A computational model predicts Xenopus meiotic spindle organization (original) (raw)
Spatially dispersed nucleation and minus end–directed transport of microtubule end disassembly activity can lead to bipolar spindle assembly.
Abstract
The metaphase spindle is a dynamic bipolar structure crucial for proper chromosome segregation, but how microtubules (MTs) are organized within the bipolar architecture remains controversial. To explore MT organization along the pole-to-pole axis, we simulated meiotic spindle assembly in two dimensions using dynamic MTs, a MT cross-linking force, and a kinesin-5–like motor. The bipolar structures that form consist of antiparallel fluxing MTs, but spindle pole formation requires the addition of a NuMA-like minus-end cross-linker and directed transport of MT depolymerization activity toward minus ends. Dynamic instability and minus-end depolymerization generate realistic MT lifetimes and a truncated exponential MT length distribution. Keeping the number of MTs in the simulation constant, we explored the influence of two different MT nucleation pathways on spindle organization. When nucleation occurs throughout the spindle, the simulation quantitatively reproduces features of meiotic spindles assembled in Xenopus egg extracts.
Introduction
The spindle is composed of microtubules and hundreds of other factors, but how these components interact to generate a dynamic, steady-state structure is unclear. Spindles assemble in Xenopus egg extracts through a self-organization pathway in which a gradient of RanGTP surrounding chromatin promotes microtubule (MT) nucleation and organization of MTs by motor proteins into an antiparallel, bipolar array. Although the spindle appears static, it is actually a steady-state structure composed of short-lived MTs. MTs align predominantly along the pole-to-pole axis and slide poleward through the action of kinesin-5, a plus end–directed tetrameric motor (Walczak and Heald, 2008). At the spindle pole, localized MT destabilization results in depolymerization from MT minus-ends. During the metaphase steady state, sliding and pole-localized disassembly are coordinated to form a spindle of constant length exhibiting “flux” (Dumont and Mitchison, 2009). Yet, how such a balance between MT assembly, sliding, and disassembly is achieved is not understood.
The contributions of various mechanisms to spindle architecture remain controversial, in part because the high density of polymers in the spindle prevents current optical imaging techniques from resolving individual MTs. Growing MT plus-ends have been observed throughout the spindle, but indirect measurements of plus-end dynamics within the spindle disagree as to whether MTs near the chromatin are stabilized relative to distal MTs (Wilde et al., 2001; Athale et al., 2008; Needleman et al., 2010). Moreover, the spatial distribution of MT minus-ends remains elusive, and speckle microscopy experiments have led to contradictory interpretations of the MT length distribution ranging from a truncated normal distribution with a mean MT length of ∼20 µm (G. Yang et al., 2007) to an exponential distribution with a mean of ∼3 µm (Verde et al., 1992; Needleman et al., 2010). Nucleation events cannot yet be observed, and multiple nucleation pathways have been proposed to contribute to spindle self-assembly. A gradient of RanGTP surrounding chromatin may induce nucleation near the midzone (Kalab et al., 2002; Caudron et al., 2005), while nucleation along existing MTs (amplification) may rapidly increase MT density within the spindle (Janson et al., 2005; Mahoney et al., 2006; Clausen and Ribbeck, 2007; Goshima et al., 2008; Zhu et al., 2008).
Computer simulations are well suited to examine the contributions and cooperativity of these mechanisms in spindle formation because a small number of well-characterized components appear responsible for the bulk of MT self-organization. We aimed to simulate important general features of Xenopus meiotic spindles, including pole formation, MT dynamics and nucleation, and spindle length regulation, and then to use this model to elucidate MT organization along the pole-to-pole axis.
Previous models of spindle assembly have not fully incorporated MT plus-end dynamics. In the most recent model of a Xenopus spindle, Burbank et al.’s slide-and-cluster model, MTs turned over with short lifetimes, but all MTs were of constant length and plus ends were nondynamic (Burbank et al., 2007). Plus-end growth is difficult to include in a model aiming to produce bipolar structures because growth is significantly faster than MT sliding. The disparity results in each MT plus-end extending three times farther than the minus end has slid poleward. If all MTs were nucleated at the midzone, as in the slide-and-cluster model, MT structures would be mainly astral: plus ends would grow far beyond the region of antiparallel overlap. In this work, we explore how dispersed nucleation of dynamic MTs resolves this issue to generate bipolar MT organization.
The mechanism aligning MT minus-ends to form a spindle pole also remains unclear. The slide-and-cluster model aligned minus ends using a dynein-like “cross-linking” motor similar to the oligomeric motors shown to cluster minus ends in asters (Nédélec et al., 1997). Yet, other work suggests that a simple balance of forces between kinesin-5 and kinesin-14/dynein motors does not readily generate MT structures of stable length (Badoual et al., 2002; Hentrich and Surrey, 2010). The necessary contribution of MT-destabilizing enzymes to pole formation (Gadde and Heald, 2004) has not yet been explored theoretically. We show here that minus end–directed transport of depolymerization activity can produce spindle poles without the help of clustering motors.
Results and discussion
Simulation of a metaphase meiotic spindle
We began by populating a simulation with 200 MTs to create a two-dimensional (2D) spindle slice of ∼5 µm in width, on the order of internal bundles observed in Xenopus spindles (Mitchison et al., 2004). Limiting the model to 2D made the simulation computationally tractable while still allowing us to address longitudinal MT organization in the spindle, which was robust against ±50% variation in MT number (Fig. S1 E). To further reduce the computation time, MTs were initialized in an antiparallel array (Fig. 1 A, color indicates MT orientation), but other initialization conditions produced similar steady-state structures. MT minus-ends were inert (Fig. 1 A, white segments), whereas plus ends underwent dynamic instability, alternating between growth and shrinkage states with parameters in the “bounded” regime based on measurements in Xenopus egg extracts (Table I; Dogterom and Leibler, 1993; Wilde et al., 2001; Brown et al., 2007). Once MTs depolymerized to a minimum length (40 nm), they were removed from the simulation. To mimic chromatin-driven spindle assembly, we delineated a zone of RanGTP activity extending 20 µm in length and 5 µm in width in which MT plus-ends were more stable, incorporating the measured downstream effects of RanGTP on MT catastrophe and rescue rates (Fig. 1 A, yellow rectangle; Wilde et al., 2001; Athale et al., 2008). The number of MTs was kept constant through two different mechanisms of nucleation. The first mimicked chromatin-mediated MT nucleation, which is known to occur in high concentrations of RanGTP (Kalab et al., 2002; Caudron et al., 2005). In the second mechanism, new MTs were nucleated along the side of randomly chosen existing microtubules, modeled after the MT amplification reported to occur within spindles (Mahoney et al., 2006; Goshima et al., 2008; Zhu et al., 2008).
