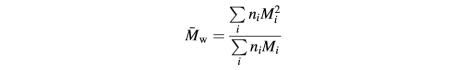Extracellular domains drive homo- but not hetero-dimerization of erbB receptors (original) (raw)
Abstract
Many different growth factor ligands, including epidermal growth factor (EGF) and the neuregulins (NRGs), regulate members of the erbB/HER family of receptor tyrosine kinases. These growth factors induce erbB receptor oligomerization, and their biological specificity is thought to be defined by the combination of homo- and hetero-oligomers that they stabilize upon binding. One model proposed for ligand-induced erbB receptor hetero-oligomerization involves simple heterodimerization; another suggests that higher order hetero-oligomers are ‘nucleated’ by ligand-induced homodimers. To distinguish between these possibilities, we compared the abilities of EGF and NRG1-β1 to induce homo- and hetero-oligomerization of purified erbB receptor extracellular domains. EGF and NRG1-β1 induced efficient homo-oligomerization of the erbB1 and erbB4 extracellular domains, respectively. In contrast, ligand-induced erbB receptor extracellular domain hetero-oligomers did not form (except for s-erbB2–s-erbB4 hetero-oligomers). Our findings argue that erbB receptor extracellular domains do not recapitulate most heteromeric interactions of the erbB receptors, yet reproduce their ligand-induced homo-oligomerization properties very well. This suggests that mechanisms for homo- and hetero-oligomerization of erbB receptors are different, and contradicts the simple heterodimerization hypothesis prevailing in the literature.
Keywords: dimerization/growth factor/scattering/signaling/tyrosine kinase
Introduction
The epidermal growth factor (EGF) receptor is the prototype of the erbB family of receptor tyrosine kinases (RTKs) that also includes erbB2 (HER-2 or Neu), erbB3 (HER-3) and erbB4 (HER-4) (Carraway and Cantley, 1994; Alroy and Yarden, 1997; Riese and Stern, 1998). Each erbB receptor contains an extracellular ligand-binding domain of 600–630 amino acids, a single transmembrane α-helix, plus an intracellular domain of ∼600 amino acids that includes the tyrosine kinase and regulatory sequences (Schlessinger and Ullrich, 1992). It was established more than a decade ago for the EGF receptor (erbB1) that growth factor-induced receptor oligomerization is critical for transmembrane signaling (Schechter et al., 1979; Schlessinger, 1979; Yarden and Schlessinger, 1987a,b). It is now generally accepted that the cytoplasmic tyrosine kinases of two (or more) RTKs in a growth factor-induced dimer (or larger oligomer) mutually activate one another through transphosphorylation (Honegger et al., 1990; Lemmon and Schlessinger, 1994; Heldin, 1995; Hubbard et al., 1998). Several downstream signaling molecules are then recruited to the phosphorylated receptor, specified by its complement of regulatory tyrosine phosphorylation sites (Songyang et al., 1993; Schlessinger, 1994).
Many cells co-express multiple members of the erbB receptor family, which can form both homo- and hetero-oligomers upon stimulation with growth factor ligands (Heldin, 1995). Oligomers containing almost every possible pairwise combination of erbB receptors have now been reported (reviewed by Carraway and Cantley, 1994; Alroy and Yarden, 1997; Riese and Stern, 1998). The earliest evidence for hetero-oligomerization of erbB receptors came from the finding that erbB2 can be activated by EGF, despite the fact that it does not bind directly to this ligand. EGF is only able to activate erbB2 when erbB1 is also present in the same cell, suggesting ‘transmodulation’ of erbB2 as a result of its EGF-induced hetero-oligomerization with erbB1 (King et al., 1988; Stern and Kamps, 1988; Goldman et al., 1990; Wada et al., 1990; Spivak-Kroizman et al., 1992).
There are >10 distinct ligands that activate erbB receptors. Three of these have been classified as ‘EGF agonists’ (Riese and Stern, 1998), since they bind directly to only erbB1 [EGF, transforming growth factor-α (TGF-α) and amphiregulin]. Four (or more) of the ligands are specific for erbB3 and/or erbB4 (the neuregulins; NRGs), while a further three have been classified as ‘bispecific’ and bind directly to both erbB1 and erbB4 [betacellulin, epiregulin and possibly heparin-binding EGF-like factor (HB-EGF)] (Riese and Stern, 1998; Harari et al., 1999; J.T.Jones et al., 1999, and references therein). The EGF agonists activate erbB1 when it is expressed alone, but also transmodulate erbB2, erbB3 and erbB4 in an erbB1-dependent manner. Similarly, the NRGs activate erbB4 directly, but can also transactivate erbB1 or erbB2 when erbB4 or erbB3 are also present (Riese et al., 1995). Finally, the bispecific ligands appear to activate erbB1 and erbB4 when either is expressed alone, and to transmodulate erbB2 and erbB3 via these receptors (reviewed by Alroy and Yarden, 1997; Riese and Stern, 1998). ErbB2, which is of particular medical interest as a target of breast cancer therapies (Sliwkowski et al., 1999), has no known ligand and can only be activated in trans by ligands in these three classes. In fact, erbB2 is considered to be a preferred hetero-oligomerization partner for all of the other erbB receptors (Karunagaran et al., 1996; Graus-Porta et al., 1997).
Several possible mechanisms for erbB receptor transmodulation have been considered. In the simplest and most often discussed, transmodulation is proposed to result from ligand-induced receptor heterodimerization (Alroy and Yarden, 1997; Burden and Yarden, 1997; Riese and Stern, 1998). According to this mechanism, a ligand stimulates two receptors to come together. If the two receptors are identical, this is homodimerization; if not, it is heterodimerization. Either way, the two receptors in the dimer become activated by transphosphorylation, and transmembrane signaling is achieved. Several studies argue that erbB receptor extracellular domains are sufficient for their hetero-oligomerization (Qian et al., 1994), and combinatorial receptor (homo- or hetero-) dimerization could be driven by simultaneous binding of bivalent erbB ligands to the extracellular domains of two receptor molecules (Lemmon et al., 1997; Tzahar et al., 1997). Different bivalent ligands could stabilize distinct receptor homo- and/or heterodimers depending on the combination of binding sites that they contain.
An alternative view is that growth factors such as EGF induce only homodimerization of the erbB receptors to which they bind directly. The resulting receptor homodimers may then activate in trans the erbB receptors to which the ligand does not bind, through quite different mechanisms. For example, transmodulation of erbB2 by EGF could simply involve phosphorylation of erbB2 as a substrate for the activated EGF receptor. Another possibility (Huang et al., 1998) is that EGF-induced erbB1 homodimers could provide an interface at which dimerization of erbB2 is promoted. ErbB2 could thus become activated by ‘proxy’ in the context of an (erbB1)2(erbB2)2 heterotetramer. A model of this sort could explain the surprising observation that a kinase-negative form of erbB1 can transmodulate erbB2 upon EGF binding (Wright et al., 1995).
In order to determine whether erbB receptor homo- and hetero-oligomerization occur through similar mechanisms, we have studied the effects of ligand binding on the assembly of isolated erbB receptor extracellular domains. We reported previously that the isolated erbB1 extracellular domain (s-erbB1) homodimerizes quantitatively upon binding to EGF or TGF-α (Lemmon et al., 1997). Here, we show that NRG1-β1 can also induce homo-oligomerization of the erbB4 extracellular domain. In contrast, ligand-induced hetero-oligomerization appears to be the exception rather than the rule for erbB receptor extracellular domains. While NRG1-β1 can induce the formation of hetero-oligomers that contain the erbB2 and erbB4 extracellular domains, no evidence could be obtained for EGF-induced formation of any extracellular domain hetero-oligomer. These findings indicate that erbB receptors form homo- and hetero-oligomers through quite different mechanisms, and that transmodulation of erbB receptors is most probably nucleated by a ligand-induced erbB1 or erbB4 homodimer.
