Identification of Therapeutic Targets for Quiescent, Chemotherapy-Resistant Human Leukemia Stem Cells (original) (raw)
. Author manuscript; available in PMC: 2010 Dec 21.
Abstract
Human acute myeloid leukemia (AML) originates from rare leukemia stem cells (LSCs). Because these chemotherapy-resistant LSCs are thought to underlie disease relapse, effective therapeutic strategies specifically targeting these cells may be beneficial. Here, we report identification of a primary human LSC gene signature and functional characterization of human LSC-specific molecules in vivo in a mouse xenotransplantation model. In 32 of 61 (53%) patients with AML, either CD32 or CD25 or both were highly expressed in LSCs. CD32- or CD25-positive LSCs could initiate AML and were cell cycle–quiescent and chemotherapy-resistant in vivo. Normal human hematopoietic stem cells depleted of CD32- and CD25-positive cells maintained long-term multilineage hematopoietic reconstitution capacity in vivo, indicating the potential safety of treatments targeting these molecules. In addition to CD32 and CD25, quiescent LSCs within the bone marrow niche also expressed the transcription factor WT1 and the kinase HCK. These molecules are also promising targets for LSC-specific therapy.
INTRODUCTION
Despite advances in cancer therapeutics and supportive care, long-term outcomes of patients with acute myeloid leukemia (AML) remain dismal (1). Even after complete remission in which the whole-body leukemia burden is reduced to nearly undetectable levels, most patients eventually succumb to disease relapse (2–4). There has been much interest, therefore, in the detection and elimination of minimal residual disease (MRD) to prevent relapse or for early treatment of relapse.
The concept of leukemia stem cells (LSCs) in human AML was proposed by Lapidot et al. (5), who reported that CD34+CD38− cells selectively engraft in CB17-SCID and nonobese diabetic–severe combined immunodeficient (NOD-SCID) mice. More recently, CD34+CD38− AML cells were shown to fulfill the criteria for cancer stem cells in vivo; that is, they have the capacity to generate AML, give rise to heterogeneous nonstem leukemic cells, and self-renew (6, 7). Moreover, human primary CD34+CD38− LSCs preferentially reside in the bone marrow (BM) endosteal region and are cell cycle–quiescent and chemotherapy-resistant in vivo (7). Together, these observations suggest that MRD can be attributed to rare quiescent CD34+CD38− LSCs remaining after therapy. Therefore, therapeutic strategies designed to eradicate LSCs may improve long-term outcomes in AML. Identification of LSC-specific molecules that could serve as drug targets would facilitate the development of therapeutic antibodies, inhibitors of LSC-specific kinases and transcription factors, or LSC-specific immunotherapy. To find possible targets for these therapeutic agents, which could possibly prevent AML relapse by eliminating LSCs, we performed global transcriptome analysis to compare human AML LSCs and human hematopoietic stem cells (HSCs) isolated from healthy individuals. These molecules were then screened for expression in primary human AML LSCs, and their functional significance was determined in vivo with a NOD-SCID-IL2rgnull primary human AML xenotransplantation model.
RESULTS
Functional definition of LSCs and HSCs
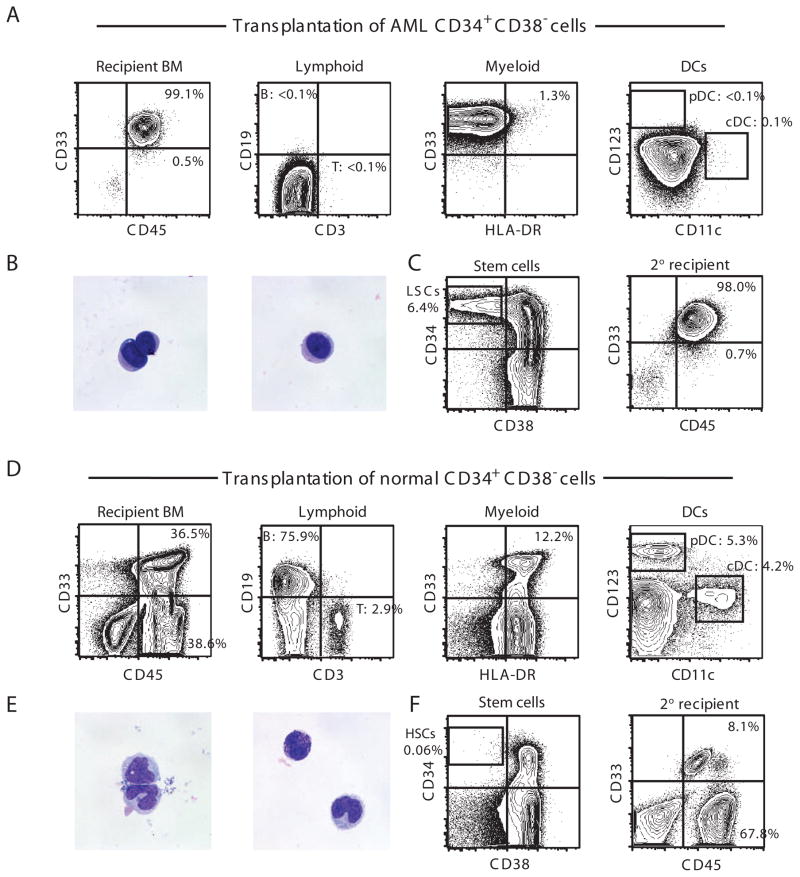
Ideally, a therapeutic strategy that targets LSCs must spare self-renewing normal HSCs to protect normal long-term hematopoiesis in the patient. Therefore, the comparison of functionally well-defined LSCs and HSCs is crucial to identify target molecules specific to LSCs. In humans, both AML stem cells and normal HSCs are enriched in CD34+CD38− blood cells (5–7). The transplantation of primary human AML BM or peripheral blood (PB) CD34+CD38− cells into newborn NOD-SCID-IL2rgnull mice results in BM repopulation by hCD45+CD33+ cells with no detectable T, B, and dendritic cells (DCs) and uniformly blast-like PB hCD45+CD33+ cells (Fig. 1, A and B). In addition, the transplantation of CD34+CD38− AML BM cells recapitulates the cellular heterogeneity in the original AML by repopulating CD34+CD38− LSCs and CD34+CD38+/CD34− nonstem AML cells (Fig. 1C). CD34+CD38− BM cells from AML-engrafted recipients can self-renew, as shown by their capacity for secondary engraftment (Fig. 1C). Finally, we have previously reported that AML patient BM CD34+CD38− cells and AML-engrafted recipient BM CD34+CD38− cells share a similar transcriptional profile (7). Therefore, we chose AML patient and AML-engrafted recipient CD34+CD38− cells as the LSC population for global expression profiling.
Fig. 1.
CD34+CD38− phenotype functionally defines primary human AML LSCs and human normal HSCs in vivo. (A to C) Xenotransplantation of CD34+CD38− primary human AML cells results in initiation of AML. pDC, plasmacytoid DC; cDC, conventional DC. In AML-engrafted recipients, (A) BM is entirely replaced with hCD45+CD33+ cells with the complete absence of the normal immune subsets such as DCs, T cells and B cells and (B) PB hCD45+CD33+ cells show uniform-appearing blast-like morphology by May-Grunwald-Giemsa staining. (C) BM of recipients of CD34+CD38− AML cells was reconstituted with CD34+CD38−, CD34+CD38+, and CD34− cells. Serial transplantation of CD34+CD38− cells results in complete replacement of the secondary recipient BM with hCD45+CD33+ cells. (D to F) Normal human CB CD34+CD38− cells are HSCs capable of long-term multilineage reconstitution of human hematopoiesis. In recipients of normal CB CD34+CD38− cells, (D) BM contains heterogeneous human hematopoietic cell types, including T cells, B cells, and DC subsets, by phenotype and (E) PB hCD45+CD33+ cells contain morphologically heterogeneous hematopoietic cells. (F) Transplantation of normal human CB CD34+CD38− cells results in the reconstitution of recipient BM with CD34+CD38−, CD34+CD38+, and CD34− cells (left). BM of secondary CD34+CD38− cell recipient contains both hCD33+CD45+ human myeloid and hCD33− CD45+ human lymphoid cells (right).
