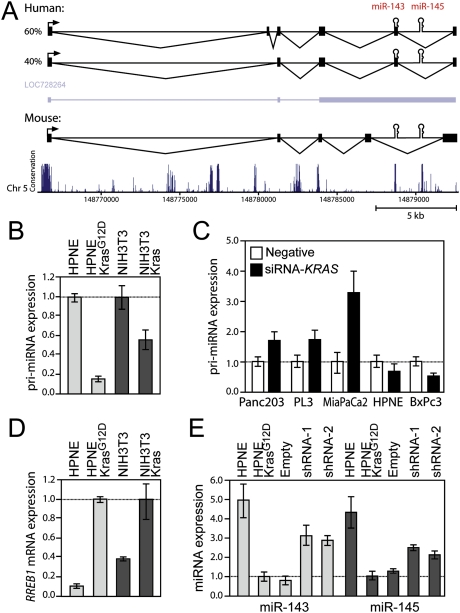
To investigate the mechanisms through which Kras signaling represses expression of the miR-143/145 cluster, we first elucidated the complete structure of the primary transcript (pri-miRNA) that encodes these miRNAs. An NCBI RNA reference sequence (RefSeq) transcript (LOC728264) encompasses the human miR-143/145 cluster. We confirmed the use of the RefSeq transcription start site (TSS) and polyadenylation signal using 5′ and 3′ rapid amplification of cDNA ends (RACE). However, RT–PCR revealed that HPNE cells express two major alternatively spliced transcripts with intron–exon structures distinct from the annotated RefSeq RNA (Fig. 3A). A similar series of experiments in NIH3T3 cells revealed that the genomic organization of the mouse miR-143/145 transcription unit is similar to the human locus, although the positions of a subset of the exons are unique to each species.
Figure 3.
Kras-mediated repression of miR-143/145 occurs at the level of transcription and requires RREB1. (A) Structures of the experimentally determined human and mouse miR-143/145 primary transcripts and the annotated human RefSeq transcript (LOC728264). The plot depicted below the transcripts shows evolutionary conservation (UCSC Genome Browser 28 species conservation track, NCBI36/hg18 assembly). (B) qPCR analysis of pri-miR-143/145 expression in HPNE and NIH3T3-derived cell lines. Error bars for this and subsequent panels represent standard deviations from three independent measurements. (C) Pri-miR-143/145 abundance in cells transfected with negative control or KRAS targeting siRNAs. (D) RREB1 expression in HPNE and NIH3T3-derived cell lines. (E) Mature miR-143 and miR-145 expression in HPNE-KrasG12D cells following knockdown of RREB1.
Close inspection of the highly conserved miR-143/145 proximal promoter using the University of California at Santa Cruz (UCSC) Genome Browser conserved transcription factor-binding site track revealed the presence of a high-scoring recognition site for RREB1 (Supplemental Fig. S4A), a transcription factor that has emerged from multiple screens as a likely oncogene (Uren et al. 2008; Starr et al. 2009). RREB1 is known to both activate and repress transcription of target genes in response to Ras pathway activity (Thiagalingam et al. 1996; Zhang et al. 1999; Date et al. 2004; Mukhopadhyay et al. 2007). These findings suggest that oncogenic Kras signaling represses transcription of these miRNAs through the direct action of RREB1 at the miR-143/145 promoter.
Consistent with transcriptional repression, pri-miR-143/145 was down-regulated in HPNE and NIH3T3 cells expressing KrasG12D (Fig. 3B). Moreover, acute knockdown of KRAS expression using siRNA partially reverses repression of pri-miR-143/145 in cell lines with activating mutations in KRAS (Panc-2.03, PL3, and MiaPaCa2) but not in cell lines with wild-type KRAS (HPNE and BxPc3) (Fig. 3C; Supplemental Fig. S4B). Expression of KrasG12D in HPNE and NIH3T3 cells also induced RREB1 expression (Fig. 3D). RREB1 knockdown in HPNE-KrasG12D cells reversed the repression of pri-miR-143/145 and the mature miRNAs and inhibited soft agar colony formation (Fig. 3E; Supplemental Fig. S4C–E). These data establish an essential role for RREB1 in Kras-mediated repression of the miR-143/145 cluster.