Results
High-affinity ligand binding by recombinant s-erbB proteins
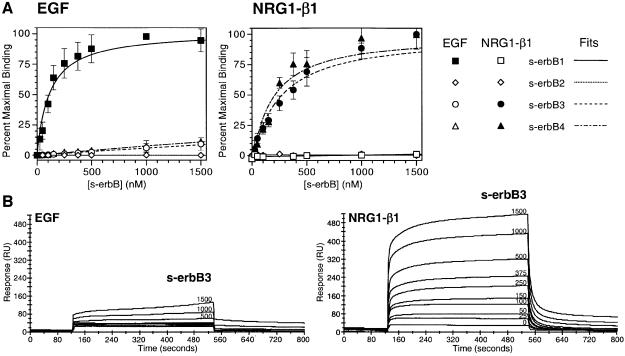
To investigate the ligand binding and dimerization properties of soluble erbB receptor extracellular domains (s-erbBs), we first established methods for their production in milligram quantities by secretion from baculovirus-infected Sf9 cells (Figure 1). Using surface plasmon resonance (BIAcore), we next measured binding of each purified s-erbB protein to both EGF and NRG1-β1 that were immobilized on BIAcore CM-5 sensor chips. The s-erbB proteins were passed across these surfaces at a variety of concentrations, and the maximum response observed was plotted against s-erbB concentration to generate the binding curves shown in Figure 2A. As anticipated, s-erbB1 bound strongly to the EGF-derivatized sensor surface (_K_D = 118 nM), but not to surfaces carrying NRG1-β1 or to surfaces with no ligand. Both s-erbB3 and s-erbB4 bound strongly to the NRG1-β1 surface (_K_D values of 249 and 179 nM, respectively; see Table I), but not to the EGF-derivatized surface. In contrast, s-erbB2 did not bind to any of the surfaces tested (Figure 2A). We repeated these experiments using 1:1 mixtures of different s-erbB proteins (e.g. s-erbB2 plus s-erbB3 or s-erbB4) to determine whether free s-erbB proteins might hetero-oligomerize, leading to significant alterations in their apparent ligand-binding affinities. In these studies, mixing s-erbB proteins had no detectable influence on their ligand-binding properties (not shown), arguing that s-erbB hetero-oligomers (if they form) do not bind the immobilized ligands with a significantly higher affinity than single s-erbB species.
Fig. 1. SDS–PAGE (7.5%) of the purified s-erbB proteins used for analysis of ligand-induced homo- and hetero-oligomerization. Purified protein (15 µl) was loaded at a concentration of 1 mg/ml, and the gel was stained with Coomassie Blue. Molecular mass standards were loaded in the left-most lane, and are marked.
Fig. 2. (A) Data for binding of s-erbB1, s-erbB2, s-erbB3 and s-erbB4 to EGF (left) and NRG1-β1 (right), immobilized on a BIAcore sensor chip. Best fits to the data, assuming a simple association model, are shown. Errors are standard deviations from the mean of at least four independent determinations at each point. _K_D values represented by the best fits are listed in Table I. (B) Representative raw BIAcore data for s-erbB3 flowed in parallel over a biosensor chip derivatized with EGF (left) and NRG1-β1 (right) at a series of different concentrations (marked on each curve in nM).
Table I. Ligand binding by s-erbB proteins.
| Ligand | _K_D (nM) | |||
|---|---|---|---|---|
| s-erbB1 | s-erbB2 | s-erbB3 | s-erbB4 | |
| EGF | 118 ± 41 | none | >104 | >104 |
| NRG-β1 | >105 | none | 249 ± 80 | 179 ± 10 |
s-erbB1 and s-erbB4 homo-oligomerize upon ligand binding, while s-erbB3 does not
To analyze ligand-induced dimerization of s-erbB proteins, we employed multi-angle laser light scattering (MALLS) and sedimentation equilibrium analytical ultracentrifugation, both of which give information on molecular mass changes that is independent of molecular shape (Cantor and Schimmel, 1980).
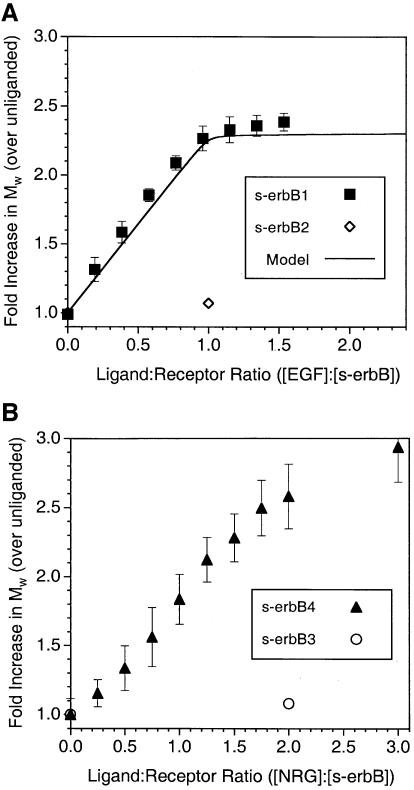
Multi-angle laser light-scattering studies. MALLS allows the weight-averaged molecular mass (_M̄_w) of proteins in solution to be measured rapidly over a wide range of protein concentrations (see Materials and methods). MALLS measurements gave an _M̄_w value of 77 ± 8 kDa for purified s-erbB1 alone. When EGF is titrated effectively into an s-erbB1 solution (with fixed s-erbB1 concentration), _M̄_w increases in a linear fashion until one molar equivalent of EGF has been added to s-erbB1 (Figure 3A). At this point, _M̄_w is 2.2-fold higher than that measured for s-erbB1 alone, suggesting EGF-induced formation of a dimeric complex containing two EGF molecules plus two molecules of s-erbB1, as we have observed with other methods (Lemmon et al., 1997). No further increase in _M̄_w is seen when EGF is added in excess, arguing that higher order oligomers of s-erbB1 do not form. The curve through the data in Figure 3A represents the results expected if EGF binds to monomeric s-erbB1 with a _K_D = 118 nM (Table I), and the resulting 1:1 (EGF:s-erbB1) complex dimerizes completely. The _K_D for this dimerization event (which is complete at 4 µM s-erbB1) appears to be <0.1 µM, based on additional MALLS studies at low concentration and gel filtration experiments (not shown).
Fig. 3. MALLS studies of EGF-induced homodimerization of s-erbB1- (A) and NRG1-β1-induced homo-oligomerization of s-erbB4 (B). The weight-averaged molecular mass (_M̄_w) of s-erbB1:EGF mixtures (relative to for s-erbB1 alone), as determined by MALLS (see Materials and methods), is plotted against the EGF:s-erbB1 ratio in the mixture. Quantitative EGF-induced s-erbB1 homodimerization is shown (filled squares). The solid line represents the expected results for a model in which EGF binds s-erbB1 with a _K_D of 118 nM, and the resulting 1:1 complex dimerizes with a _K_D of 100 nM (see text). The single open diamond in (A) shows one point for a similar experiment with s-erbB2, demonstrating that s-erbB2 does not dimerize when EGF is added (see also Figure 4B). In (B), the same experiment is shown for NRG1-β1 binding to s-erbB4 (filled triangles), which it causes to oligomerize. Also in (B), a single point (open circle) shows the failure of NRG1-β1 to induce s-erbB3 homo-oligomerization. Error bars correspond to the standard deviations for the mean of three or more experiments. The concentration of s-erbB protein was 4 µM in each experiment.