In contrast, the transplantation of normal human CD34+CD38− cells results in the repopulation of T cells, B cells, and DCs in the BM and morphologically heterogeneous hematopoietic cells in the PB (Fig. 1, D and E). Furthermore, transplanted normal hCD34+CD38− cells give rise to hCD34+CD38−, hCD34+CD38+, and hCD34− cells in recipient BM, and serial transplantation of purified BM hCD34+CD38− cells from normal HSC-engrafted recipients results in the reconstitution of hCD45+CD33+ myeloid and hCD45+CD33− lymphoid lineages in the secondary recipients (Fig. 1F). We therefore selected healthy human cord blood (CB) and BM CD34+CD38− cells for gene expression profiling.
Identification of human AML LSC-specific targets
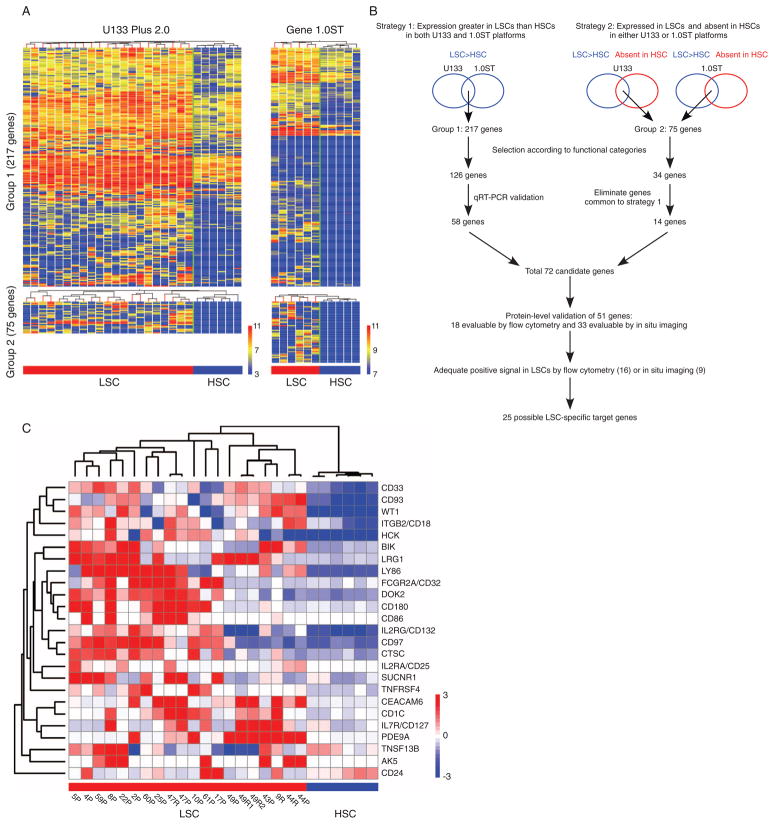
To increase the accuracy of candidate identification, we performed gene expression analyses with two independent array platforms: (i) the Human Genome U133 Plus 2.0 GeneChips with probes close to 3′ end for each gene and (ii) the Human Gene 1.0ST GeneChips with probes across the full length of each gene (Fig. 2A). We analyzed LSC populations from 21 AML specimens [AML: M1, n = 3; M2, n = 7; M4, n = 4; myelodysplastic syndrome (MDS)/AML: n = 7] and HSC populations from 2 normal BM and 4 CB specimens with the U133 Plus 2.0 platform, and LSC populations from 6 AML specimens (AML: M1, n = 1; M2, n = 3; M4, n = 2) and HSC populations from 4 normal BM and 1 CB specimens with the Gene 1.0ST platform (table S1).
Fig. 2.
LSC-specific gene candidates were derived from genes overrepresented in LSCs relative to HSCs. (A) Hierarchical clustering of genes overrepresented in AML LSCs relative to normal human HSCs identified by global expression pattern analyses with two microarray platforms. (B) Schematic representation of the analysis strategies integrating expression profiles obtained from two independent array platforms. (C) The expression of 25 candidate LSC-specific genes discriminate LSCs from HSCs. Patient ID and sample source (P, patient; R, recipient engrafted with the patient LSCs) are indicated below each column. The heat maps represent microarray signal intensity on a log2 scale.
Two independent strategies that we used to integrate the gene expression data obtained with the two platforms are summarized in Fig. 2B. First, the RankProd method was performed to extract genes with expression levels significantly higher in LSCs than in HSCs [P < 0.01; percentage of false positive (pfp) < 0.05] in both platforms (strategy 1, group 1, 217 genes; table S2) (8–10). Second, we identified genes that met the following criteria in all HSC samples tested in either array platform: (i) a pfp of <0.005 and (ii) expression lower than the median levels. Thereby, we extracted genes that were highly expressed in LSCs but showed minimal expression in HSCs (strategy 2, group 2, 75 genes; table S2).
Validation of putative LSC-specific targets
Among the 217 genes in group 1, 126 genes encoding molecules in the following categories were chosen for further evaluation as candidates for drug targeting: (i) proteins that localize in the plasma membrane or extracellular space; (ii) cytokines, growth factors, transmembrane receptors, protein kinases, phosphatases, transcriptional modulators, and/or other signaling molecules; and (iii) regulators of immunity, cell cycle, apoptosis, and/or cell adhesion. Of these, LSC-specific expression was validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) for 58 genes by using LSCs from five AML-engrafted recipient BM and four healthy BM HSCs (fig. S1 and table S1). Among the 75 genes in group 2, 34 fit the categories listed above. Of these, 20 genes were common to group 1 and had already undergone validation by qRT-PCR, leaving 14 genes for further analysis. Of the 72 candidate genes thus identified, cellular expression of 16 molecules could be adequately evaluated by flow cytometry (FCGR2A/CD32, ITGB2/CD18, CD93, CD97, CD33, IL2RG/CD132, LY86/MD1, TNFRSF4/CD134, TNFSF13B/CD257, IL2RA/CD25, CD127, BDCA-1, CD86, CD24, CD66c, and CD180) and 9 molecules by immunofluorescence imaging (WT1, SUCNR1, PDE9A, HCK, AK5, DOK2, LRG1, BIK, and CTSC). Therefore, overall, 25 genes were identified as possible LSC-specific target genes (Fig. 2C).
We next analyzed the expression of 25 LSC-specific target genes in 20 AML LSC samples (AML: M0, n = 1; M1, n = 5; M2, n = 3; M5, n = 1; MDS/AML: n = 10) and 6 normal BM HSC samples that were previously unexamined (table S1). By hierarchical clustering based on the expression patterns of the 25-gene signature, LSCs were successfully segregated from healthy HSCs (Fig. 2C).
LSC-specific targets in cell cycle–quiescent LSCs in situ
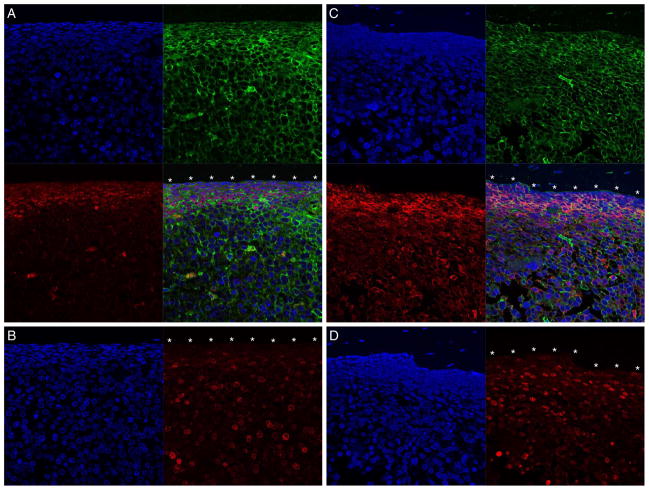
As we have previously reported that human AML LSCs residing in the BM endosteal region are cell cycle–quiescent and chemotherapy-resistant, we examined the expression of the nine candidate molecules evaluable by immunofluorescence labeling (WT1, SUCNR1, PDE9A, HCK, AK5, DOK2, LRG1, BIK, and CTSC) in situ in the BM endosteal region (7). Among them, WT1 and HCK were the most promising LSC targets, with genes encoding these molecules overrepresented in the greatest proportions of LSC samples by microarray analysis (fig. S2). These two molecules were expressed in the Ki67-negative cells lining the endosteum, indicating that cell cycle–quiescent LSCs express these molecules in situ (Fig. 3). SUCNR1, PDE9A, AK5, DOK2, LRG1, BIK, and CTSC were also detected in the endosteal region at varying levels of expression (fig. S3). Each bone section was prepared from recipients that exhibited >98.0% hCD45+hCD33+ cells in BM when they were killed, indicating that nearly all the cells in the BM were human AML cells. These findings confirm that these molecules, specifically WT1 and HCK, are promising targets for immunotherapy and kinase inhibition against chemotherapy-resistant LSCs within the BM endosteal niche.