Direct repression of the miR-143/145 promoter by RREB1
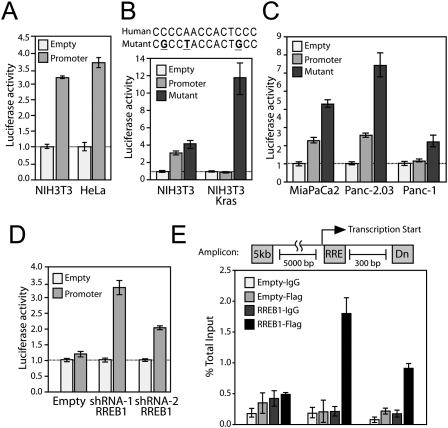
To determine whether RREB1 directly represses the miR-143/145 promoter, we constructed a luciferase reporter plasmid with a 1-kb human genomic segment encompassing the highly conserved TSS (the region shown in Supplemental Fig. S4A), which was active in multiple cell lines (Fig. 4A). Like the endogenous promoter, this reporter was repressed in NIH3T3-Kras relative to NIH3T3 cells, and mutations in the RREB1-binding site reversed this repression (Fig. 4B). Similarly, the mutant promoter construct was derepressed relative to the wild-type promoter when introduced into PDAC cell lines harboring activating mutations in KRAS (Fig. 4C). Additionally, shRNA-mediated RREB1 knockdown in NIH3T3-Kras cells induced activity of the promoter (Fig. 4D). Lastly, we performed chromatin immunoprecipitation (ChIP), which documented that Flag-tagged RREB1 interacts directly with the endogenous miR-143/145 promoter in NIH3T3 cells (Fig. 4E). These experiments demonstrate that RREB1 directly represses the activity of the miR-143/145 promoter in cells expressing activated Kras.
Figure 4.
RREB1 negatively regulates the miR-143/145 promoter. (A) Activity of the miR-143/145 promoter reporter construct in NIH3T3 and HeLa cells. For this and subsequent panels, values represent firefly luciferase activity produced from each plasmid normalized to renilla luciferase activity produced from a cotransfected control vector. Error bars represent standard deviations from three independent transfections, each measured in triplicate. (B,C) Activity of the wild-type and mutant miR-143/145 promoter reporter constructs in NIH3T3 and NIH3T3-Kras cells (B) or PDAC cell lines (C). (D) RREB1 knockdown reverses repression of the miR-145/145 promoter reporter construct in NIH3T3-Kras cells. (E) qPCR analysis of Flag-RREB1 chromatin immunoprecipitates in NIH3T3 cells. Signal obtained from the miR-143/145 promoter amplicon containing the RREB1-binding site (RRE), an amplicon 300 base pairs (bp) downstream (Dn), and a negative control amplicon 5 kb upstream (5kb) are shown. Error bars represent standard deviations derived from three independent measurements.