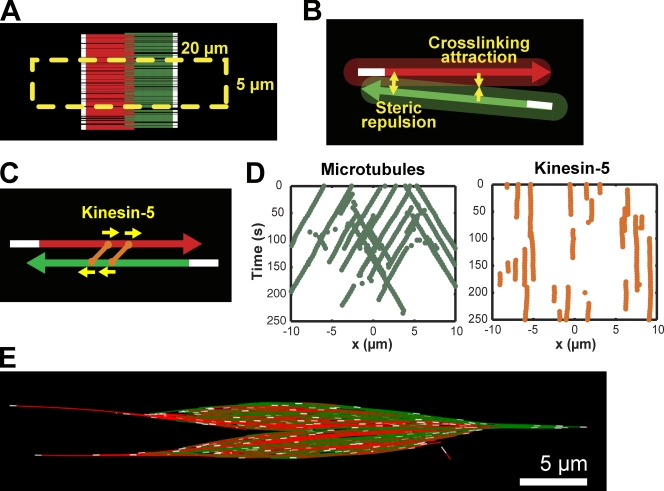
Figure 1.
Self-assembly of a fluxing bipolar MT array with dynamic MTs and kinesin-5. (A) Simulations were initiated with MTs in an antiparallel array centered in the zone of high RanGTP concentration (yellow dashed line). MT color indicates orientation (MTs with plus-ends pointed to the right in red, to the left in green) and region (minus-end region in white). (B) MTs experience repulsion at close range (0–50 nm) and attraction between 50 and 90 nm. The shaded area represents the interaction zone. (C) Homotetrameric kinesin-5 (orange) are simple plus end–directed motors, and slide apart antiparallel MTs. (D) Kymograph of selected MT minus-ends (green) and kinesin-5 motors (orange) shows that kinesin-5 remains stationary while bound MTs slide poleward. (E) Bipolar MT structure formed by MTs, cross-linking force, and kinesin-5. Single MTs extend far from the midzone, and poles are not defined.
Table I.
Parameters of the simulation
| Parameter | Value | Note/reference |
|---|---|---|
| Total time | 5,000 s | |
| Viscosity | 0.1 pN s/µm2 | 100x water |
| MTs | ||
| Amount | 200 | Kept constant by nucleation |
| Steric repulsion | Radius 0.025 µm | (Mitchison et al., 2004) |
| Stiffness 500 pN/µm every 0.25 µm | ||
| Bundling attraction | Radius 0.045 µm | |
| Stiffness 50 pN/µm every 0.25 µm | ||
| Plus-end assembly speeds | Growth 0.18 µm/s (10.8 µm/min) | (Carazo-Salas et al., 2001; |
| Shrinkage 0.33 µm/s (19.8 µm/min) | Tirnauer et al., 2004) | |
| Plus-end rates, low RanGTP | Catastrophe 0.1 s−1 | (Wilde et al., 2001; |
| Rescue 0.004 s−1 | Brown et al., 2007) | |
| Plus-end rates, high RanGTP | Catastrophe 0.024 s−1 | (Wilde et al., 2001) |
| Rescue 0.018 s−1 | ||
| Kinesin-5 motors | ||
| Amount | 6,000 | |
| Motor domain | Max speed 0.04 µm/s (2.4 µm/min) | (Valentine et al., 2006; |
| Stall force 5 pN | Korneev et al., 2007) | |
| Binding range 0.05 µm | ||
| Binding rate 2.5 s−1 | ||
| Unbinding rate 0.05 s−1 | ||
| Dual-motor complex | Stiffness 100 pN/µm | Has no binding specificity |
| Resting length 0.05 µm | ||
| NuMA sphere | ||
| Amount | 3,000 | |
| Radius | 0.025 µm | |
| Turnover rate | 0.1 s−1 | Allows dissociation of aggregates |
| Binding to MT minus end | Max distance 0.5 µm | |
| Binding rate 1 s−1 | ||
| Dynein driven motility | Max speed 0.8 µm/s (48 µm/min) | (Toba et al., 2006) |
| Stall force 2.5 pN | ||
| Binding range 0.05 µm | ||
| Binding rate 0.5 s−1 | ||
| Unbinding rate 0.05 s−1 | ||
| Depolymerization of MT minus-ends | −0.6 µm/min per sphere | |
| Discretization parameters | ||
| Time step | 0.005 s | |
| MT section length | 0.5 µm | For bending elasticity |
| MT initial length | 0.1045 µm | Length when created |
| MT minimal length | 0.04 µm | Shorter MTs are deleted |
In the spindle, many proteins cross-link adjacent MTs, resulting in an average MT spacing of 50 nm (Mitchison et al., 2004). Rather than explicitly modeling individual cross-linking proteins, we specified that MTs closer than 50 nm repelled each other, whereas MTs between 50 and 90 nm apart attracted each other (Fig. 1 B). Under these conditions, MTs exhibited Brownian motion and aligned without crossing one another, but did not organize along the horizontal axis (unpublished data). Next we implemented kinesin-5, a well-characterized tetrameric, plus end–directed molecular motor essential for bipolar spindle organization in Xenopus (Fig. 1 C; G. Yang et al., 2007; Kapitein et al., 2008). Simulations with as few as five kinesin-5 motors per MT generated antiparallel arrays in which MTs underwent poleward sliding at rates near those measured for kinesin-5 motility, while the motor itself was static in the spindle (Fig. 1 D; Videos 1 and 2). However, structures lacked any high density of aligned MT minus-ends that would be characteristic of spindle poles (Fig. 1 E).