Fig. 3.
LSC target molecules are expressed on cell cycle–quiescent cells in BM endosteal region. (A and C) WT1 (red)–positive hCD45 (green)–positive (A) and HCK (red)–positive hCD45 (green)–positive (C) AML cells are adjacent to the endosteum (*). (B and D) The cells adjacent to the endosteum (*) are also Ki67 (red)–negative. Nuclei are labeled with 4′,6-diamidino-2-phenylindole (DAPI) (blue). The BM contained >98% hCD45+CD33+ cells in each recipient. Scale bar, 20 μm.
Expression of LSC-specific targets in primary human AML
Among the 16 candidate molecules adequately evaluable by flow cytometry, CD32, CD93, CD18, CD97, CD33, CD132, and CD25 were highly expressed in LSCs in at least two of eight patients chosen as pilot cases (AML: M1, n = 1; M2, n = 3; M4, n = 2; MDS/AML: n = 2; table S1). However, CD97, CD33, CD132, and CD24 were also expressed in normal BM HSCs. Therefore, we proceeded to evaluate CD32, CD25, CD18, and CD93 as candidate human LSC-specific targets.
We first examined the expression of these markers in normal HSCs by flow cytometry. The mean fluorescence intensity (MFI) ± SD of CD32, CD25, CD18, and CD93 on normal human BM HSCs was 103.6 ± 34.4, 162.8 ± 32.6, 127.8 ± 14.3, and 98.6 ± 17.2, respectively (n = 5). A MFI of 800 was chosen as the threshold value for determining positive expression of these molecules because the value of mean MFI + 10 × SD for the four molecules in normal BM HSCs was below 800.
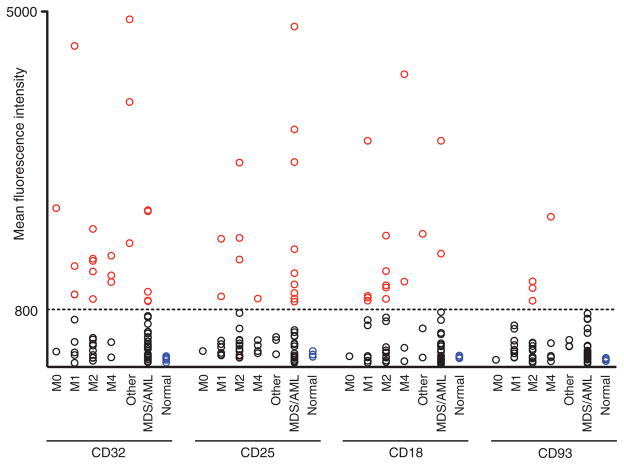
Then, we examined the expression of CD32, CD25, CD18, and CD93 on the surface of LSCs in 53 additional AML patient samples, for a total of 61 cases (AML: M0, n = 2; M1, n = 8; M2, n = 15; M4, n = 4; M5, n = 1; M7, n = 1; subtype undetermined, n = 1; MDS/AML: n = 29; table S1). In general, 52 of 61 (85.2%) of the cases were poor-risk (advanced age, induction failure, relapse, and/or unfavorable cytogenetics). Among the 61 patients, 39 (63.9%) showed high expression (defined by MFI > 800) of at least one of the four molecules in LSCs. The MFI for each molecule in LSCs is listed in table S1, and the frequency of marker positivity is summarized in Fig. 4 and Table 1. CD32 and CD25 were the most frequently expressed molecules in LSCs at 34.4% and 24.6%, respectively. In 32 of 61 (52.5%) patients, CD32, CD25, or both molecules were highly expressed in LSCs.
Fig. 4.
Target molecules are present on the cell surface of primary AML LSCs. MFI of expression for CD32, CD25, CD18, and CD93 in 61 primary AML patient BM LSCs with normal human BM HSCs as controls. Cutoff MFI = 800 (indicated as a horizontal line). Red, black, and blue circles represent marker-positive AML LSCs, marker-negative AML LSCs, and normal BM HSCs, respectively. AML: M0, n = 2; M1, n = 8; M2, n = 15; M4, n = 4; other AML (M5, M7, and undetermined), n = 1 each; MDS/AML: n = 29; normal human BM: n = 5.
Table 1.
Frequency of CD32-, CD25-, CD18-, and CD93-positive cells in 61 primary AML patient BM LSCs. Samples were collected from AML patients with the French-American-British (FAB) subtypes indicated.
| FAB | n | Any marker | CD32 or CD25 | CD32 | CD25 | CD18 | CD93 |
|---|---|---|---|---|---|---|---|
| M0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 |
| % Positive | 50.0 | 50.0 | 50.0 | 0.0 | 0.0 | 0.0 | |
| M1 | 8 | 7 | 6 | 4 | 2 | 4 | 0 |
| % Positive | 87.5 | 75.0 | 50.0 | 25.0 | 50.0 | 0.0 | |
| M2 | 15 | 10 | 6 | 5 | 3 | 5 | 3 |
| % Positive | 66.7 | 40.0 | 33.3 | 20.0 | 33.3 | 20.0 | |
| M4 | 4 | 4 | 4 | 3 | 1 | 2 | 1 |
| % Positive | 100.0 | 100.0 | 75.0 | 25.0 | 50.0 | 25.0 | |
| Other | 3 | 3 | 3 | 3 | 0 | 1 | 0 |
| % Positive | 100.0 | 100.0 | 100.0 | 0.0 | 33.3 | 0.0 | |
| MDS/AML | 29 | 14 | 12 | 5 | 9 | 3 | 0 |
| % Positive | 48.3 | 41.4 | 17.2 | 31.0 | 10.3 | 0.0 | |
| All cases | 61 | 39 | 32 | 21 | 15 | 15 | 4 |
| % Positive | 63.9 | 52.5 | 34.4 | 24.6 | 24.6 | 6.6 |
Because CD32 and CD25 were the most frequently expressed molecules, we examined their functional significance in primary AML. CD25-positive cells (CD25+CD34+CD38−) from AML patients were able to initiate AML when transplanted into mice [103 to 104 sorted cells transplanted, four of four recipients engrafted, PB hCD45+ = 18.1 ± 4.7% (SEM) at 13 to 14 weeks after transplantation]. Immunofluorescence labeling of CD25-positive AML-engrafted recipient BM demonstrated the preferential and extensive expression of hCD25 in the endosteal region of the BM (fig. S4). The CD25+ cells in the endosteal region were in the G0 phase of the cell cycle as shown by their negative Ki67 staining. Nearly all cells in the bone sections were human AML cells because the recipient BM contained 98.4% and 99.6% hCD45+CD33+ cells when the animals were killed.