A feed-forward circuit established by direct targeting of KRAS and RREB1 by miR-143 and miR-145
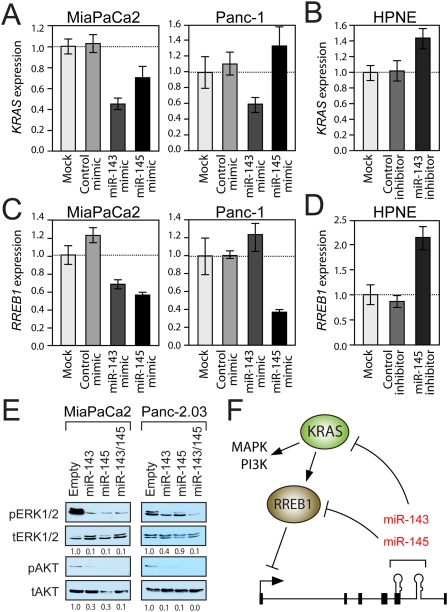
Interestingly, KRAS and RREB1 are identified by TargetScan (Grimson et al. 2007) as potential miR-143 and miR-145 targets, respectively. Moreover, the direct targeting of KRAS by miR-143 in colon cancer cells was recently experimentally validated (Chen et al. 2009). Overexpression of miR-143 reduces KRAS mRNA and protein abundance in MiaPaCa2 and Panc-1 cells (Fig. 5A; Supplemental Fig. S5A). Conversely, inhibition of miR-143 increases KRAS expression in HPNE cells (Fig. 5B). These observations confirm that miR-143 regulates KRAS in pancreatic cancer cells.
Figure 5.
miR-143 and miR-145 target KRAS and RREB1, establishing a feed-forward circuit that potentiates Ras signaling. (A–D) qPCR analysis of KRAS (A,B) or RREB1 (C,D) transcript abundance in cells transfected with negative control, miR-143, or miR-145 mimics (A,C) or inhibitors (B,D). Error bars represent standard deviations from three independent measurements. (E) Western blot analysis of phospho-ERK1/2 (pERK1/2), total ERK1/2 (tERK1/2), phospho-AKTS473 (pAKT), and total AKT (tAKT) in retrovirally infected cell lines. (F) Model of feed-forward regulatory circuit in which Kras signaling, via RREB1, represses miR-143/145 transcription, thereby relieving KRAS and RREB1 negative regulation and potentiating signaling through Ras effector pathways.
RREB1 expression was reduced upon miR-145 overexpression in MiaPaCa2 and Panc-1 cells and was increased upon miR-145 inhibition in HPNE cells (Fig. 5C,D), suggesting that this transcript is a bona fide miR-145 target. The RREB1 3′ untranslated region (UTR) contains one predicted miR-145-binding site, which is highly conserved in vertebrates (Supplemental Fig. S5B). Reporter assays verified that this site, but not a mutant version, resulted in miR-145-mediated repression when placed in the 3′ UTR of luciferase (Supplemental Fig. S5C).
The ability of miR-143 and miR-145 to negatively regulate KRAS and RREB1 expression suggests the existence of a feed-forward circuit through which Kras-mediated repression of these miRNAs further potentiates Kras signaling. Consistent with this model, miR-143 reduces RREB1 expression in MiaPaCa2 cells (Fig. 5C), possibly indirectly through targeting of KRAS. Similarly, miR-145 expression results in reduced KRAS levels in this cell line (Fig. 5A), likely by inhibiting RREB1, thereby increasing miR-143, which can target KRAS. Interestingly, this circuit is not fully active in Panc-1 cells, since miR-143 did not repress RREB1 and miR-145 did not repress KRAS (Fig. 5A,C). Although the molecular basis for this defect in the circuit in this cell line remains to be elucidated, this might explain why Panc-1 is insensitive to individual expression of the miRNAs (Fig. 2F).
To further test this model, we examined the activity of two major Kras effectors—the MAPK and the PI3K pathways—in pancreatic cancer cells with enforced miR-143/145 (Fig. 5E). Expression of these miRNAs reduced phosphorylation of ERK1/2 (indicative of MAPK signaling) and AKT (indicative of PI3K signaling). Thus, expression of miR-143/145 suppresses Ras pathway activity.