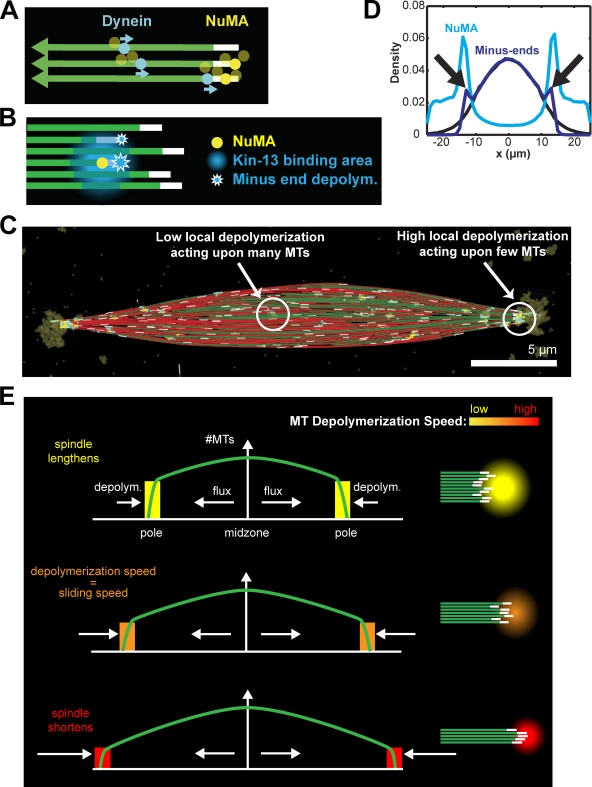
To generate poles, we modeled the effects of Xenopus kinesin-13s, kinesin superfamily protein 2a (Kif2a) and mitotic centromere-associated kinase (MCAK), which can depolymerize MTs from the minus end and are recruited to spindle poles by other pole-localized proteins in a dynein-dependent manner (Walczak et al., 1996; Desai et al., 1999; Gaetz and Kapoor, 2004; Jang et al., 2008). To localize the activity, we implemented the pole cohesion factor NuMA as an oligomeric MT cross-linker delivered to minus ends by dynein (Dionne et al., 1999; Haren and Merdes, 2002). NuMA could bind MTs within 0.5 µm of a minus end (Fig. 2 A, yellow), or it could bind anywhere along a microtubule, travel toward its minus end at the speed of dynein, and remain bound there (Fig. 2 A, cyan; Toba et al., 2006). Each NuMA protein recruited a fixed amount of kinesin-13, which formed a gradient of depolymerization activity around NuMA with maximal strength within a 0.5-µm radius, dropping to 10% of the maximal activity between 0.5 and 1.5 µm (Fig. 2 B, blue zone). As observed for kinesin-13s, the activity could bind to the MT lattice (Fig. 2 B, blue area of MTs), but could only depolymerize MTs from the minus end (Fig. 2 B, stars; Helenius et al., 2006). In the model, all kinesin-13 bound some portion of a MT, generating a minus-end depolymerization rate that depended on local MT density. The result was slow minus-end depolymerization in the midzone, where activity was low and spread over many MTs, and fast depolymerization at the periphery, where activity was concentrated on a small number of MTs (Fig. 2 C). Simulations generated steady-state spindles with focused poles in which minus ends and NuMA converged sharply, hallmarks of spindle pole formation (Fig. 2, C and D, blue and cyan lines, black arrows; Video 3; Merdes et al., 1996; Burbank et al., 2006).
Figure 2.
Depolymerization recruited by dynein-transported NuMA generates spindle poles. (A) NuMA (yellow) is transported to MT minus-ends by dynein (cyan). Oligomerization of NuMA leads to its accumulation at the spindle poles. (B) NuMA (yellow) recruits kinesin-13, whose binding (blue gradient) is spread among adjacent MTs. However, depolymerization (blue stars) only occurs at minus ends. Depolymerization is proportional to the amount of NuMA and inversely proportional to the local MT density (see Materials and methods). (C) Steady-state spindle structure with oligomerized NuMA (yellow) delivered to the pole by dynein (cyan). In the midzone, small amounts of kinesin-13 activity produce only slow depolymerization of minus ends. At the poles, accumulated NuMA recruits a large amount depolymerization activity distributed over few MTs, resulting in high depolymerization rates. (D) Minus-end density shows no polar accumulation (black) unless kinesin-13 depolymerization is added to the model (dark blue). With depolymerization, NuMA (light blue) accumulates at spindle poles (black arrows; average of 100, 95 simulations). (E) Schematic of metaphase steady state. MT density decreases from the midzone to the pole (green). Because depolymerization activity is spread over MTs, the depolymerization rate per MT increases with the distance from the midzone.
No model captures all features of the Xenopus meiotic spindle, and ours in particular lacks elements that determine spindle width. However, the model reproduced many key features of the organization of MTs along the longitudinal axis. The spindle length was well defined at 28 ± 0.86 µm (Table S1; Fig. S1 A), and pole-to-pole MT organization appeared similar to Xenopus meiotic spindles (Heald et al., 1997; Z. Yang et al., 2007). Spindle length ranged from 24.4 to 32.0 µm when the number of MTs in the spindle was varied ± 50% (Fig. S1 E), similar to experimental observations when the number of nonkinetochore MTs was reduced in Xenopus extract (Houghtaling et al., 2009). Plots of MT density based on MT orientation (Fig. S1 B) compared favorably to those interpolated from flux experiments (Burbank et al., 2006; G. Yang et al., 2007), as did fractional MT end density (Fig. S1 C; Burbank et al., 2006). MT end density was also similar to experimental interpolations (Tirnauer et al., 2004; Burbank et al., 2006), with plus and minus ends enriched in the midzone and pole, respectively, but with a lower MT minus-end density at the poles in the simulation (Fig. S1 D). This discrepancy suggests that nucleation near the spindle poles is higher in vivo than in the model.
Kinesin-5 operated in the low load limit, near its maximum speed and similar to experimentally measured values (Table S1; Video 4). MT flux was spatially homogeneous (see discussion below) and occurred at experimentally observed rates (2.28 ± 0.07 µm/min [mean ± SEM]; Ohi et al., 2007; G. Yang et al., 2007). For simplicity, the simulated kinesin-5 did not have a preference for antiparallel MTs (van den Wildenberg et al., 2008), and was not transported by dynein (Kapoor and Mitchison, 2001; Uteng et al., 2008). The latter effect has been suggested to boost kinesin-5 bundling activity near the spindle poles (Uteng et al., 2008). This was not necessary in our model because the cross-linking force bundled MTs independently of the simulated kinesin-5–like motor.