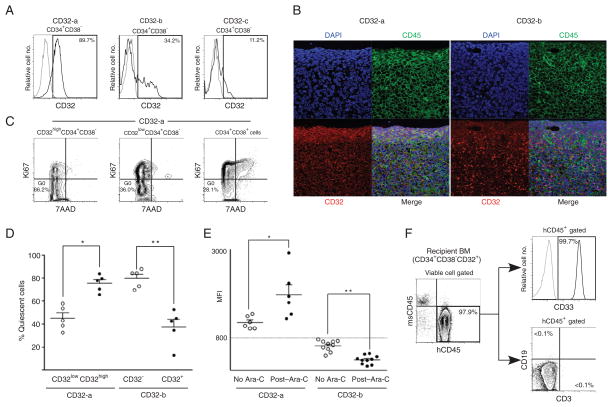
We saw three patterns of CD32 expression in the LSCs of the 61 patients with AML (Fig. 5A): CD32-positive cells with a single positive peak (category CD32-a); CD32-negative cells (MFI < 800) with a bimodal expression, showing a minor but separable CD32-positive population (category CD32-b); and CD32-negative cells with a single negative peak (category CD32-c). To determine the functional significance of these CD32 expression patterns, we performed xenotransplantation with CD32-a and CD32-b CD32+CD34+CD38− AML cells. The CD32+CD34+CD38− cells from the CD32-a AML patients initiated AML when transplanted to mice [7 × 102 to 1 × 104 sorted cells transplanted, 13 of 13 recipients engrafted, BM hCD45+ = 90.0 ± 4.2% (SEM) at 9 to 15 weeks after transplantation]. We could not test whether CD32− CD34+CD38− cells initiated AML because we could not isolate a sufficient number of viable cells; nearly all CD34+CD38− cells are CD32+ in CD32-a AML patients. In contrast, in CD32-b AML, CD32-negative CD34+CD38− but not CD32-positive CD34+CD38minus; cells initiated AML [1 × 104 sorted cells transplanted, n = 3 each, BM hCD45+ = 78.5 ± 9.7% (SEM) and 0.0 ± 0.0% (SEM) for recipients of CD32-negative and CD32-positive cells, respectively, at 15 to 21 weeks after transplantation]. Immunofluorescence labeling of recipient femurs engrafted with CD32-a AML demonstrated that hCD32 and hCD45 were coexpressed in AML cells lining the endosteum (Fig. 5B, left), whereas CD32-b–engrafted recipient femur shows scattered CD32 expression in the central region of the BM (Fig. 5B, right). At the time of killing, 98.2% and 98.5% of the BM in the femurs shown in Fig. 5 were positive for hCD45 and hCD33, indicating that nearly all the cells in the sections were human AML cells. In addition, the CD32highCD34+CD38minus; population of cells was significantly enriched for quiescent cells in cells from CD32-a AML patients [representative flow cytometry plots are shown in Fig. 5C; %G0 = 75.5 ± 3.3 (SEM) and 45.2 ± 4.6 (SEM), respectively, n = 5 each, P = 0.0007 by two-tailed t test, Fig. 5D]. In contrast, in CD32-b AML, the CD32-negative CD34+CD38minus; population was significantly enriched for quiescent cells [%G0 = 79.8 ± 3.6 (SEM) and 37.6 ± 6.8 (SEM), respectively, _n_ = 5 each, _P_ = 0.0006 by two-tailed _t_ test; Fig. 5D]. These findings demonstrate that CD32+CD34+CD38minus; cells are the cell cycle–quiescent, AML-initiating cells residing in the BM endosteal region in CD32-a CD32-positive AML.
Fig. 5.
CD32 expression is associated with cell cycle quiescence and chemotherapy resistance of LSCs. (A) In CD32-positive AML, a single peak of CD32+ expression is present in LSCs (CD32-a, left panel). CD32-negative AML LSCs show two patterns of CD32 expression (CD32-b and CD32-c, middle and right panels). (B) In CD32-a AML-engrafted recipient (left), CD32 (red) is co-expressed with hCD45 (green) on AML cells abutting the BM endosteum (*). In CD32-b AML-engrafted recipient (right), there is scattered CD32 expression in the central region of the BM but no CD32 expression in the endosteal region. Nuclei are labeled with DAPI (blue). Scale bar, 20 μm. (C) Representative flow cytometry plots demonstrating enrichment of quiescent cells in CD32highCD34+CD38− population in CD32-positive AML. (D) In CD32-a LSCs, CD32-high LSCs are cell cycle–quiescent, whereas CD32-negative LSCs are cell cycle–quiescent in CD32-b LSCs (n = 5 for each group). *P = 0.0034, **P = 0.0006 by two-tailed t test. (E) CD32 expression in LSCs after in vivo Ara-C treatment. In vivo Ara-C treatment increases CD32 expression in CD32-positive (CD32-a) BM LSCs, indicating that CD32-low LSCs are preferentially eliminated by Ara-C (n = 6 each for control and Ara-C–treated group). *P = 0.0121. In contrast, CD32 expression decreases in CD32-negative (CD32-b) BM LSCs with Ara-C treatment (n = 10 each). **P < 0.0001 by two-tailed t test. (F) CD32+CD34+CD38− cells retain in vivo AML-initiating capacity after in vivo Ara-C treatment. Flow cytometry plots showing engraftment of hCD45+ cells in the BM of a representative recipient of purified CD34+CD38− CD32+ cells from an Ara-C–treated recipient. All hCD45+ human hematopoietic cells are also hCD33+ and there are no CD3+ T or CD19+ B cells present.
Because LSC-targeting therapeutic agents will likely be used in conjunction with conventional chemotherapy, an LSC marker molecule must show stable expression even after chemotherapy. To test this, we examined the expression of CD32 and CD25 in BM of AML-engrafted recipients after in vivo treatment with cytarabine (Ara-C). In CD32-b AML patients in whom CD32-negative CD34+CD38minus; cells are the LSCs, nearly all CD32-positive CD34+CD38minus; cells were eliminated by Ara-C, resulting in decreased overall CD32 expression [CD32+CD34+CD38minus; = 19.2 ± 1.6% (SEM) and 0.5 ± 0.1 (SEM), respectively, for control and Ara-C–treated recipients, _n_ = 5 for each group, _P_ < 0.0001 by two-tailed _t_ test; MFI = 600.1 ± 49.0 to 259.9 ± 34.3 (SD), _n_ = 10 each, _P_ < 0.0001 by two-tailed _t_ test; Fig. 5E]. In contrast, CD32 expression was augmented in CD32-a LSCs after in vivo treatment with Ara-C with the elimination of cell cycle–active, CD32-negative CD34+CD38minus; AML cells [MFI = 1180.3 ± 63.3 to 1871.5 ± 244.4 (SD), _n_ = 6 each, _P_ = 0.0121 by two-tailed _t_ test; Fig. 5E]. Serial transplantation of purified CD34+CD38minus; CD32+ cells from Ara-C–treated, CD32-positive AML recipient mice resulted in in vivo AML initiation, as evidenced by the engraftment of hCD45+ cells, all of which were CD33+ AML blasts, with no development of normal human immunohematopoietic subsets (11 of 11 secondary recipients from Ara-C–treated recipients of two independent CD32-positive AML cases engrafted; representative flow cytometry plots shown in Fig. 5F). This finding functionally demonstrates that CD32-positive CD34+CD38minus; AML cells retain LSC function with no contaminating normal HSCs after in vivo Ara-C treatment. The stable expression of CD32 after treatment with a chemotherapeutic agent in LSCs would allow the targeting of CD32-positive LSCs concurrently or after chemotherapy. Similarly, in the recipients engrafted with CD25-positive AML, CD25 expression in BM was maintained after in vivo treatment with Ara-C (MFI = 1664.8 ± 250.8 to 2137.6 ± 336.5, n = 5 each, P = not significant by two-tailed t test).