miR-143 and miR-145 have well-documented anti-proliferative and proapoptotic effects through their ability to negatively regulate a host of target genes, including MYC, Insulin Receptor Substrate-1 (IRS-1), and Extracellular Signal-Regulated Kinase 5 (ERK5) (Esau et al. 2004; Shi et al. 2007; Chen et al. 2009; Sachdeva et al. 2009). Repression of this miRNA cluster thereby contributes to Ras-mediated cellular transformation by promoting the coordinated activation of other important oncogenic signals. In addition, our studies uncovered the existence of a feed-forward loop established by the direct targeting of KRAS and RREB1 by miR-143 and miR-145, respectively (Fig. 5F). This circuit is reminiscent of feed-forward loops involving miRNAs in other key oncogenic pathways that establish irreversible switches that drive cellular transformation. This is well illustrated by the recently described NF-κB/IL-6/Lin28/let-7 pathway (Iliopoulos et al. 2009), in which NF-κB signaling activates expression of Lin28, an inhibitor of let-7 biogenesis, thereby increasing expression of IL-6 (a let-7 target), which further stimulates NF-κB activity. Similarly, Myc is another let-7 target that activates Lin28 expression (Chang et al. 2009; Dangi-Garimella et al. 2009), constituting an intersecting positive feedback loop. These observations demonstrate that major oncoproteins—including Ras, Myc, and NF-κB—use similar regulatory circuitry to establish stable cellular transformation programs, and suggest that use of analogous positive feedback loops may be a widespread property of oncogenic pathways.
The ability of miR-143/145 expression to completely abrogate tumorigenesis by pancreatic cancer cells is likely the result of repression of a large set of oncogenic targets. Nevertheless, the ability of these miRNAs to robustly suppress downstream Ras signaling, as evidenced by suppression of the MAPK and PI3K pathways, likely plays a central role in suppressing pancreatic tumorigenesis, since activating KRAS mutations are nearly ubiquitous in this setting (Caldas and Kern 1995). In light of the importance of loss of miR-143/145 in sustaining Kras pathway activity and the exquisite sensitivity of pancreatic cancer cells to restored miR-143/145 expression, delivery of these miRNAs represents a promising therapeutic approach for this tumor type.
Materials and methods
Cell culture
Proliferation rates were measured using the CKK-8 kit (Dojindo). For anchorage-independent growth assays, cells were plated in 4% agarose in culture media; for tumorigenesis assays, 5 × 106 cells were subcutaneously injected into immunodeficient (BALB/c nu/nu) mice.
Measurement of miRNA expression
A custom microarray was used to profile miRNA expression as described previously (Chang et al. 2009). Following DROSHA knockdown (HPNE cells) (Y Lee et al. 2003) or Dgcr8 knockdown (NIH3T3 cells) with siRNA, the GeneRacer kit (Invitrogen) was used to characterize the miR-143/145 primary transcript.
Luciferase assays
Cells were transfected with pGL3-IRES-promoter reporter constructs or pGL3-control 3′ UTR reporter constructs using Lipofectamine 2000 (Invitrogen), and luciferase activity was assayed using the Dual-Luciferase Reporter Assay System (Promega). Where appropriate, control or miR-145 mimics (Dharmacon) were cotransfected with 3′ UTR reporters at 15 nM final concentration.
ChIP
A mouse N-terminal Flag-tagged RREB1 cDNA expression construct was transfected into NIH3T3 cells using PolyFECT (Qiagen), and ChIP was performed as described (Boyd et al. 1998). Immunoprecipitation was performed with anti-Flag-M2 antibody (Sigma) or control mouse IgG1 antibody (Dako).
Additional Materials and Methods are provided in the Supplemental Material.
Acknowledgments
We thank Nils Habbe for assistance with xenograft assays and Michel Ouelette at the University of Nebraska for HPNE cells. We also thank Alex Amiet and Devin Leake at Thermo Fisher Scientific (Dharmacon) for miRNA inhibitors, transfection reagents, and consultation. This work was supported by grants from The Lustgarten Foundation for Pancreatic Cancer Research (to J.T.M. and S.D.L.), The Sol Goldman Center for Pancreatic Cancer Research (to J.T.M), The Michael Rolfe Foundation for Pancreatic Cancer Research (to A.M.), and the NIH (R01CA120185 to J.T.M., R01CA113669 and P50CA062924 to A.M., and P01CA134292 to J.T.M., A.M., and S.D.L.). J.T.M. is an HHMI Early Career Scientist and a Rita Allen Foundation Scholar. O.A.K. is a Life Sciences Research Foundation Fellow supported by Pfizer.