At spindle poles, the mean NuMA residency was 230 ± 26 s (SEM) with an average half life of 167 ± 30 s (SEM), similar to the half life of the mobile fraction of NuMA observed in cells (176 ± 46 s; Kisurina-Evgenieva et al., 2004). The simulation did not contain an immobile fraction of NuMA because NuMA de-oligomerization occurred at a constant rate. In agreement with in vitro observations of kinesin-13 activity on single MTs (Helenius et al., 2006), more than 99% of individual MT minus-end depolymerization rates were less than 4 µm/min. As observed experimentally, flux rates were unchanged upon kinesin-13 inhibition (2.22 ± 0.04 µm/min [mean ± SEM]; Ohi et al., 2007).
Inhibition of individual activities in the simulation reproduced experimental findings. For example, immobilizing a fraction of kinesin-5 in the simulation reproduced the inhibitory effects of the small molecule monastrol, slightly decreasing spindle length (Fig. S2 A; Burbank et al., 2007), and decreasing poleward flux rates (Fig. S2 C; Miyamoto et al., 2004) as long as the structure did not collapse. Keeping all other parameters intact but removing the ability of NuMA to oligomerize, or removing dynein-dependent transport of NuMA, disrupted spindle poles and lengthened structures, consistent with experimental results (Fig. S2, D–F; Merdes et al., 1996, 2000).
Certain features, specifically those dependent on the MT-clustering ability of dynein, were not replicated by the model. In simulations, flux was spatially homogeneous (2.26 ± 0.06 µm/min [mean ± SEM]; unpublished data) and did not decrease near the spindle poles as seen in vivo (Burbank et al., 2007; Yang et al., 2008). At high monastrol concentrations, Xenopus extract spindles collapse in a dynein-dependent manner into a radially symmetric aster (Kapoor et al., 2000). In simulations as well, bipolar arrays collapse to form an astral structure (Fig. S2 A), although this pattern is not radially symmetric due to the residual bundling interactions (Fig. S2 B). Furthermore, monastral spindle formation is mostly due to kinesin-13 activity. In the absence of kinesin-5–mediated flux, distal MT minus-ends are quickly depolymerized, causing NuMA oligomers to collapse into a single, central cluster, where dynein and depolymerization activities work together to align MT minus-ends. In addition, because the zone of primary MT nucleation and stabilization remains always centered in the simulations, the astral pattern more closely resembles the effects of kinesin-5 inhibition on chromatin bead spindles (Walczak et al., 1998).
Mechanisms of spindle pole formation
The establishment of a steady-state spindle requires polar MT disassembly to balance poleward MT flux (Fig. 2 E; Video 3). Such a balance is not obtained by fine-tuning the enzymatic rates of kinesin-5 and kinesin-13 (Goshima et al., 2005). Rather, Xenopus egg extract spindles exhibit robustness, forming over a range of flux and depolymerization rates, at the expense of changing spindle length (Shirasu-Hiza et al., 2004; Ohi et al., 2007). A simple feedback mechanism confers a similar robustness in the simulation (Fig. S2 G, arrows): accumulated kinesin-13 activity is spread over all MTs at the pole, so that the depolymerization rate per MT depends on the number of MTs at the pole (Fig. 2 E).
When the simulation begins, poles initially separate while depolymerization activity is spread over so many MTs that each MT slides faster than it depolymerizes (Fig. 2 E, top). As the poles extend farther, the number of MTs arriving at the pole decreases due to plus-end catastrophes. Eventually, the number of MTs arriving at the pole results in a depolymerization rate per MT that perfectly balances the flux rate (Fig. 2 E, middle). This feedback mechanism can also cause spindle poles to contract if MTs extend too far (Fig. 2 E, bottom), creating a stable equilibrium for pole position (Fig. 2 E, middle; Video 5). Due to the stochasticity of MT dynamics and NuMA localization, spindle length fluctuates around the mean value (Fig. S1 A). However, the kinesin-13 mechanism requires that MT density increase toward the midzone. In spindles composed of a constant number of MTs midzone-to-pole, such as in yeast, alternative feedback mechanisms must be used, such as MT length-dependent depolymerization by yeast kinesin-8 (Varga et al., 2006).
The MT depolymerization mechanism aligning minus ends is in principle independent from a motor-driven MT clustering (Burbank et al., 2006), yet both mechanisms seem to use the same motor in vivo: dynein. Theoretically, both mechanisms can work in concert, and neither alone fully explains spindle dynamics. Decreased rates of MT sliding have been measured for a “polar array” of MTs near the spindle pole in Xenopus spindles (Burbank et al., 2007; Yang et al., 2008), and arise in the slide-and-cluster model by antagonism between the dynein- and kinesin-5–like motors. The kinesin-13 mechanism does not replicate this phenomenon because dynein acts primarily to transport NuMA and is not abundant enough to oppose kinesin-5. On the other hand, the slide-and-cluster model incorporates only uncoordinated stochastic MT disappearance. In our model, flux is possible because minus-end depolymerization is linked to the arrival of MTs at the pole, where kinesin-13 is localized.