Effect of CD32 and CD25 inhibition on normal hematopoiesis
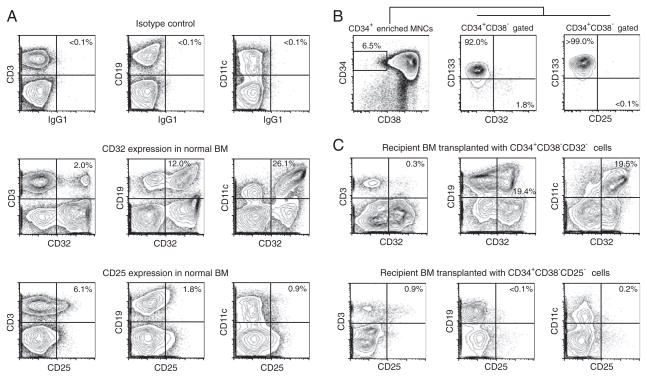
For CD32 and CD25 to serve as therapeutic targets against LSCs, the elimination of CD32- or CD25-positive cells from normal human BM must not disrupt normal hematopoiesis. In normal human BM, CD32 is expressed in B cells, T cells, and monocytes, and CD25 is expressed in a portion of T cells (Fig. 6A). However, neither antigen was expressed in CD34+CD38minus; CD133+ HSCs (Fig. 6B).
Fig. 6.
Depletion of LSC marker-positive cells does not affect normal human multilineage hematopoietic reconstitution in vivo. (A) CD32 and CD25 expression in normal human BM T, B, and myeloid cells with their corresponding isotypes. (B) In normal human CB, CD32+ and CD25+CD34+CD38− cells do not express CD133, a positive marker for normal HSCs. (C) Recipients of human normal CB depleted of CD32-positive cells are repopulated with both CD32-positive and CD32-negative human T, B, and myeloid cells. Similarly, human CB depleted of CD25-positive cells repopulated both CD25-positive and CD25-negative human T, B, and myeloid cells.
We directly examined the capacity of CD32- and CD25-negative healthy human HSCs to repopulate BM by xenotransplantation. The transplantation of 4 × 103 to 2 × 104 CD32-negative human CB CD34+CD38− cells resulted in long-term engraftment of human CD45+ cells with multilineage differentiation demonstrated by the presence of both CD32-positive and CD32-negative human T, B, and myeloid cells in the BM [hCD45 = 31.7 ± 4.4% (SEM) at 17 to 22 weeks after transplantation, n = 7]. Similarly, recipients of 104 CD25minus; normal HSCs resulted in multilineage repopulation with human T, B, and myeloid cells, including CD25+ T cells in the BM [hCD45 = 10.2 ± 3.5% (SEM) at 18 weeks, n = 5]. Representative sets of flow cytometry plots are shown in Fig. 6C. Moreover, neither CD32 nor CD25 mRNA was expressed at significant levels in human nonhematopoietic organs (fig. S5). These findings suggest that normal HSCs would be functionally preserved and that the potential for nonhematopoietic adverse effects is relatively low with anti-LSC therapeutic strategies targeting CD32 and/or CD25.
DISCUSSION
Although most of younger and 40 to 60% of elderly patients with AML achieve complete remission at the time of initial diagnosis, overall survival for AML patients remains low, at ~20% at 5 years (1, 3, 4, 11–14). This discrepancy between successful remission induction and poor long-term outcome is due to the difficulty of preventing and overcoming disease relapse (2–4). Rare cell cycle–quiescent, chemotherapy-resistant LSCs that survive chemotherapy are likely responsible for relapse (7).
To begin to develop anti-LSC therapies, we identified human primary AML LSC-specific molecules by comparing global mRNA expression patterns in LSC and HSC populations functionally defined by in vivo serial engraftment and long-term multilineage repopulation. The analysis of global gene expression profiles in bulk AML blasts by Valk et al. and others have linked expression profiles with prognosis of AML (15–18). Similarly, gene expression signatures associated with prognosis have been identified in breast cancer (19). In a comparison of human AML LSCs and HSCs, 3005 differentially expressed genes were analyzed by correspondence with pathway information from publicly accessible databases, generating candidate pathways dysregulated in LSCs (20). Our approach is distinct from these previous studies in four aspects. First, the gene expression comparisons were restricted to specifically identify genes that are overrepresented in LSCs and underrepresented in HSCs. Previous studies [except that by Majeti _et al_. (20)] compared larger populations containing bulk AML blasts and more differentiated normal hematopoietic progenitors. Second, we validated the mRNA expression of candidate LSC targets by qRT-PCR and the presence of protein by flow cytometry and/or immunofluorescence imaging. This step was crucial, as expression (or lack thereof) of RNA did not necessarily correspond with protein abundance. Third, we demonstrated the functional significance of candidate LSC target molecules by linking their expression with cell cycle quiescence, in situ localization, and in vivo AML initiation capacity. Last, we confirmed that the candidate LSC target molecules are not required for normal HSC function by showing that normal HSCs that lack these molecules can repopulate the hematopoietic system in vivo.
Although both AML LSCs and normal HSCs show the Linminus; CD34+CD38− phenotype, we identified 259 genes overrepresented in LSCs by analyzing global gene expression patterns using two independent array platforms. Of these, 25 candidate molecules with favorable characteristics for potential drug development were chosen for detailed analyses. Although molecules such as CD33, CD97, and CD132 are present at higher concentrations in LSCs than HSCs, we did not include them in our candidate list because a therapeutic antibody administered in vivo might bind normal HSCs, even if they have relatively low amounts of antigen, leading to clearance of normal HSCs as well as LSCs. The functional properties of CD34+CD38− primary human LSCs are maintained in the xenogeneic BM microenvironment of the NOD-SCID-IL2rgnull mouse recipients, allowing the analysis of candidate LSC-specific molecules in situ, a procedure difficult to perform directly in AML patients (7). By dual labeling of the candidate marker and Ki67 (which marks actively cycling cells), we determined the cell cycle status of marker-positive AML cells in the BM. In particular, WT1 and HCK were highly expressed in quiescent AML cells at the BM endosteum.
Overexpression of WT1 in acute leukemia has been well described, with expression in 73 to 100% and 32% of myeloblasts in newly diagnosed and relapsed adult AML patients, respectively (21). The expression of WT1 has been linked to poor prognosis in newly diagnosed patients and relapse in patients that achieve complete remission, and it has been suggested as a target molecule for immunotherapy of AML (22–27). Here, we found that WT1 gene is expressed in cell cycle–quiescent primary AML LSCs at the endosteal region, suggesting that the association of high WT1 expression with poor prognosis and relapse may be related to its expression in LSCs.
A member of the Src family of tyrosine kinases (SFKs), HCK was also identified as a potential target molecule in cell cycle–quiescent, chemotherapy-resistant LSCs in the BM endosteal region. HCK is expressed in myeloid cells, particularly in granulocytes and monocytes (28, 29). HCK and other SFKs interfere with the maturation of Flt3 in a kinase-dependent manner (30). The overexpression of HCK in LSCs may therefore be linked to abnormal Flt3 signaling. Expression of WT1 and HCK in quiescent AML cells within the BM endosteal niche suggests that immunotherapeutic approaches targeting these molecules may be effective in eliminating LSCs.
Among the surface markers identified as differentially expressed in LSCs, 14 molecules, including CD33, CD97, and CD132, showed expression in normal HSCs by flow cytometry, albeit at lower values than in LSCs. Although these molecules may be helpful in segregating HSCs from LSCs (for instance, in the setting of graft purging in autologous HSC transplantation), they are probably not ideal as targets for LSC-specific therapy.
The remaining four surface markers, confirmed to be absent from normal human BM HSCs, were screened in 61 primary AML patient samples by flow cytometry. Most of these were from poor-risk patients with a high likelihood for relapse after standard treatment protocols. In these patients, LSC-specific therapy may be particularly effective in improving long-term outcomes. Of these, CD32 and CD25 were most frequently expressed on AML LSCs (52.5% expressing CD32, CD25, or both). Of note, CD25 expression on bulk AML blasts in patients has recently been reported to be associated with poor prognosis and MRD, but its expression on primary AML LSCs is yet unexamined (31). Both molecules were functionally confirmed as markers for human AML LSCs by their expression on quiescent LSCs within the BM endosteal niche and by the capacity of CD32+CD34+CD38− and CD25+CD34+CD38− AML cells to initiate AML in vivo in mice.