Similar MALLS studies of s-erbB4 gave a _M̄_w of 82 ± 6 kDa that increased by a factor of >2 as NRG1-β1 was added (Figure 3B). In this case, the maximum _M̄_w value was not reached until more than two equivalents of NRG1-β1 had been added. Furthermore, the final _M̄_w value (∼235 kDa) was higher than expected for a dimeric s-erbB4–NRG1-β1 complex. These data therefore suggest that NRG1-β1 is able to induce formation of s-erbB4 oligomers that are larger than dimers. Without more detailed analysis at significantly higher protein concentrations and at larger excesses of ligand, we cannot determine the maximum oligomeric state. However, an increase of nearly 3-fold in _M̄_w (at an NRG1-β1:s-erbB4 ratio of 3:1) is equally consistent with the formation of s-erbB4 trimers and with the formation of a mixture that contains 50% of the s-erbB4 as dimers plus 50% as tetramers.
We also used MALLS to analyze the ability of NRG1-β1 to induce s-erbB3 oligomerization. As shown by a single data point in Figure 3B (and confirmed in centrifugation studies described below), addition of a 2-fold excess of NRG1-β1 did not increase the _M̄_w measured for s-erbB3 above that measured for s-erbB3 alone (90 ± 4 kDa). This finding is consistent with a previous report (Horan et al., 1995), and does not reflect a lack of NRG1-β1 binding by s-erbB3 (see Figure 2B and Table I). Addition of neither EGF (Figure 3A) nor NRG1-β1 (not shown) altered the value measured for s-erbB2 (78 ± 10 kDa), as was expected since neither ligand binds to this protein (Figure 2A).
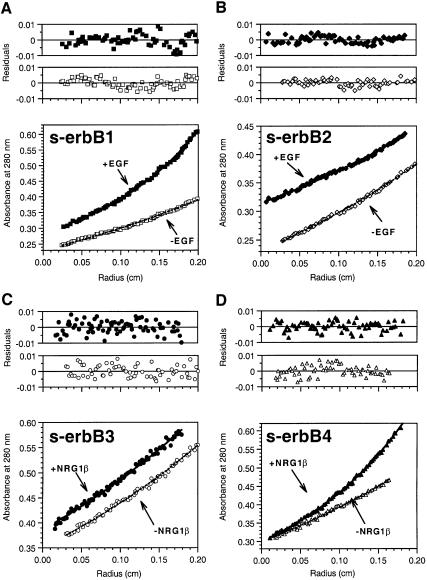
Analytical ultracentrifugation. Sedimentation equilibrium experiments gave the same results for ligand-induced s-erbB protein homo-oligomerization. Figure 4 shows typical data from sedimentation equilibrium experiments (at 6000 r.p.m.) in which 5 µM samples of each s-erbB protein were centrifuged both with (filled symbols) and without (open symbols) a 2-fold molar excess of the most relevant growth factor ligand. Data obtained with the ligand-free s-erbB proteins can be fit, using a model that assumes a single non-ideal species, to give molecular mass estimates of 81 ± 1 kDa (s-erbB1), 80 ± 3 kDa (s-erbB2), 82 ± 7 kDa (s-erbB3) and 81 ± 3 kDa (s-erbB4). The residuals for these fits, plotted above the data in Figure 4, are both small and random, indicating good fits. When EGF is added to s-erbB1 (Figure 4A), or NRG1-β1 is added to s-erbB4 (Figure 4D), the radial distribution plots suggest a substantial increase in molecular mass (with material accumulating at higher radii). Since the molecular masses of EGF and NRG1-β1 are only 6 and 8 kDa, respectively (Lemmon et al., 1997; data not shown), and those of s-erbB1 and s-erbB4 are ∼80 kDa, this effect can only be explained by homo-oligomerization of the s-erbB proteins upon addition of the relevant growth factor. The data for s-erbB:ligand mixtures can be fit using a model that assumes two ideal species: the ligand–receptor complex and excess ligand. Using this model, the masses of s-erbB1–EGF and s-erbB4–NRG1-β1 complexes are estimated as 159 ± 10 kDa and 146 ± 18 kDa, respectively (residuals for these fits are shown in Figure 4A and D), consistent with the ligand-induced oligomerization of these extracellular domains seen by MALLS. In other sedimentation experiments (not shown), TGF-α and HB-EGF were also found to induce formation of s-erbB1 homo-oligomers (assumed dimers). As with MALLS, sedimentation equilibrium studies of s-erbB4:NRG1-β1 mixtures at higher s-erbB4 concentrations and larger ligand excesses (not shown) suggested that NRG1-β1 induces formation of s-erbB4 oligomers larger than dimers. However, we have not yet been able to determine whether these are trimers or mixtures of different oligomers.
Fig. 4. Representative sedimentation equilibrium analytical ultracentrifugation data for analysis of s-erbB homo-oligomerization induced by EGF (A and B) or NRG1-β1 (C and D). In each case, open symbols represent s-erbB protein without added ligand, which is fit as a single non-ideal species. Filled symbols represent samples to which a 2-fold molar excess of the noted ligand has been added. As discussed in the text, fits to these data are with two ideal species (complex plus excess free ligand)—fixing the mass of the ligand and floating the mass of the complex. Purified s-erbB protein was used at 5 µM for each sample. All experiments shown were performed at 6000 r.p.m. Repeats at 9000 and 12 000 r.p.m. gave the same results. Residuals for the fits described above are shown, and are seen to be both small and random, indicative of a good fit. EGF induced homo-oligomerization of s-erbB1 only, while NRG1-β1 induced homo-oligomerization of s-erbB4 only. Radius is plotted as (_r – r_o), where r is the radial position in the sample, and _r_o the radial position of the meniscus.
In contrast to the findings for s-erbB1 and s-erbB4, no indication of ligand-induced oligomerization was seen when EGF was added to s-erbB2 (Figure 4B), or when NRG1-β1 was added to either s-erbB3 (Figure 4C) or s-erbB2 (see below). The data for the s-erbB2:EGF mixture were best fit as a combination of free EGF and free s-erbB2 (82 ± 12 kDa), and those for the s-erbB3:NRG1-β1 mixture fit best as free NRG1-β1 (8 kDa) plus a 1:1 s-erbB3–NRG1-β1 complex of 83 ± 17 kDa.
ErbB1 and erbB2 extracellular domains do not heterodimerize upon EGF binding
Having confirmed that EGF induces s-erbB1 homodimerization, and that NRG1-β1 induces s-erbB4 homo-oligomerization, we next investigated the ability of erbB ligands to induce heterodimerization of erbB receptor extracellular domains. As described in the Introduction, the most well-studied example of erbB receptor transmodulation involves erbB1 and erbB2. Since EGF induces complete homodimerization of s-erbB1, we expected from the simple heterodimerization model for erbB receptor transmodulation that EGF should also induce the formation of s-erbB1–s-erbB2 heterodimers.