MT amplification is essential for bipolarity
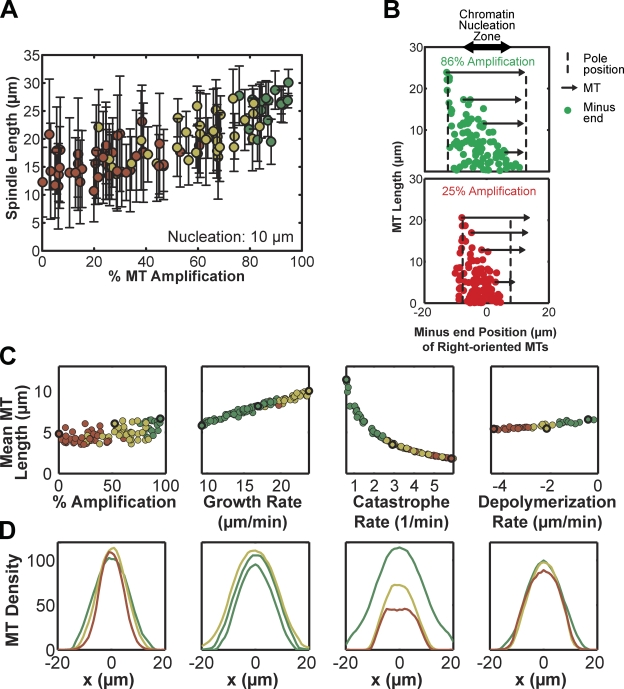
How MT nucleation mechanisms contribute to spindle architecture is unclear. Nucleation within Xenopus spindles is not directly observable, as current reagents cannot distinguish between nucleation and plus-end rescue events, but γ-tubulin–mediated MT nucleation along existing MTs has been reported in Drosophila and human mitotic spindles (Goshima et al., 2008; Zhu et al., 2008). A similar MT-amplification mechanism in Xenopus would explain the exponential increase in MT mass during RanGTP-stimulated aster formation (Clausen and Ribbeck, 2007). We therefore tested the sensitivity of the simulation to the percentage of RanGTP-mediated nucleation versus MT amplification. We found the two sources of nucleation in the spindle to be highly redundant (Fig. S1 F), likely reflecting the fact that the 20-µm breadth of the RanGTP zone nearly matched that of MT amplification throughout the 28-µm spindle. Restricting RanGTP-mediated nucleation from 20 to 10 µm increased sensitivity to the nucleation mechanism, as structures required at least 75% MT amplification for bipolarity (Fig. 3 A, color indicates bipolarity). With greater than 25% RanGTP nucleation, MT structures were primarily astral, with minus ends close to the midzone, plus ends growing past minus ends (Fig. 3 B, bottom), and disrupted NuMA localization, illustrating the argument that plus-end growth disrupts bipolar MT organization if most nucleation occurs near the midzone. MT amplification also allowed large structures to achieve the “tiled array” composition proposed by G. Yang et al. (2007), in which short MTs exist throughout the spindle (Fig. 3 B, top).
Figure 3.
Spindle assembly is sensitive to mechanisms affecting internal MT organization but not to MT length distribution. (A) When the area of chromatin-mediated nucleation was decreased to span 10 µm, the bipolarity (green, yellow, and red) of MT structures was sensitive to the nucleation pathway. Color indicates MT ”bipolarity index” defined in Materials and methods: astral (red), bipolar (yellow), strongly bipolar (green). MT structures remained bipolar only for percentages of chromatin-mediated nucleation up to 25%, while spindle length in the bipolar regime scaled slightly with nucleation pathway (92 simulations). (B) Position of the minus ends of right-oriented MTs at a single time point with a 10-µm zone of chromatin-mediated MT nucleation (double arrow). In a simulation with 86% MT amplification (green), the bipolar MT structure contained short MTs throughout while long MTs were positioned with their minus ends at the pole (dashed lines). Four sample MTs are illustrated (arrows). In a simulation with only 25% MT amplification (red), the astral MT structure also contained small MTs throughout, but minus ends did not extend far from the midzone, and plus ends grew past minus ends. (C) Mean MT length and bipolarity index (color) for simulations in which parameters for the nucleation pathway, MT plus-end growth rate, MT plus-end catastrophe rate, and kinesin-13 rate were varied. Bold circles indicate the simulations represented in D (92, 99, 95, 99 simulations). (D) Total MT density along the pole-to-pole axis from representative simulations (bold circles in C), where color indicates bipolarity index. Spindles could form and fail in a variety of sizes, indicating a strong dependence for bipolarity on assembly mechanism rather than the exact organization of the MT structure.
Spindles are robust to MT length distribution but sensitive to pole formation mechanism
Estimates of the MT length distribution in Xenopus spindles vary significantly. Centrosomal assays have indicated an exponential length distribution dependent primarily on the plus-end dynamic instability parameters, whereas a statistical model from tubulin speckle measurements indicated a truncated normal distribution within the spindle (Table S1). The simulation generates a truncated exponential distribution with a mean MT length of 6.51 ± 0.19 µm (mean ± SE; Fig. S1 G).
When screening for sensitivity to different parameters, we noted that the mean MT length could change dramatically without disrupting spindle structure (Fig. 3, C and D), implying that spindle bipolarity is insensitive to shifts in the length distribution. However, spindles were highly sensitive to minus-end depolymerization, even though this mechanism accounted for only 3.2 ± 0.1% (mean ± SE) of the total MT depolymerization in a simulation. Almost all MT depolymerization occurred at the plus end, as previously predicted (Ohi et al., 2007). Thus, the simulation demonstrates that a bipolar spindle could be composed of MTs with nearly exponentially distributed lengths.
Length of simulated spindles scales with MT destabilization activity
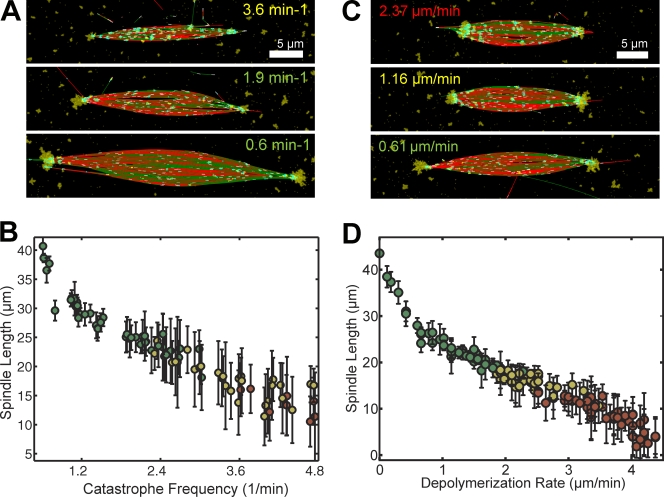
Next, we used the spindle simulation to examine which parameters affected spindle length most. Varying the MT growth rate and nucleation mechanism did not have a strong effect (Fig. 3 D; Fig. S1 F), but spindle length varied dramatically with plus-end catastrophe frequency and minus-end depolymerization rate. Interestingly, these two parameters determine how far a MT could slide before disappearing. Bipolar spindles with lengths from 40 to 25 µm formed within a relatively wide range of catastrophe frequencies between 0.6 and 3.0 min−1 (Fig. 4, A and B), which correspond to measured values (Wilde et al., 2001). Higher catastrophe frequencies led to aberrant structures with very short microtubules, whereas low frequencies were tolerated due to the constant number of MTs in the simulation. Altering the rate of MT minus-end depolymerization from 0.1 to 1.2 µm/min gave bipolar structures with an even wider range of lengths, from 45 to 17 µm (Fig. 4, C and D), although simulations with very low activity did not form high MT density spindle poles. Interestingly, Xenopus spindle length has been documented to scale with titration of the kinesin-13s MCAK and Kif2a (Ohi et al., 2007) and the tubulin-sequestering protein stathmin/Op18 (Budde et al., 2001; Houghtaling et al., 2009), which can act on both plus and minus ends to promote catastrophe or minus-end depolymerization.