CD32 and CD25 are receptors known to be expressed in subsets of mature immune cells in mammals. As expected, CD32- and CD25-null mice showed deficiencies in myeloid and lymphoid development (32–35). In humans, allelic variants of FCGR2A have been associated with susceptibility to systemic lupus erythematosus and to bacterial infections in certain populations (36–38). These findings are attributed to differential function of phagocytes bearing the particular polymorphisms. However, the association between aberrant hematopoiesis and CD32 expression in human has not been reported to date.
A truncated mutation of human CD25 results in a primary immunodeficiency syndrome (39). In addition, the IL2RA locus and its polymorphisms have been associated with autoimmune disorders, including type 1 diabetes mellitus, autoimmune vasculitides, multiple sclerosis, and Graves’ disease (40–44). Although these reports raise concerns for the development of autoimmune diseases when CD25 is inhibited, clinical experiences with monoclonal antibody to Tac directed toward CD25 for the prevention of organ allograft rejection is reassuring (45, 46). In this patient population, there have been no hematopoietic or nonhematopoietic adverse effects other than those related to the expected immunosuppression.
We have demonstrated that neither CD32 nor CD25 is expressed on normal human BM and CB HSCs. Nevertheless, the lack of detectable expression is insufficient to ensure that normal hematopoiesis is completely unaffected by therapeutic strategies targeting these molecules. The transplantation of CD32- or CD25-negative normal human HSCs resulted in long-term multilineage hematopoietic reconstitution, including the development of CD32+ or CD25+ myeloid and lymphoid progeny, giving some reassurance that anti-LSC therapies targeting CD32 and CD25 will not significantly affect normal hematopoiesis.
In remission induction, the goal is a rapid clearance of total body AML burden, mainly highly proliferative mature AML blasts. Because LSCs are a rare population of quiescent cells, specific anti-LSC therapy may be best used in postremission therapy to eliminate the MRD that eventually leads to disease relapse. This suggestion is consistent with a study showing that the timing of in vivo administration of a neutralizing antibody to CD123 influences its efficacy (47). Therefore, anti-LSC therapy may prove most effective during consolidation therapy or in pre-transplantation conditioning. In any case, anti-LSC therapy would most likely be instituted in patients subsequently and/or concurrently with chemotherapy. Our data show that both CD32 and CD25 expression on LSCs is maintained after chemotherapy in vivo.
In summary, we identified human AML LSC-specific molecules by comparing human LSC and HSC transcriptomes followed by the demonstration of the functional significance of the differentially expressed genes with a multifaceted analysis including xenogeneic transplantation—the current gold standard for the demonstration of LSC function. Two of the identified molecules, CD32 and CD25, are particularly promising as targets of anti-LSC therapy because (i) they are expressed in a large fraction of primary human AML LSCs, (ii) they are stably located on the LSC cell surface after chemotherapy, (iii) they are present on the cell cycle–quiescent, AML-initiating cells residing within the endosteal niche that may be responsible for AML relapse, and (iv) elimination of normal human HSCs expressing these molecules does not compromise normal hematopoietic development. Therapeutic strategies targeting these molecules may be effective because they were frequently expressed in an AML patient population in which 85% were poor-risk and in need of effective treatments to improve outcomes. The identification of other target molecules with similar approaches will facilitate the development of drugs and therapeutic strategies in other malignancies in which patient outcomes are now poor.
MATERIALS AND METHODS
Human samples
All experiments were performed with authorization from the Institutional Review Board for Human Research at RIKEN Research Center for Allergy and Immunology. Samples were collected with written informed consent from AML patients with French-American-British (FAB) classification subtypes M0 (n = 2), M1 (n = 8), M2 (n = 16), M4 (n = 4), M5 (n = 1), M7 (n = 1), and undetermined (n = 1) and from patients with MDS/AML (n = 29). CB samples were collected by the Tokyo Cord Blood Bank with written informed consent. BM mononuclear cells (MNCs) from healthy donors were obtained from Cambrex. AML BM MNCs and CB MNCs were isolated with density gradient centrifugation.
Animals
NOD.Cg-PrkdcscidIl2rgtmlWjl/Sz (NOD-SCID-IL2rgnull) mice were developed at The Jackson Laboratory by backcrossing a complete null mutation at the Il2rg locus onto the NOD.Cg-Prkdcscid (NOD-SCID) strain (48). Mice were bred and maintained under defined flora at the animal facility of RIKEN and at The Jackson Laboratory according to guidelines established by the Institutional Animal Committees at each institution.
Xenogeneic transplantation
Newborn NOD-SCID-IL2rgnull recipients received 150 cGy of total body irradiation followed by intravenous injection of sorted cells. To generate AML-engrafted recipients, we injected 103 to 105 sorted 7-aminoactinomycin D (7AAD)− lineage (hCD3/hCD4/hCD8)− hCD34+ hCD38− AML patient BM cells into each recipient as described (7). To generate normal human HSC-engrafted recipients, we injected 104 sorted 7AAD− lineage (hCD3/hCD4/hCD8)− hCD34+hCD38− CB cells per recipient as described (49). PB human cell engraftment was assessed by retro-orbital phlebotomy. When PB hCD45 chimerism reached 50 to 90% (15 to 20 weeks after transplantation), the recipients were killed for analysis.
Flow cytometry and fluorescence-activated cell sorting
To determine human cell engraftment and marker expression, we labeled cells with fluorochrome-conjugated rat monoclonal antibody to mCD45 and mouse monoclonal antibodies to hCD45, hCD33, hCD34, hCD38, hCD32, hCD25, hCD18, hCD93, hCD33, hCD132, hCD97, hCD86, hCD150 (BD Biosciences), hCD66c (R&D Systems), hCD180 (eBioscience), hCD24 (BioLegend), hCD127 (Beckman Coulter), hCD133, and hCD1c (Miltenyi Biotec). For cell cycle analysis, cells were labeled with mouse monoclonal antibodies to hKi67 (BD Biosciences), hCD45, hCD34, hCD38, and hCD32 (where indicated). Analyses were performed with FACSAria and FACSCanto II (BD Biosciences). To obtain cells for microarray analyses and xenogeneic transplantation, we labeled human MNCs with fluorochrome-conjugated mouse monoclonal antibodies to hCD3, hCD4, hCD8, hCD34, hCD38, hCD32, or hCD25 (where indicated), and recipient BM MNCs were labeled with mouse monoclonal antibodies to hCD45, hCD34, hCD38, hCD32, and hCD25 (where indicated) followed by cell sorting with FACSAria. The purity of sorted cells was >98%.
Cytospin preparations
Thin-layer preparations were made with Shandon Cytospin 4 cytocentrifuge (Thermo Electric). May-Grunwald-Giemsa staining was performed with standard procedures. Light microscopy was performed with Zeiss Axiovert 200 (Carl Zeiss).
Microarray analyses
Twenty-one LSC samples (see table S1) and 6 HSC samples (BM, n = 2; CB, n = 4) were evaluated with Human Genome U133 Plus 2.0 GeneChips (Affymetrix). Hierarchical clustering by 25-gene LSC signature was performed on 20 additional LSC samples (see table S1) and 6 additional BM HSC samples. The U133 Plus 2.0 GeneChips expression data for these additional samples were newly obtained and were not included when generating the 25-gene LSC signature. Six LSC samples (see table S1) and five HSC samples (BM, n = 4; CB, n = 1) were evaluated with Human Gene 1.0ST GeneChips (Affymetrix). From total RNA extracted with TRIzol (Invitrogen) from >104 sorted cells, biotinylated complementary RNA was synthesized with Two-Cycle Target Labeling kit (Affymetrix) for Human Genome U133 Plus 2.0 GeneChips and with MessageAmp Premier RNA Amplification kit (Applied Biosystems) and WT cDNA Synthesis and Terminal Labeling kits (Affymetrix) for Human Gene 1.0ST GeneChips. Microarray data were analyzed with the Bioconductor package (10). The signal intensities of the probe sets were normalized with the GC-RMA program (10). RankProd program was used to select differentially expressed genes with a cutoff P value of 0.01 and an estimated false-positive rate of 0.05 (8). Gene annotation was obtained from Ingenuity Pathway Analysis and Gene Ontology Annotation databases (50, 51).