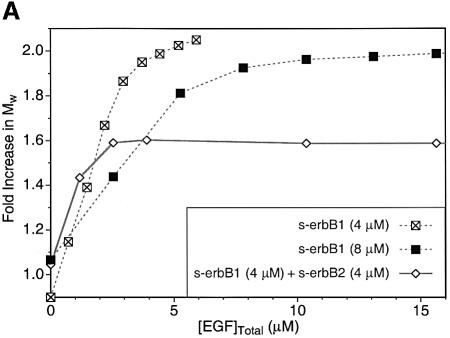
Contrary to these expectations, heterodimer formation could not be observed in MALLS studies when EGF was added to a 1:1 mixture of s-erbB1 and s-erbB2. Instead, EGF induced homodimerization of s-erbB1 in the mixture, while s-erbB2 remained monomeric. As shown in Figure 5A, titration of EGF into a solution containing 4 µM (crossed-squares) or 8 µM (filled squares) s-erbB1 alone caused complete dimerization. _M̄_w reached a maximum value (∼2-fold) after addition of EGF to ∼4 and 8 µM, respectively, as expected for the formation of a 2:2 EGF:s-erbB1 dimer. If EGF-induced heterodimerization of s-erbB1 with s-erbB2 were similarly strong, MALLS data for a 1:1 s-erbB1:s-erbB2 mixture (8 µM total receptor) should resemble that seen for 8 µM s-erbB1 alone. However, EGF addition to such a 1:1 mixture (diamonds in Figure 5A) induced a maximum _M̄_w increase of only 1.6-fold, and this maximum was reached at 4 µM, not 8 µM, total EGF. Homodimerization of just s-erbB1 (at 4 µM) in this mixture would be maximal at 4 µM EGF according to the data in Figure 3A. Furthermore, a 1.6-fold increase in _M̄_w is exactly what is expected if s-erbB1 homodimerizes (yielding 174 kDa s-erbB1 dimers at 2 µM) while s-erbB2 remains monomeric (80 kDa s-erbB2 monomers at 4 µM). Therefore, EGF does not induce heterodimerization of s-erbB1 with s-erbB2—or at least the _K_D for this heterodimerization event is sufficiently weak to be undetectable under these conditions (where s-erbB1 homodimerization is complete).
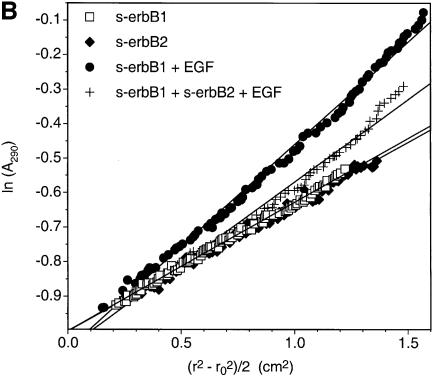
Fig. 5. (A) A MALLS experiment demonstrating that, while EGF induces complete homo-dimerization of s-erbB1 at 4 µM (crossed squares) or 8 µM (filled squares), it does not induce the formation of heterodimers between s-erbB2 and s-erbB1 (open diamonds). The experiment was performed as described for Figure 3. With 4 µM s-erbB1, complete dimerization is seen after addition of 4 µM EGF (note that the horizontal axis here is EGF concentration, and not ligand:receptor ratio). With 8 µM s-erbB1, addition of 8 µM EGF is required for complete dimerization. When the 1:1 s-erbB1:s-erbB2 mixture is studied, with a total s-erbB protein concentration of 8 µM, only 4 µM EGF is required for maximal dimerization, and the maximum fold increase in _M_w is consistent only with a mixture of s-erbB1 homodimers and s-erbB2 monomers. Lines are drawn to guide the eye, and do not represent fits to the data. (B) Plots of the natural logarithm of absorbance at 290 nm (monitoring protein concentration) against a function of the radius squared (_r_2 – _r_o2)/2 (see text for explanation) for sedimentation equilibrium analytical ultracentrifugation data obtained at 6000 r.p.m. with s-erbB1 and s-erbB2. For an ideal single species, this representation of the data should appear as a straight line with a gradient proportional to the molecular mass (see text). When analyzed alone, both s-erbB1 (open squares) and s-erbB2 (filled diamonds) yield good straight lines, with gradients proportional to their monomeric molecular masses (see also fits in Figure 4). Each sample contained a total s-erbB concentration of 10 µM. The increase in gradient for the s-erbB1/s-erbB2/EGF mixture (crosses) is consistent with the formation of s-erbB1 homodimers only.
Sedimentation equilibrium experiments also argue strongly against EGF-induced s-erbB1–s-erbB2 heterodimerization. For a set of experiments performed at 6000 r.p.m., the natural logarithm of absorbance at 290 nm (proportional to protein concentration) is plotted in Figure 5B against (_r_2 – _r_o2)/2, where r is the radial position in the sample, and _r_o the radial position of the meniscus. For an ideal single species, this plot is linear and the gradient of the line [_M_ω2(1 – _V̄_2ρ)/RT] is proportional to the molecular mass (M) of the ideal species (Cantor and Schimmel, 1980). The data for s-erbB1 or s-erbB2 alone fit well to a straight line with a gradient that suggests a molecular mass of ∼80 kDa in each case. When two molar equivalents of EGF were added to s-erbB1, the gradient of the best straight line (Figure 5B, filled squares) was increased substantially over that for s-erbB1 alone, because of EGF-induced s-erbB1 homodimerization. When the same excess of EGF was added to a 1:1 s-erbB1:s-erbB2 mixture (two EGFs added per s-erbB molecule), the data fit less well to a straight line (indicating multiple species), and the gradient of the best line was increased only slightly over that for s-erbB1 or s-erbB2 alone. Similar experiments at substantially higher receptor concentrations also failed to provide evidence for erbB1–erbB2 hetero-oligomerization. Thus, as seen with MALLS, analytical ultracentrifugation studies suggest that EGF induces homodimerization of s-erbB1 in a s-erbB1: s-erbB2 mixture, while s-erbB2 remains monomeric.
These biophysical studies show that the isolated extracellular domains of erbB1 and erbB2 do not associate with one another in a heterodimer (or any other oligomer) upon EGF addition, whereas s-erbB1 homodimerizes efficiently upon EGF binding (Figures 3–5) and EGF-dependent co-immunoprecipitation of intact erbB1 and erbB2 has been reported by many groups. In studies not shown, we attempted to detect s-erbB1–s-erbB2 interactions using chemical cross-linking and co-immunoprecipitation approaches, and obtained only negative results (although s-erbB1 homodimers could be seen readily by chemical cross-linking). We therefore suggest that, while the extracellular domain is sufficient for EGF-induced homodimerization of erbB1, extracellular domains are not capable of driving receptor hetero-oligomerization. Before concluding this, however, an important caveat must be considered. Since erbB2 has no known ligand, we cannot validate the functional integrity of Sf9 cell-derived s-erbB2 by virtue of its ligand binding, as was possible with s-erbB1, s-erbB3 and s-erbB4 (Figure 2). However, we believe that s-erbB2 is functional, since it appears to form NRG1-induced hetero-oligomers with s-erbB4 (see below).
ErbB1 and erbB4 extracellular domains do not hetero-oligomerize upon EGF or NRG1-β1 binding
Evidence for hetero-oligomerization (or transmodulation) of erbB1 and erbB4 upon treatment of cells with either EGF or NRG has been reported by several groups (Riese et al., 1995, 1996; Cohen et al., 1996; Zhang et al., 1996; F.E.Jones et al., 1999). We therefore used analytical ultracentrifugation to investigate whether EGF and NRG1-β1 induce s-erbB1–s-erbB4 heterodimerization. Since we know that s-erbB1 and s-erbB4 are both competent to homo-oligomerize upon binding of EGF and NRG1-β1, respectively, we can be confident that these proteins are functionally active.