Figure 4.
Length of simulated spindles scales with catastrophe rate and minus-end depolymerization activity. (A and B) Range of steady-state spindle structures obtained with different MT plus-end catastrophe frequencies (mean ± SD, 95 simulations). (C and D) Range of steady-state spindle structures obtained with different MT minus-end depolymerization rates (mean ± SD, 126 simulations). In A–D, the text or symbol color designates bipolarity index: astral (red), bipolar (yellow), strongly bipolar (green).
Conclusion
The model discussed here offers a coherent picture of how MT dynamic instability, flux, and nucleation contribute to the self-organization of a steady-state bipolar MT structure. Simulations reconstituted a steady-state spindle composed of dynamic MTs using basic mechanisms: antiparallel MT sliding by a tetrameric motor, MT amplification, and minus-end cross-linking and depolymerization. Because MT growth speeds exceed the speed of poleward motion, dispersed nucleation in the form of nucleation along existing MTs was necessary to organize dynamic MTs into a structure where MT plus-ends point inward. The simulation also illustrated that spindles can be composed of MTs with a truncated exponential length distribution, with short MTs present throughout the structure. Finally, whereas previous theoretical models proposed that spindle length is determined by a balance of forces between motors of opposite polarity, the length of simulated spindles in our model was determined by a balance between the speed of MT sliding associated with flux and the speed of MT depolymerization at the poles.
The model makes several predictions that can be tested experimentally. For instance, simulations showed that plus- and minus-end dynamics independently influence spindle length, which may be verifiable once a specific inhibitor becomes available, or more is discovered about the mechanisms of kinesin-13 localization to the pole. Similarly, the contribution of MT amplification to bipolar structure assembly can be explored when the Xenopus homologues of other known amplification proteins are characterized. Finally, the accuracy of our predicted spindle architecture and MT length distribution could be assessed using cryo-electron tomography and reconstruction of meiotic spindles. Although the model addresses pole-to-pole MT organization, construction of the spindle in the perpendicular plane remains to be explored. Furthermore, the model could be extended to examine the contributions of additional elements, such as chromatin and kinetochores, to the assembly of a functional spindle.
Materials and methods
Simulation
We used Cytosim to solve the Langevin equation for the motion of flexible MTs and NUMA spheres at low Reynolds number (Nedelec and Foethke, 2007). Parameter values (Table I) were taken from experimental measurements when possible and otherwise broadly screened to identify regimes resulting in appropriate spindle morphology. Parameters varied include: strength of steric repulsion and attraction (not depicted), plus-end growth speed (Fig. 3, C and D), plus-end catastrophe (Fig. 3, C and D; Fig. 4, A and B) and rescue rates (not depicted), number of kinesin-5 motors (not depicted), speed/mobility of kinesin-5 motors (Fig. S2, A–C, G and H), proportions and area of nucleation mechanism (Fig. 3 A, Fig. S1 F), amount of NuMA (not depicted), NuMA binding and transport efficiency (not depicted), kinesin-13 depolymerization rates (Fig. 4, C and D), and MT and NuMA initialization conditions (not depicted).
Simulations were run with a time step of 5 ms (similar results were obtained for smaller time steps), and iterations reached a total of 5,000 s unless otherwise noted. Measurements at steady state included only statistics from 2,500–5,000 s at 100-s intervals, except for kinesin-5 and dynein inhibition, where statistics were taken from 2,000–2,500 s. Spindle longitudinal axes were defined by the average MT orientation weighted by MT length. MT position and NuMA density were projected along this axis for line density measurements. Spindle length was calculated as the distance between the points of maximal NuMA density on each side of the midzone (i.e., the spindle poles) with 1 µm resolution, and time-derived to yield the spindle elongation rate. Spindle width was calculated as the distance spanned by MT ends at the midzone. MT density and MT end density were normalized by the number of MTs, and fractional MT end density was normalized by MT density. MT flux speed was calculated from the distance travelled along the longitudinal spindle axis by MTs during their lifetime and only included measurements for MTs with lifetimes >40 s.
A bipolarity index in [−1,1] is defined from P(x) and A(x), the numbers of MTs pointing to and away from the midzone, both functions of longitudinal position x:
B.I.=[∫sdx]−1∫sP(x)−A(x)P(x)+A(x)dx
where
is the segment covered by MTs. The bipolar index provides a quantitative measure of bipolarity for a MT structure: B.I. < 0 (red) indicates that MTs are predominantly oriented away from the midzone. On the contrary, B.I. > 0 for a spindle-like organization, in which MT minus ends are distant from the midzone while plus ends overlap in the midzone. We further differentiated between weak (0 < B.I. < 0.1, yellow) and strong (0.1 < B.I., green) bipolar structures.
MTs and cross-linking force
MT plus-ends switched stochastically between growth and shrinkage, which occurred at constant speed for all MTs (Table I). The rates of switching depended on the position of the plus end: either inside or outside the RanGTP zone (Table I). We obtained similar results with either a rectangular or circular (Fig. S1 E) RanGTP zone. MT minus-ends were stable unless depolymerized by the NuMA/kinesin-13 mechanism (see below). Any MT shorter than 0.04 µm was deleted from the simulation and replaced immediately by a new growing MT of length ∼0.1 µm with a random orientation. The positions of new MTs were determined according to two nucleation pathways. In the first, MTs were nucleated at random positions within the RanGTP zone. In the second, MTs were nucleated at random locations on the side of existing microtubules. For each nucleation event, a random number was used to choose between the primary and secondary pathways, following their proportions (80% and 20%, respectively, except for Fig. 3 A).