Quantitative RT-PCR analyses
Quantitative RT-PCR was performed on complementary DNA amplified from total RNA with the WT-Ovation RNA Amplification System (Nugen) with Platinum Quantitative PCR SuperMix (Invitrogen) on LightCycler 480 (Roche). Sequences of dual-labeled fluorogenic probes and gene-specific primers (Sigma-Aldrich) are listed in table S3. Abundance of each transcript was calculated by the standard curve method (52). The difference in expression levels was considered significant if P < 0.05 using either Kruskal-Wallis, Wilcoxon-Mann-Whitney, or Student’s t test in KaleidaGraph (Synergy Software). For the analysis of nonhematopoietic tissues, total RNA samples from normal human tissues (Clontech and Ambion), small airway epithelial cells (Lonza), and alveolar epithelial cells (ScienCell) were amplified with WT-Ovation RNA Amplification kit for qRT-PCR analysis.
Immunofluorescence labeling and imaging
Paraformaldehyde-fixed, decalcified, paraffin-embedded femoral sections were labeled with mouse monoclonal antibodies to hCD45 (DAKO), hCD32 (Abcam), and hCD25 (Imgenex); rabbit monoclonal antibody for hWT1 (Abcam); rabbit polyclonal antibodies to hKi67 (Spring Bioscience), hPDE9A (Atlas Antibodies), hHCK (Novus Biologicals), hAK5 (Abgent), hDOK2 (Spring Bioscience), hLRG1 (Atlas Antibodies), and hBIK (Abcam); and goat polyclonal antibodies to hSUCNR1 (Santa Cruz Biotechnology) and hCTSC (Santa Cruz Biotechnology). Laser-scanning confocal imaging was obtained with Zeiss LSM 710 (Carl Zeiss).
In vivo chemotherapy treatment
AML-engrafted recipients at 16 to 20 weeks after transplantation and at 40 to 50% PB hCD45 engraftment received Ara-C (Biogenesis) (1 g/kg, intraperitoneally, daily for 2 days) and were killed 16 hours after the second injection for analysis. For serial transplantation after Ara-C treatment, CD34+CD38− CD32+ cells were purified from the BM or spleen of AML-engrafted recipients treated with Ara-C and transplanted into 11 recipients at 9 × 103 to 1.2 × 105 cells per recipient.
Statistical analysis
Statistical methods used in the microarray analyses are described in the microarray analysis section of the Materials and Methods. MFI data are presented as means ± SD. Other data are presented as means ± SEM. The differences in %G0 cells, CD32 MFI, and %CD32 cells were examined with two-tailed t test (GraphPad Prism, GraphPad). A P value of <0.05 was deemed significant.
Supplementary Material
Supplemental Data
Table S1. Summary of analyses performed on AML patient samples.
Table S2. List of LSC signature genes.
Table S3. List of primers and probes for qRT-PCR.
Fig. S1. LSC-specific expression of 58 genes was validated by qRT-PCR.
Fig. S2. WT1 and HCK are frequently overexpressed in LSCs.
Fig. S3. Candidate LSC target molecules show expression in human AML cells lining the BM endosteum.
Fig. S4. CD25 is expressed in cell cycle–quiescent AML cells in the BM endosteal region.
Fig. S5. Expression of CD32 and CD25 in nonhematopoietic tissues.
Acknowledgments
We thank A. Kobayashi, T. Hasegawa, S. Hayashi, S. Matsuyama, M. Mori, S. Takahashi, M. Ito, and M. Yamagishi for technical assistance.
Funding: This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, National Institute of Biomedical Innovation, Uehara Foundation, and Takeda Science Foundation (F.I.) and NIH and Juvenile Diabetes Research Foundation (L.D.S.).
Footnotes
Author contributions: Y.S., O.O., L.D.S., and F.I. designed the study, analyzed the data, and wrote the manuscript. H.K. and A.H. performed the experiments, analyzed the data, and wrote the manuscript. N.U., A.W., and S. Taniguchi provided clinical samples, information, and discussion. Y.S., S. Tanaka, M.T.-M., and F.I. performed the experiments. Y.N., S. Takagi, and H.O. analyzed the data.
Competing interests: The authors have no competing interests to declare.
Accession numbers: The microarray data can be found in CIBEX (http://cibex.nig.ac.jp/) with accession ID CBX21 and CBX111.
REFERENCES AND NOTES
- 1.Horner MJ, Ries LAG, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK. SEER Cancer Statistics Review 1975–2006. National Cancer Institute; Bethesda: 2009. [accessed 2 July 2009]. http://seer.cancer.gov/csr/1975_2006/, based on November 2008 SEER data submission. [Google Scholar]
- 2.Breems DA, Van Putten WL, Huijgens PC, Ossenkoppele GJ, Verhoef GE, Verdonck LF, Vellenga E, De Greef GE, Jacky E, Van der Lelie J, Boogaerts MA, Löwenberg B. Prognostic index for adult patients with acute myeloid leukemia in first relapse. J Clin Oncol. 2005;23:1969–1978. doi: 10.1200/JCO.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, Harrison CJ, Wheatley K, Burnett AK, Goldstone AH Medical Research Council Adult Leukemia Working Party. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): Analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 5.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 6.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, Nakamura R, Tanaka T, Tomiyama H, Saito N, Fukata M, Miyamoto T, Lyons B, Ohshima K, Uchida N, Taniguchi S, Ohara O, Akashi K, Harada M, Shultz LD. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 8.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 9.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 10.Bioconductor. http://www.bioconductor.org/
- 11.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD Cancer and Leukemia Group B (CALGB 8461) Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 12.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 13.Gardin C, Turlure P, Fagot T, Thomas X, Terre C, Contentin N, Raffoux E, de Botton S, Pautas C, Reman O, Bourhis JH, Fenaux P, Castaigne S, Michallet M, Preudhomme C, de Revel T, Bordessoule D, Dombret H. Postremission treatment of elderly patients with acute myeloid leukemia in first complete remission after intensive induction chemotherapy: Results of the multicenter randomized Acute Leukemia French Association (ALFA) 9803 trial. Blood. 2007;109:5129–5135. doi: 10.1182/blood-2007-02-069666. [DOI] [PubMed] [Google Scholar]
- 14.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, Forman SJ, Appelbaum FR. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 15.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B, Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 16.Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, Heinecke A, Radmacher M, Marcucci G, Whitman SP, Maharry K, Paschka P, Larson RA, Berdel WE, Büchner T, Wörmann B, Mansmann U, Hiddemann W, Bohlander SK, Buske C Cancer and Leukemia Group B; German AML Cooperative Group. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 18.Langer C, Radmacher MD, Ruppert AS, Whitman SP, Paschka P, Mrózek K, Baldus CD, Vukosavljevic T, Liu CG, Ross ME, Powell BL, de la Chapelle A, Kolitz JE, Larson RA, Marcucci G, Bloomfield CD Cancer and Leukemia Group B (CALGB) High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: A Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 20.Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, Chiang JH, Hood L, Clarke MF, Weissman IL. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci USA. 2009;106:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld C, Cheever MA, Gaiger A. WT1 in acute leukemia, chronic myelogenous leukemia and myelodysplastic syndrome: Therapeutic potential of WT1 targeted therapies. Leukemia. 2003;17:1301–1312. doi: 10.1038/sj.leu.2402988. [DOI] [PubMed] [Google Scholar]
- 22.Ommen HB, Nyvold CG, Braendstrup K, Andersen BL, Ommen IB, Hasle H, Hokland P, Ostergaard M. Relapse prediction in acute myeloid leukaemia patients in complete remission using WT1 as a molecular marker: Development of a mathematical model to predict time from molecular to clinical relapse and define optimal sampling intervals. Br J Haematol. 2008;141:782–791. doi: 10.1111/j.1365-2141.2008.07132.x. [DOI] [PubMed] [Google Scholar]