Footnotes
Supplemental material is available for this article.
References
- Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T 2007. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci 98: 1914–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM 2007. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297: 1901–1908 [DOI] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci 95: 13887–13892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas C, Kern SE 1995. K-ras mutation and pancreatic adenocarcinoma. Int J Pancreatol 18: 1–6 [DOI] [PubMed] [Google Scholar]
- Calhoun ES, Hucl T, Gallmeier E, West KM, Arking DE, Maitra A, Iacobuzio-Donahue CA, Chakravarti A, Hruban RH, Kern SE 2006. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res 66: 7920–7928 [DOI] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. 2009. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci 106: 3384–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, et al. 2009. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28: 1385–1392 [DOI] [PubMed] [Google Scholar]
- Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, Avances C, Villalba M, Culine S, Fajas L 2009. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS ONE 4: e7542 doi: 10.1371/journal.pone.0007542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR 2009. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 28: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date S, Nibu Y, Yanai K, Hirata J, Yagami K, Fukamizu A 2004. Finb, a multiple zinc finger protein, represses transcription of the human angiotensinogen gene. Int J Mol Med 13: 637–642 [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et al. 2004. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279: 52361–52365 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ 2008. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7: 759–764 [DOI] [PubMed] [Google Scholar]
- Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, Fendrich V, Beaty R, Mullendore M, Karikari C, Bardeesy N, et al. 2008. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut 57: 1420–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Tuveson DA 2003. Ras redux: Rethinking how and where Ras acts. Curr Opin Genet Dev 13: 6–13 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4: 437–450 [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K 2009. An epigenetic switch involving NF-κB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Mullendore M, Wentzel EA, Lopez-Romero P, Tan AC, Alvarez H, West K, Ochs MF, Hidalgo M, Arking DE, et al. 2009. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol Ther 8: 2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T 2008. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci 105: 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM 2003. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun 301: 1038–1044 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD 2007. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120: 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, et al. 2010. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 120: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterman CD, Kent OA, Mendell JT 2008. Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle 7: 2493–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Hruban RH 2008. Pancreatic cancer. Annu Rev Pathol 3: 157–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1: 882–891 [PubMed] [Google Scholar]
- Mukhopadhyay NK, Cinar B, Mukhopadhyay L, Lutchman M, Ferdinand AS, Kim J, Chung LW, Adam RM, Ray SK, Leiter AB, et al. 2007. The zinc finger protein ras-responsive element binding protein-1 is a coregulator of the androgen receptor: Implications for the role of the Ras pathway in enhancing androgenic signaling in prostate cancer. Mol Endocrinol 21: 2056–2070 [DOI] [PubMed] [Google Scholar]
- Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD 2008. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology 134: 2080–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, Shi C, Danenberg K, Danenberg PV, Kuramochi H, et al. 2006. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res 12: 4652–4661 [DOI] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY 2009. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci 106: 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R 2007. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem 282: 32582–32590 [DOI] [PubMed] [Google Scholar]
- Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G 2003. A comprehensive characterization of pancreatic ductal carcinoma cell lines: Towards the establishment of an in vitro research platform. Virchows Arch 442: 444–452 [DOI] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, et al. 2009. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323: 1747–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA 2007. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26: 4442–4452 [DOI] [PubMed] [Google Scholar]
- Thiagalingam A, De Bustros A, Borges M, Jasti R, Compton D, Diamond L, Mabry M, Ball DW, Baylin SB, Nelkin BD 1996. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol Cell Biol 16: 5335–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, et al. 2008. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao J, Edenberg HJ 1999. A human Raf-responsive zinc-finger protein that binds to divergent sequences. Nucleic Acids Res 27: 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]