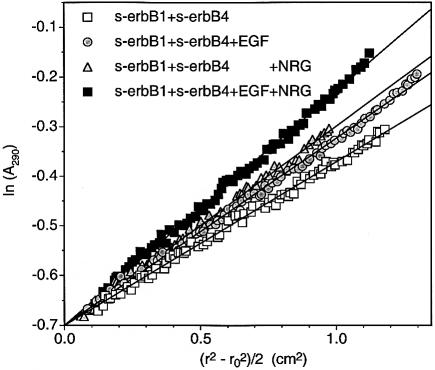
A series of sedimentation equilibrium experiments was performed with 1:1 mixtures of s-erbB1 and s-erbB4, with the same total receptor concentration (8 µM) in each case (Figure 6). With no ligand added, the gradient of the straight line through the data gives an average monomeric molecular mass of ∼80 kDa. Addition of EGF to a concentration twice that of total receptor (i.e. two EGF molecules per s-erbB1 molecule plus two EGF molecules per one s-erbB4) increases the gradient of the straight line only slightly (circles in Figure 6), suggesting that some oligomerization is induced. Addition of only NRG1-β1 to the same final concentration gives a similar result (triangles in Figure 6). Since ligand is not limiting in either of these cases, we hypothesized that these small increases in gradient result from homo-oligomerization of just s-erbB1 when EGF is added, and of just s-erbB4 when NRG1-β1 is added. If this is true, an identical sample containing the same total ligand concentration, but as a 1:1 mixture of EGF and NRG1-β1 (i.e. with two EGF molecules per s-erbB1 molecule plus two NRG1-β1 molecules per one s-erbB4), should give a substantially steeper gradient by inducing independent homo-oligomerization of both s-erbB1 and s-erbB4. Indeed, the steepest line in Figure 6 (filled squares) shows this to be the case, arguing that s-erbB1 and s-erbB4 do not form hetero-oligomers under these conditions with either EGF or NRG1-β1.
Fig. 6. Analytical ultracentrifugation data, presented as ln(Abs) against (_r_2 – _r_o2)/2 plots, to study s-erbB1–s-erbB4 hetero-oligomerization. The s-erbB1:s-erbB4 mixture (8 µM total [s-erbB]) without ligand gives a straight line with the gradient expected for monomeric protein (open squares). Addition of EGF alone (16 µM) or NRG alone (16 µM) results in a modest increase in molecular mass that is consistent with homo-oligomerization of one species only (gray circles and triangles, respectively). Addition of both EGF and NRG (8 µM each) results in a substantially larger increase in the gradient (black squares), indicating that both species homo-oligomerize independently, and do not form hetero-oligomers (see text for explanation).
Evidence for NRG1-β1-induced hetero-oligomerization of s-erbB4 and s-erbB2
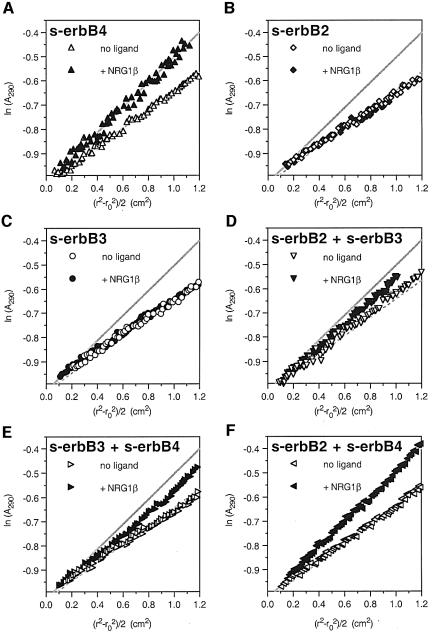
The experiments described above show that EGF does not induce hetero-oligomerization of s-erbB1 with s-erbB2 or s-erbB4. Other experiments showed that EGF does not induce the formation of s-erbB1–s-erbB3 or s-erbB2–s-erbB3 hetero-oligomers, and that NRG1-β1 does not drive the interaction of s-erbB1 with s-erbB3 (not shown). Therefore, although EGF-induced s-erbB1 homodimerization is highly efficient, s-erbB1 does not participate in formation of any s-erbB hetero-oligomer. Furthermore, EGF cannot induce the formation of any s-erbB hetero-oligomer. To compare these properties of EGF with those of NRG1-β1, we next tested the ability of NRG1-β1 to induce formation of a series of s-erbB dimers (Figure 7). Using linearized sedimentation equilibrium data as a qualitative guide, Figure 7A, B and C shows that NRG1-β1 induces homo-oligomerization of s-erbB4 (see also Figures 3B and 4D), but not of s-erbB2 or s-erbB3. The data for s-erbB4 homo-oligomerization (from Figure 7A) are superimposed upon all other graphs in Figure 7 to aid comparison. NRG1-β1 addition to an s-erbB2:s-erbB3 mixture caused a slight increase in the gradient of the best straight line through the data (Figure 7D), suggesting that there may be very weak hetero-oligomerization of these proteins (although much weaker than s-erbB4 homo-oligomerization). The data obtained with a s-erbB3:s-erbB4 mixture (Figure 7E) are most consistent with NRG1-β1 inducing independent homo-oligomerization of s-erbB4, with no effect on s-erbB3 (as seen for NRG1-β1 addition to a s-erbB1/s-erbB4 mixture) and therefore do not suggest a hetero-oligomerization event.
Fig. 7. Plots of ln(Abs) against (_r_2 – _r_o2)/2 for different pairwise mixtures of s-erbB2, s-erbB3 and s-erbB4 with (open symbols) and without (filled symbols) added NRG1-β1. (A) The increase in gradient of the ln(Abs) against (_r_2 – _r_o2)/2 plot that results from NRG1-β1-induced homo-oligomerization of s-erbB4. Lines corresponding to these data are superimposed in gray on each other graph in the figure. (B and C) NRG1-β1 fails to induce homo-oligomerization of s-erbB2 or s-erbB3. The data in (D) suggest that s-erbB2 and s-erbB3 may form very weak hetero-oligomers upon NRG1-β1 addition. As seen for s-erbB1 and s-erbB2 in Figure 5B, the data in (E) argue that s-erbB3 does not form hetero-oligomers with s-erbB4. The correspondence (F) of the line for the s-erbB2/s-erbB4 + NRG1-β1 sample with that for NRG1-β1-induced s-erbB4 oligomers shown in (A) indicates that NRG1-β1 can induce formation of s-erbB2–s-erbB4 hetero-oligomers (see text for details). Experiments were performed with a total s-erbB concentration of 10 µM, to which was added a 2-fold molar excess of NRG1-β1.
Figure 7F shows the most interesting of these results, and represents the only data in this study that argue for ligand-induced s-erbB hetero-oligomerization. In the absence of NRG1-β1, sedimentation of the s-erbB2:s-erbB4 mixture is indistinguishable from that of unliganded s-erbB4. When NRG1-β1 is added, sedimentation of the s-erbB2:s-erbB4 mixture is almost identical to that seen with s-erbB4 alone (at the same total s-erbB concentration). This argues that NRG1-β1 addition induces the same increase in average molecular mass regardless of whether all of the s-erbB molecules in the sample are s-erbB4, or half of them are s-erbB2. There are two possible explanations for this. One is that NRG1-β1 can induce homo-oligomerization of s-erbB2 (as well as that of s-erbB4), which Figure 7B shows to be false. The other explanation is that hetero-oligomers containing s-erbB2 plus s-erbB4 are induced by NRG1-β1 with an efficiency similar to s-erbB4 homo-oligomerization. Independent MALLS studies (not shown) also showed that the addition of 1.5-fold molar excess of NRG1-β1 induces the same increase in weight-averaged molecular mass for a 1:1 s-erbB2:s-erbB4 mixture as it does for a solution of s-erbB4 alone, again suggesting NRG1-β1-induced s-erbB2–s-erbB4 hetero-oligomerization.