In the 2D simulations, MTs were not prevented from crossing, but a steric repulsive spring force with a resting length of 0.05 µm separated any MTs that came closer than this distance. The attractive cross-linking force also had a rest length of 0.05 µm but acted on any MTs closer than 0.09 µm. For repulsion and attraction, MTs interacted through points spaced at 0.25 µm along their length (Table I), with repulsion 10 times stronger than attraction. The attractive force represented approximately one cross-link of the strength of kinesin-5 per 0.5 µm of MT length. Forces produced by these interactions were projected perpendicular to the MT axis, so as to not generate any MT sliding forces, and thus did not impede kinesin-5–driven MT sliding.
Motors
Motor–MT attachment occurred at a constant rate, whereas the detachment rate increased exponentially with applied load:
Motors could bind and walk along MTs processively at a speed linearly dependent on load (Valentine et al., 2006; Gennerich et al., 2007):
Kinesin-5
Kinesin-5 was modeled as two separate motors connected by a linear spring of nonzero resting length, each walking toward MT plus-ends. Binding occurred independently of the orientation of MTs, and binding of the two motors to the same MT was forbidden. Unbinding was immediate upon reaching a MT end. The concentration of unbound kinesin-5 motors was considered to be uniform, and thus unbound diffusing motors were not simulated.
Dynein and NuMA
A single dynein was bound to each free NuMA and could also bind anywhere along MTs, traveling toward the minus end (Toba et al., 2006) and stalling at the final 0.5 µm of the MT. Because NuMA bound either a MT or a single dynein, only NuMA oligomers could cross-link MTs. Adjacent NuMA proteins could oligomerize via a Hookean force to form clusters with a 50-nm bond length. Turnover was implemented by displacing a fraction of the oligomerized NuMA to a random position in the simulation space at each time step.
Minus-end depolymerization
The depolymerization gradient was calculated at every time point by using a square grid size of 1 µm. The resolution of the grid was varied without affecting the simulation results. We first computed for each grid box a total amount of depolymerization activity proportional to the amount of NuMA within the region. Each NuMA recruited 0.01 µm/s depolymerization activity within 0.5 µm, and 0.001 µm/s activity within 0.5–1.5 µm. Then, the depolymerization activity present in a grid box was shared equally among all MT segments present in the grid box. When a MT segment corresponded to the minus end of a MT, it depolymerized at the calculated rate. This procedure is inspired by the experimental observation that kinesin-13s can bind and diffuse along the MT lattice (Helenius et al., 2006). Activity from multiple NuMA proteins was additive, meaning that increased NuMA localization resulted in increased depolymerization rates. There is no mechanism to protect minus ends from this depolymerization activity.
Initialization
To minimize computation costs, simulations were initialized with horizontal microtubules (Fig. 1 A) and with 25% of MT plus-ends in the shrinking state. Simulations with random initial configurations eventually reached the same steady states (not depicted). NuMA could be initialized randomly throughout the simulation space or at the minus ends of microtubules, reaching similar steady states.
Online supplemental material
Fig. S1 presents simulated elements that are directly comparable with figures published elsewhere (see text for references), which contain measurements of unperturbed meiotic spindles. Fig. S2 illustrates standard perturbations such as inhibitions of essential molecular components of the spindle. Five videos display different aspects of the simulated spindles. A table compares spindle length, microtubule distribution, length, and flux speed, in the simulation and in Xenopus laevis and tropicalis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006076/DC1.
Acknowledgments
We thank members of the Heald and Nédélec laboratories for helpful discussions, and in particular Michael Mitchell and Jonathon Ward for critically reading the manuscript.
R. Heald is supported by the National Institutes of Health Director’s Pioneer Award (DP1 OD000818) and The Miller Institute for Basic Research in Science. R. Loughlin acknowledges support from the National Science Foundation and the Cancer Research Coordinating Committee. F. Nédélec is supported by BioMS (Center for Modeling and Simulation in the Biosciences), the Volkswagenstiftung, Human Frontier Science Program Grant HFSP RGY084, and E.U. project MitoSys.
Footnotes
Abbreviations used in this paper:
Kif2a
kinesin superfamily protein 2a
MCAK
mitotic centromere-associated kinase
MT
microtubule
References
- Athale C.A., Dinarina A., Mora-Coral M., Pugieux C., Nedelec F., Karsenti E. 2008. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 322:1243–1247 10.1126/science.1161820 [DOI] [PubMed] [Google Scholar]
- Badoual M., Jülicher F., Prost J. 2002. Bidirectional cooperative motion of molecular motors. Proc. Natl. Acad. Sci. USA. 99:6696–6701 10.1073/pnas.102692399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.S., Blower M.D., Maresca T.J., Grammer T.C., Harland R.M., Heald R. 2007. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol. 176:765–770 10.1083/jcb.200610043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde P.P., Kumagai A., Dunphy W.G., Heald R. 2001. Regulation of Op18 during spindle assembly in Xenopus egg extracts. J. Cell Biol. 153:149–158 10.1083/jcb.153.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank K.S., Groen A.C., Perlman Z.E., Fisher D.S., Mitchison T.J. 2006. A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol. 175:369–375 10.1083/jcb.200511112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank K.S., Mitchison T.J., Fisher D.S. 2007. Slide-and-cluster models for spindle assembly. Curr. Biol. 17:1373–1383 10.1016/j.cub.2007.07.058 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Gruss O.J., Mattaj I.W., Karsenti E. 2001. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3:228–234 10.1038/35060009 [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. 2005. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 309:1373–1376 10.1126/science.1115964 [DOI] [PubMed] [Google Scholar]
- Clausen T., Ribbeck K. 2007. Self-organization of anastral spindles by synergy of dynamic instability, autocatalytic microtubule production, and a spatial signaling gradient. PLoS One. 2:e244 10.1371/journal.pone.0000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78 10.1016/S0092-8674(00)80960-5 [DOI] [PubMed] [Google Scholar]
- Dionne M.A., Howard L., Compton D.A. 1999. NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil. Cytoskeleton. 42:189–203 [DOI] [PubMed] [Google Scholar]
- Dogterom M., Leibler S. 1993. Physical aspects of the growth and regulation of microtubule structures. Phys. Rev. Lett. 70:1347–1350 10.1103/PhysRevLett.70.1347 [DOI] [PubMed] [Google Scholar]