- 23.Inoue K, Sugiyama H, Ogawa H, Nakagawa M, Yamagami T, Miwa H, Kita K, Hiraoka A, Masaoka T, Nasu K, Kyo T, Dohy H, Nakauchi H, Ishidate T, Akiyama T, Kishimoto T. WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood. 1994;84:3071–3079. [PubMed] [Google Scholar]
- 24.Bergmann L, Miething C, Maurer U, Brieger J, Karakas T, Weidmann E, Hoelzer D. High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997;90:1217–1225. [PubMed] [Google Scholar]
- 25.Keilholz U, Letsch A, Busse A, Asemissen AM, Bauer S, Blau IW, Hofmann WK, Uharek L, Thiel E, Scheibenbogen C. A clinical and immunologic phase 2 trial of Wilms tumor gene product 1 (WT1) peptide vaccination in patients with AML and MDS. Blood. 2009;113:6541–6548. doi: 10.1182/blood-2009-02-202598. [DOI] [PubMed] [Google Scholar]
- 26.Kitawaki T, Kadowaki N, Kondo T, Ishikawa T, Ichinohe T, Teramukai S, Fukushima M, Kasai Y, Maekawa T, Uchiyama T. Potential of dendritic-cell immunotherapy for relapse after allogeneic hematopoietic stem cell transplantation, shown by WT1 peptide- and keyhole-limpet-hemocyanin-pulsed, donor-derived dendritic-cell vaccine for acute myeloid leukemia. Am J Hematol. 2008;83:315–317. doi: 10.1002/ajh.21127. [DOI] [PubMed] [Google Scholar]
- 27.Rezvani K, Yong AS, Mielke S, Savani BN, Musse L, Superata J, Jafarpour B, Boss C, Barrett AJ. Leukemia-associated antigen-specific T-cell responses following combined PR1 and WT1 peptide vaccination in patients with myeloid malignancies. Blood. 2008;111:236–242. doi: 10.1182/blood-2007-08-108241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quintrell N, Lebo R, Varmus H, Bishop JM, Pettenati MJ, Le Beau MM, Diaz MO, Rowley JD. Identification of a human gene (HCK) that encodes a protein-tyrosine kinase and is expressed in hemopoietic cells. Mol Cell Biol. 1987;7:2267–2275. doi: 10.1128/mcb.7.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziegler SF, Marth JD, Lewis DB, Perlmutter RM. Novel protein-tyrosine kinase gene (hck) preferentially expressed in cells of hematopoietic origin. Mol Cell Biol. 1987;7:2276–2285. doi: 10.1128/mcb.7.6.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitina O, Warmuth M, Krause G, Hallek M, Obermeier A. Src family tyrosine kinases phosphorylate Flt3 on juxtamembrane tyrosines and interfere with receptor maturation in a kinase-dependent manner. Ann Hematol. 2007;86:777–785. doi: 10.1007/s00277-007-0344-0. [DOI] [PubMed] [Google Scholar]
- 31.Terwijn M, Feller N, van Rhenen A, Kelder A, Westra G, Zweegman S, Ossenkoppele G, Schuurhuis GJ. Interleukin-2 receptor alpha-chain (CD25) expression on leukaemic blasts is predictive for outcome and level of residual disease in AML. Eur J Cancer. 2009;45:1692–1699. doi: 10.1016/j.ejca.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 32.Takai T, Ono M, Hikida M, Ohmori H, Ravetch JV. Augmented humoral and anaphylactic responses in FcγRII-deficient mice. Nature. 1996;379:346–349. doi: 10.1038/379346a0. [DOI] [PubMed] [Google Scholar]
- 33.Bolland S, Ravetch JV. Spontaneous autoimmune disease in FcγRIIB-deficient mice results from strain-specific epistasis. Immunity. 2000;13:277–285. doi: 10.1016/s1074-7613(00)00027-3. [DOI] [PubMed] [Google Scholar]
- 34.Sharma R, Bagavant H, Jarjour WN, Sung SS, Ju ST. The role of Fas in the immune system biology of IL-2Rα knockout mice: Interplay among regulatory T cells, inflammation, hemopoiesis, and apoptosis. J Immunol. 2005;175:1965–1973. doi: 10.4049/jimmunol.175.3.1965. [DOI] [PubMed] [Google Scholar]
- 35.Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW. Interleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 36.Salmon JE, Millard S, Schachter LA, Arnett FC, Ginzler EM, Gourley MF, Ramsey-Goldman R, Peterson MG, Kimberly RP. FcγRIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, Bacino D, Dietz M, Hogue R, Koelsch G, Nightingale L, Shaver T, Abdou NI, Albert DA, Carson C, Petri M, Treadwell EL, James JA, Harley JB. Genome scan of human systemic lupus erythematosus: Evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci USA. 1998;95:14869–14874. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rose V, Arduino C, Cappello N, Piana R, Salmin P, Bardessono M, Goia M, Padoan R, Bignamini E, Costantini D, Pizzamiglio G, Bennato V, Colombo C, Giunta A, Piazza A. Fcγ receptor IIA genotype and susceptibility to P. aeruginosa infection in patients with cystic fibrosis. Eur J Hum Genet. 2005;13:96–101. doi: 10.1038/sj.ejhg.5201285. [DOI] [PubMed] [Google Scholar]
- 39.Sharfe N, Dadi HK, Shahar M, Roifman CM. Human immune disorder arising from mutation of the α chain of the interleukin-2 receptor. Proc Natl Acad Sci USA. 1997;94:3168–3171. doi: 10.1073/pnas.94.7.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brand OJ, Lowe CE, Heward JM, Franklyn JA, Cooper JD, Todd JA, Gough SC. Association of the interleukin-2 receptor alpha (IL-2Rα)/CD25 gene region with Graves’ disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf) 2007;66:508–512. doi: 10.1111/j.1365-2265.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- 41.Carr EJ, Clatworthy MR, Lowe CE, Todd JA, Wong A, Vyse TJ, Kamesh L, Watts RA, Lyons PA, Smith KG. Contrasting genetic association of IL2RA with SLE and ANCA-associated vasculitis. BMC Med Genet. 2009;10:22. doi: 10.1186/1471-2350-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 43.Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, Bourget K, Plagnol V, Field S, Atkinson M, Clayton DG, Wicker LS, Todd JA. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 44.Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, Jones R, Ring SM, McArdle W, Pembrey ME, Strachan DP, Dunger DB, Twells RC, Clayton DG, Todd JA. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincenti F, Kirkman R, Light S, Bumgardner G, Pescovitz M, Halloran P, Neylan J, Wilkinson A, Ekberg H, Gaston R, Backman L, Burdick J. Interleukin-2-receptor blockade with daclizumab to prevent acute rejection in renal transplantation. Daclizumab Triple Therapy Study Group. N Engl J Med. 1998;338:161–165. doi: 10.1056/NEJM199801153380304. [DOI] [PubMed] [Google Scholar]
- 46.Nashan B, Light S, Hardie IR, Lin A, Johnson JR. Reduction of acute renal allograft rejection by daclizumab. Daclizumab Double Therapy Study Group. Transplantation. 1999;67:110–115. doi: 10.1097/00007890-199901150-00019. [DOI] [PubMed] [Google Scholar]
- 47.Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, Guthridge MA, Thomas D, Barry EF, Boyd A, Gearing DP, Vairo G, Lopez AF, Dick JE, Lock RB. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor α chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5:31–42. doi: 10.1016/j.stem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 48.Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, Greiner DL, Handgretinger R. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 49.Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, Watanabe T, Akashi K, Shultz LD, Harada M. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chainnull mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingenuity Systems. http://www.ingenuity.com.
- 51.Gene Ontology Annotation. http://www.ebi.ac.up/GOA/
- 52.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
Table S1. Summary of analyses performed on AML patient samples.
Table S2. List of LSC signature genes.
Table S3. List of primers and probes for qRT-PCR.
Fig. S1. LSC-specific expression of 58 genes was validated by qRT-PCR.
Fig. S2. WT1 and HCK are frequently overexpressed in LSCs.
Fig. S3. Candidate LSC target molecules show expression in human AML cells lining the BM endosteum.
Fig. S4. CD25 is expressed in cell cycle–quiescent AML cells in the BM endosteal region.
Fig. S5. Expression of CD32 and CD25 in nonhematopoietic tissues.