Discussion
Using analytical ultracentrifugation and MALLS, we have shown that EGF induces efficient homodimerization of the EGF receptor extracellular domain (s-erbB1), but does not induce formation of any detectable hetero-oligomers (or other homo-oligomers) of erbB receptor extracellular domains. Similar studies with NRG1-β1 showed that this ligand induces efficient homo-oligomerization of the erbB4 extracellular domain (s-erbB4), but no other s-erbB homo-oligomers. The s-erbB4 oligomers induced by NRG1-β1 appear to be larger than dimers, although we have not yet established their maximum size. As well as inducing s-erbB4 homo-oligomerization, NRG1-β1 appears to stabilize the formation of hetero-oligomers containing both s-erbB4 and s-erbB2. The qualitative results of our studies are summarized in Table II.
Table II. Summary of ligand-induced s-erbB oligomers observed.
| | s-erbB1 | s-erbB2 | s-erbB3 | s-erbB4 | | | | | | | ---------- | ------- | ------- | ------- | --- | ------- | ------------- | ------- | ------ | | | EGF | NRG1-β1 | EGF | NRG1-β1 | EGF | NRG1-β1 | EGF | NRG1-β1 | | | s-erbB1 | homo | – | – | – | – | – | – | – | | s-erbB2 | | | – | – | – | hetero (weak) | – | hetero | | s-erbB3 | | | | | – | – | – | – | | s-erbB4 | | | | | | | – | homo |
Comparisons with previous studies
The _K_D value reported in Table I for EGF binding by s-erbB1 (118 nM) is comparable with values previously reported (100–500 nM) for EGF binding by monomeric s-erbB1 (Greenfield et al., 1989; Günther et al., 1990; Hurwitz et al., 1991; Lax et al., 1991; Zhou et al., 1993; Brown et al., 1994; Lemmon et al., 1997). However, the data in Figure 3A suggest that the s-erbB1 used here dimerizes at least 15-fold more strongly upon EGF binding than material used in our earlier studies. Whereas the _K_D for dimerization of a 1:1 EGF:s-erbB1 complex was estimated previously as 3.3 µM (Lemmon et al., 1997), in which case it would be <50% dimeric in Figure 3A, the protein used in this study remained completely dimeric at concentrations as low as 250 nM (not shown). This difference may reflect the fact that, rather than using chaotropes to elute the protein from immunoaffinity columns, s-erbB1 produced for this study was purified under milder conditions, using metal affinity chromatography (see Materials and methods).
The _K_D value reported for s-erbB3 binding to the EGF domain of NRG1-β1 (249 nM; Table I) is ∼10-fold weaker than the value reported for its binding to full-length NRG1-β2 in analytical ultracentrifugation studies (Horan et al., 1995). This difference may reflect the use of alternative NRG1-β isoforms in the two studies or, more likely, a contribution to s-erbB3 binding by regions of full-length NRG1-β2 outside the EGF domain (although the EGF domain is sufficient for all known biological activities of NRG1; Holmes et al., 1992). In agreement with our findings (Figures 4C and 7), Horan et al. (1995) did not detect s-erbB3 homodimerization or s-erbB2–s-erbB3 heterodimerization upon NRG1-β2 binding.
Implications for erbB receptor oligomerization
As stated in the Introduction, we set out to test the hypothesis that the mechanism of erbB receptor transmodulation involves simple formation of receptor heterodimers upon binding to one or another bivalent ligand (Alroy and Yarden, 1997; Lemmon et al., 1997; Tzahar et al., 1997). We found that, in common with almost every other RTK extracellular domain that has been studied (Lemmon and Schlessinger, 1994; Heldin, 1995), the erbB1 and erbB4 extracellular domains form homo-oligomers upon binding to their respective ligands (EGF and NRG1-β1). As with other well characterized examples, this homo-oligomerization may be driven by bivalent erbB ligand binding. However, we could only detect the formation of one of the six possible pairwise s-erbB hetero-oligomers; s-erbB4 forming co-oligomers with s-erbB2 upon NRG1-β1 binding. EGF did not induce any s-erbB oligomer other than s-erbB1 homodimers, and our data suggest that the one hetero-oligomer that we could detect (s-erbB2–s-erbB4) is likely to be larger than a dimer.
These observations suggest that the simple erbB receptor heterodimerization hypothesis, in which ligand binding drives the heteromeric association of two different erbB receptors through their extracellular ligand-binding domains, is false. Instead, our findings argue that the mechanisms of ligand-induced erbB receptor homo- and hetero-oligomerization must be fundamentally different. In particular, the fact that ligand-induced erbB1 and erbB4 homo-oligomerization can be recapitulated with the isolated extracellular domains of these receptors, while hetero-oligomerization cannot, suggests that regions outside the extracellular domain are required for heteromeric, but not homomeric, interactions of the intact forms of these receptors.
A model for ‘homodimer-nucleated’ erbB receptor transmodulation
There are ∼104–105 erbB1 or erbB4 receptors on the surface of a typical EGF- or NRG-responsive cell. For a cell with a radius of 8 µm, this receptor density translates to an effective local concentration of 0.1–3 µM at the very least. More reasonable estimates that account for orientation effects would be 10–100 times higher (Grasberger et al., 1986). All experiments presented herein were performed with s-erbB proteins at concentrations of 4–10 µM, mimicking the effective erbB receptor concentration at the cell surface. Since liganded s-erbB1 and s-erbB4 homo-oligomerize so strongly under these conditions, we suggest that homo-oligomerization of the intact membrane-anchored receptors is likely to be the first response to ligand binding in vivo. It seems unlikely that ligand-induced hetero-oligomerization events that we cannot detect in the studies described here (driven by regions outside the extracellular domains) would compete with these strong, directly ligand-induced, homomeric interactions. We therefore suggest that the ligand-induced erbB receptor hetero-oligomers seen in many studies of intact erbB receptors are ‘nucleated’ by ligand-induced erbB1 or erbB4 homo-oligomers, and most probably represent something larger than a heterodimer. Huang et al. (1998) have suggested a similar model, as outlined in the Introduction, in which a ligand-induced homodimer of one receptor (e.g. erbB1) transactivates a second receptor (e.g. erbB2) by inducing its dimerization. In the resulting heterotetramer, the two molecules of the second (unliganded) receptor could activate one another through trans-autophosphorylation, and may be identical or different [if different, the ‘secondary dimerization’ observations made by Gamett et al. (1997) could be explained]. A ‘homodimer-nucleated’ hetero-tetramer model of this sort could explain the initially surprising finding that a kinase-negative mutant of erbB1 is nonetheless able to mediate EGF-induced transmodulation of erbB2 (Wright et al., 1995). According to the model, an EGF-induced homodimer of the erbB1 mutant would transactivate erbB2 by inducing erbB2 homodimerization (and consequent activation) within the context of a heterotetramer—the kinase activity of erbB1 would not be required. The model could also explain how an erbB2 mutant with its intracellular domain deleted can inhibit transmodulation of endogenous erbB2 in a dominant-negative manner (Jones and Stern, 1999).