- Dumont S., Mitchison T.J. 2009. Force and length in the mitotic spindle. Curr. Biol. 19:R749–R761 10.1016/j.cub.2009.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde S., Heald R. 2004. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14:R797–R805 10.1016/j.cub.2004.09.021 [DOI] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T.M. 2004. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166:465–471 10.1083/jcb.200404015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A., Carter A.P., Reck-Peterson S.L., Vale R.D. 2007. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 131:952–965 10.1016/j.cell.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., Vale R.D. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R.D. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Merdes A. 2002. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 115:1815–1824 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., Hyman A. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J. 2006. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 441:115–119 10.1038/nature04736 [DOI] [PubMed] [Google Scholar]
- Hentrich C., Surrey T. 2010. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J. Cell Biol. 189:465–480 10.1083/jcb.200910125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling B.R., Yang G., Matov A., Danuser G., Kapoor T.M. 2009. Op18 reveals the contribution of nonkinetochore microtubules to the dynamic organization of the vertebrate meiotic spindle. Proc. Natl. Acad. Sci. USA. 106:15338–15343 10.1073/pnas.0902317106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.-Y., Wong J., Coppinger J.A., Seki A., Yates J.R., III, Fang G. 2008. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J. Cell Biol. 181:255–267 10.1083/jcb.200711032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson M.E., Setty T.G., Paoletti A., Tran P.T. 2005. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J. Cell Biol. 169:297–308 10.1083/jcb.200410119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 295:2452–2456 10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Kwok B.H., Weinger J.S., Schmidt C.F., Kapoor T.M., Peterman E.J. 2008. Microtubule cross-linking triggers the directional motility of kinesin-5. J. Cell Biol. 182:421–428 10.1083/jcb.200801145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Mitchison T.J. 2001. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154:1125–1133 10.1083/jcb.200106011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., Mitchison T.J. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisurina-Evgenieva O., Mack G., Du Q., Macara I., Khodjakov A., Compton D.A. 2004. Multiple mechanisms regulate NuMA dynamics at spindle poles. J. Cell Sci. 117:6391–6400 10.1242/jcs.01568 [DOI] [PubMed] [Google Scholar]
- Korneev M.J., Lakämper S., Schmidt C.F. 2007. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur. Biophys. J. 36:675–681 10.1007/s00249-007-0134-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney N.M., Goshima G., Douglass A.D., Vale R.D. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569 10.1016/j.cub.2006.01.053 [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W.C., Cleveland D.W. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J., Maddox P., Groen A., Cameron L., Perlman Z., Ohi R., Desai A., Salmon E.D., Kapoor T.M. 2004. Bipolarization and poleward flux correlate during Xenopus extract spindle assembly. Mol. Biol. Cell. 15:5603–5615 10.1091/mbc.E04-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto D.T., Perlman Z.E., Burbank K.S., Groen A.C., Mitchison T.J. 2004. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 167:813–818 10.1083/jcb.200407126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec F., Foethke D. 2007. Collective Langevin dynamics of flexible cytoskeletal fibers. N. J. Phys. 9:427 10.1088/1367-2630/9/11/427 [DOI] [Google Scholar]
- Nédélec F.J., Surrey T., Maggs A.C., Leibler S. 1997. Self-organization of microtubules and motors. Nature. 389:305–308 10.1038/38532 [DOI] [PubMed] [Google Scholar]
- Needleman D.J., Groen A., Ohi R., Maresca T., Mirny L., Mitchison T. 2010. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol. Biol. Cell. 21:323–333 10.1091/mbc.E09-09-0816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Burbank K., Liu Q., Mitchison T.J. 2007. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol. 17:953–959 10.1016/j.cub.2007.04.057 [DOI] [PubMed] [Google Scholar]
- Shirasu-Hiza M., Perlman Z.E., Wittmann T., Karsenti E., Mitchison T.J. 2004. Eg5 causes elongation of meiotic spindles when flux-associated microtubule depolymerization is blocked. Curr. Biol. 14:1941–1945 10.1016/j.cub.2004.10.029 [DOI] [PubMed] [Google Scholar]
- Tirnauer J.S., Salmon E.D., Mitchison T.J. 2004. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol. Biol. Cell. 15:1776–1784 10.1091/mbc.E03-11-0824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba S., Watanabe T.M., Yamaguchi-Okimoto L., Toyoshima Y.Y., Higuchi H. 2006. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 103:5741–5745 10.1073/pnas.0508511103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng M., Hentrich C., Miura K., Bieling P., Surrey T. 2008. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 182:715–726 10.1083/jcb.200801125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M.T., Fordyce P.M., Krzysiak T.C., Gilbert S.P., Block S.M. 2006. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 8:470–476 10.1038/ncb1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg S.M.J.L., Tao L., Kapitein L.C., Schmidt C.F., Scholey J.M., Peterman E.J.G. 2008. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 18:1860–1864 10.1016/j.cub.2008.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., Howard J. 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962 10.1038/ncb1462 [DOI] [PubMed] [Google Scholar]
- Verde F., Dogterom M., Stelzer E., Karsenti E., Leibler S. 1992. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118:1097–1108 10.1083/jcb.118.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Heald R. 2008. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265:111–158 10.1016/S0074-7696(07)65003-7 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Mitchison T.J., Desai A. 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 84:37–47 10.1016/S0092-8674(00)80991-5 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., Heald R. 1998. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Wilde A., Lizarraga S.B., Zhang L., Wiese C., Gliksman N.R., Walczak C.E., Zheng Y. 2001. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3:221–227 10.1038/35060000 [DOI] [PubMed] [Google Scholar]
- Yang G., Houghtaling B.R., Gaetz J., Liu J.Z., Danuser G., Kapoor T.M. 2007. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat. Cell Biol. 9:1233–1242 10.1038/ncb1643 [DOI] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., Rieder C.L. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17:973–980 10.1016/j.cub.2007.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Cameron L.A., Maddox P.S., Salmon E.D., Danuser G. 2008. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J. Cell Biol. 182:631–639 10.1083/jcb.200801105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Coppinger J.A., Jang C.Y., Yates J.R., III, Fang G. 2008. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 183:835–848 10.1083/jcb.200807046 [DOI] [PMC free article] [PubMed] [Google Scholar]