While this homodimer-nucleated heterotetramer model may explain transmodulation mediated by erbB1 or erbB2, it cannot readily explain the formation of erbB2–erbB3 hetero-oligomers. We and others (Horan et al., 1995; Tzahar et al., 1997) have failed to detect NRG-induced homodimerization of the erbB3 extracellular domain using biophysical or cross-linking methods. However, NRG-induced homo-oligomerization of intact (or truncated) erbB3 in cells has been detected in chemical cross-linking studies (Sliwkowski et al., 1994; Tzahar et al., 1997). Unlike erbB1 or erbB4, erbB3 appears to require more than just the extracellular domain for its ligand-induced homo-oligomerization. Tzahar et al. (1997) have presented evidence suggesting that transmembrane domain interactions may be important for both homo- and hetero-oligomeric interactions of erbB3. An NRG-induced erbB3 oligomer, stabilized by such interactions, could transmodulate erbB2 by inducing its ‘proxy’ dimerization in the model discussed above (see also Huang et al., 1998).
Relationship of hetero-oligomer formation to ligand binding
Despite the fact that it does not bind either ligand independently, overexpression of erbB2 increases the NRG-binding affinity of cells that express erbB3 (Sliwkowski et al., 1994; Karunagaran et al., 1996) and the EGF-binding affinity of cells that express erbB1 (Karunagaran et al., 1996). In an effort to understand these effects, Sliwkowski and colleagues investigated how forced heterodimerization of erbB receptor extracellular domains alters their ligand-binding properties. Hetero- (and homo-) dimerization was forced by fusing erbB receptor extracellular domains to the (dimeric) hinge and Fc portions of IgG1 heavy chain. Heterodimeric IgG fusions containing the erbB2 extracellular domain alongside that of erbB3 or erbB4 bound NRG1-β significantly more strongly than erbB3 or erbB4 homodimer fusion proteins (Fitzpatrick et al., 1998; J.T.Jones et al., 1999). In contrast, a heterodimer containing the extracellular domains of erbB2 and erbB1 was indistinguishable from the equivalent erbB1 homodimer in its binding to EGF, TGF-α, HB-EGF or betacellulin (J.T.Jones et al., 1999). This difference suggests that erbB2 enhances NRG and EGF binding through distinct mechanisms. While NRG binding may be enhanced simply by receptor extracellular domain heteromerization, some other mechanism must be invoked for the enhancement of cellular EGF binding by overexpression of erbB2 (Karunagaran et al., 1996). Our studies of s-erbB oligomerization suggest a similar distinction: while the isolated extracellular domains cannot recapitulate ligand-induced erbB1–erbB2 hetero-oligomerization, at least NRG-induced erbB2–erbB4 heteromerization could be reproduced with the soluble s-erbB proteins studied here.
Conclusions
Regardless of the precise mechanism of ligand-induced erbB receptor hetero-oligomerization, the results presented here show that isolated extracellular domains reproduce ligand-induced homomeric interactions of erbB receptors more faithfully than their reported heteromeric interactions. This finding alone argues that the mechanisms for homo- and hetero-oligomerization of the erbB receptors must differ. Our data therefore provide strong evidence against the simple heterodimerization hypothesis that we set out to test. Rather, in agreement with suggestions made by other groups (Gamett et al., 1997; Huang et al., 1998), we suggest that the ligand-induced erbB homo-oligomers that can be formed with isolated extracellular domains nucleate larger erbB hetero-oligomers through interactions that may also involve other regions of the receptor. Transphosphorylation within these larger ‘homodimer-nucleated’ hetero-oligomers may be responsible for erbB receptor transmodulation.
Materials and methods
Generation of s-erbB constructs
A fragment of human erbB1 cDNA directing expression of residues 1–642 (1–618 of the mature sequence), followed by a hexahistidine tag and stop codon, was subcloned into pFastBac1 (Life Technologies Inc). The 1955 bp fragment was generated by PCR, introducing a unique _Bgl_II site immediately before the initiation codon and a unique _Xba_I site that follows the introduced stop codon. The 1955 bp _Bgl_II–_Xba_I-digested PCR product was ligated into _Bam_HI–_Xba_I-digested pFastBac I. To minimize the risk of PCR artifacts, a 1260 bp _Eco_RI–_Apa_I fragment of this PCR-derived clone was swapped for the equivalent region from the original erbB1 cDNA. A fragment of human erbB2 cDNA, directing expression of residues 1–647 (1–628 of the mature sequence), was generated similarly. In this case, a unique _Xba_I site was introduced before the initiation codon, and a unique _Hin_dIII site was introduced after the histidine tag and stop codon. The 1980 bp _Xba_I–_Hin_dIII-digested PCR product was ligated into _Xba_I–_Hin_dIII-digested pFastBac I. An 1880 bp internal fragment of this PCR product, extending from an _Nco_I site at the initiation codon to a unique _Sph_I site, was then swapped for the equivalent fragment from the original erbB2 cDNA.
Fragments encoding human erbB3 residues 1–639 (1–620 of the mature protein) and human erbB4 residues 1–649 (1–624 of the mature protein), with a unique _Bam_HI site at one end and an _Xba_I site at the other, were generated by PCR, and ligated into _Bam_HI–_Xba_I-digested pFastBac I. The sequence of all PCR-derived fragments and their cloning boundaries were confirmed by automated dideoxynucleotide sequencing methods.
Protein production
Typically, 5–10 l of Sf9 cells were grown as a suspension culture in Sf900-II medium (Gibco-BRL) using multiple 1 l spinner flasks that each contained <500 ml of medium (to ensure adequate aeration). At a cell density of 2.5 × 106 cells/ml (viability >98%), freshly amplified high-titer virus stock was added to a multiplicity of infection (m.o.i.) of ∼5. Cultures were incubated at 27°C for a further 96 h. Clarified conditioned medium was concentrated 2-fold, and then diafiltered against 3.5 vols of 25 mM Tris–HCl, 150 mM NaCl, pH 8.0 (buffer A), using a Millipore Prep/Scale-TFF 30 kDa cartridge. The solution was concentrated further to ∼300 ml prior to loading onto a 5 ml Ni-NTA Superflow column (Qiagen). After extensive washing with buffer A, the column was washed sequentially with two column volumes of buffer A containing 30, 50, 75, 100 and 300 mM imidazole, pH 8.0. Typically, most s-erbB protein eluted in the 75 and 100 mM fractions. Fractions were concentrated in a Centriprep 30 (Amicon), and loaded onto a Pharmacia Superose 6 gel filtration column in 25 mM HEPES pH 8.0, 100 mM NaCl, from which they eluted as ∼85 kDa species. For s-erbB1 and s-erbB4, appropriate gel filtration fractions were pooled, diluted 1.5-fold with 50 mM MES pH 6.0, and were loaded on to an BioScale-S2 cation exchange column (Bio-Rad) pre-equilibrated with 25 mM MES pH 6.0. Protein was eluted with a gradient in NaCl, s-erbB1 eluting at ∼200 mM NaCl and s-erbB4 at ∼300 mM NaCl. Attempts to purify s-erbB2 and s-erbB3 by ion exchange led to precipitation of the proteins at the low salt concentration required for column binding. Purified s-erbB proteins were buffer exchanged into 25 mM HEPES, 100 mM NaCl, pH 8.0, concentrated to between 20 and 100 µM, and stored at 4°C. Purity was checked by SDS–PAGE (Figure 1), and concentrations were determined by absorbance at 280 nM using extinction coefficients calculated as described (Mach et al., 1992) of 56 920/M/cm (s-erbB1), 62 460/M/cm (s-erbB2), 63 940/M/cm (s-erbB3) and 74 300/M/cm (s-erbB4). We previously had used quantitative amino acid analysis to measure a value of 58 500/M/cm for s-erbB1 from mammalian cells (Lemmon et al., 1997); this value is within 3% of that calculated according to Mach et al. (1992). Calculated extinction coefficients of 18 780/M/cm (EGF) and 5920/M/cm (NRG1-β1) were also used for determination of ligand concentration.
Approximate final yields of purified protein from 1 l of conditioned medium were 1 (s-erbB1), 0.2 (s-erbB2), 1 (s-erbB3) and 0.5 mg (s-erbB4). Ligands used for this study were purchased from Intergen (human EGF) or R & D Systems (human NRG1-β1).
Multi-angle laser light-scattering (MALLS) studies
A DAWN DSP laser photometer from Wyatt Technologies (Santa Barbara, CA) was used for MALLS studies (Wyatt, 1993). The instrument was used in micro-batch mode, with samples being introduced into the flow cell via a 0.1 µm filter using a syringe pump. To avoid introduction of air bubbles, concentrated protein solutions were diluted to working concentrations in degassed buffer, and samples were introduced into the flow cell via a low dead volume multi-port valve that was loaded with several samples and purged of air prior to a series of measurements. Scattering data at all 17 angles were collected until maximum stable scattering for a sample was seen, which can be achieved at flow rates of 2 ml/h with samples of ∼300 µl. Scattering data were collected and analyzed using ASTRA software (Wyatt Technologies) supplied with the instrument. Relative weight-averaged molecular masses were determined from the scattering data collected for a given ligand:receptor mixture (once stabilized) using Debye plots, in which R(θ)/K*c is plotted against sin2(θ/2), where θ is the scattering angle; R(θ) is the excess intensity (I) of scattered light at that angle; c is the concentration of the sample; and K* is a constant equal to 4π2_n_2(d_n/d_c)2/λ04_N_A (where n = solvent refractive index, d_n/d_c = refractive index increment of scattering sample, λ0 = wavelength of scattered light and _N_A = Avogadro’s number). Extrapolation of a Debye plot to zero angle gives an estimate of the weight-averaged molecular mass (_M̄_w) (Wyatt, 1993). _M̄_w is defined as:
for n moles of i different species with molecular weight Mi.
In ligand titration experiments, the contribution of added ligand to the mass concentration was neglected (see also Lemmon et al., 1997). Since we are interested in dimerization, i.e. only the ‘fold increase’ in _M̄_w, our results are not affected by the value of K*, of which we are uncertain since we have not determined the extent of glycosylation of the s-erbB proteins accurately. MALLS data are therefore discussed in terms of ‘fold increase’ in _M̄_w over that measured for s-erbB protein alone. Where estimates for _M̄_w are reported, mass concentrations were converted from molar concentrations using the molecular weight suggested by the amino acid sequence, and assuming that s-erbB glycoproteins are 20% carbohydrate by mass.
Analytical ultracentrifugation studies
Sedimentation equilibrium experiments employed the XL-A analytical ultracentrifuge (Beckman). Samples were loaded into six-channel epon charcoal-filled centerpieces, using quartz windows. Experiments were performed at 20°C, detecting at 280–300 nm, using three different speeds (6000, 9000 and 12 000 r.p.m.), with very similar results. Solvent density was taken as 1.003 g/ml, and the partial specific volumes of the s-erbB proteins were approximated from their amino acid compositions and the assumption of ∼20% carbohydrate as 0.71 ml/g for the purposes described here. Experiments were performed at 5–10 µM protein. Data were fit using the Optima XL-A data analysis software (Beckman/MicroCal) to models assuming a single non-ideal species for unliganded s-erbB proteins. When ligand was added, a two-species fit was used, in which one of the species was the excess ligand (partial specific volume 0.74 ml/g), which sediments as a 6 kDa (EGF) or 8 kDa (NRG) species (not shown). The molecular mass of the ligand species was fixed in these fits, while the mass and concentration of the receptor species were allowed to float. Goodness of fit was judged by the occurrence of randomly distributed residuals, examples of which are shown in Figure 4. For more complicated mixtures of receptors and ligands, simple qualitative interpretations of analytical ultracentrifugation experiments were made by inspection when possible (see Figures 5B, 6 and 7).
BIAcore studies
BIAcore binding experiments employed a BIAcore 2000 instrument, and were performed in 10 mM HEPES buffer, pH 7.4, that contained 150 mM NaCl, 3.4 mM EDTA and 0.005% Tween-20 at 25°C. The hydrogel matrix of BIAcore CM5 Biosensor chips was activated with _N_-hydroxysuccinimide (NHS) and _N_-ethyl-_N_′-[3-(diethylamino)propyl] carbodiimide (EDC). EGF (at 200 µg/ml) in 10 mM sodium acetate, pH 4.0, or NRG1-β1 (at 200 µg/ml) in 10 mM sodium acetate, pH 4.8, was then flowed over the activated surface at 5 µl/min for 10 min. Non-cross-linked ligand was removed, and unreacted sites were blocked with 1 M ethanolamine, pH 8.5. The signal contributed by immobilized EGF or NRG1-β1 ranged from 150 to 400 RU, depending on the specific chip.
Purified s-erbB proteins at a series of concentrations were each flowed simultaneously over the EGF and NRG1-β1 (and mock/control) surfaces at 5 µl/min for 7 min, by which time binding had reached a plateau in each case. The RU value corresponding to this plateau was taken as a measure of s-erbB protein binding, and was corrected for background non-specific binding and bulk refractive index effects by subtraction of data obtained in parallel using the mock-coupled hydrogel surface. RU values were then converted into percentage maximal binding. This conversion was performed separately for each surface (since levels of immobilization varied); 100% binding was defined for an NRG surface as the highest corrected signal seen with s-erbB3 and s-erbB4 (which were always the same to within 10%), and for an EGF surface the highest corrected signal seen with s-erbB1. Buffer washes between runs were sufficient to bring the RU value back down to baseline. Data were plotted as s-erbB concentration against percentage maximal binding, and fit to a simple binding equation in ORIGIN (MicroCal) to estimate the _K_D.
Acknowledgments
Acknowledgements
We thank Irit Lax, Yossi Schlessinger, Greg Van Duyne and members of the Lemmon laboratory for many valuable discussions and comments on the manuscript. BIAcore experiments were performed in the Biosensor Interaction Analysis Core at Penn. This work was supported by grants from the National Institutes of Health (CA79992 to M.A.L.), the US Army Breast Cancer Research Program (DAMD17-98-1-8232 to M.A.L., and DAMD17-98-1-8228 to K.M.F.), a Scholar Award from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation (to M.A.L.) and the Research Foundation of the University of Pennsylvania.
References
- Alroy I. and Yarden,Y. (1997) The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand–receptor interactions. FEBS Lett., 410, 83–86. [DOI] [PubMed] [Google Scholar]
- Brown P.M., Debanne,M.T., Grothe,S., Bergsma,D., Caron,M., Kay,C. and O’Connor-McCourt,M.D. (1994) The extracellular domain of the epidermal growth factor receptor. Studies on the affinity and stoichiometry of binding, receptor dimerization and a binding-domain mutant. Eur. J. Biochem., 225, 223–233. [DOI] [PubMed] [Google Scholar]
- Burden S. and Yarden,Y. (1997) Neuregulins and their receptors: a versatile signaling module in organogenesis and oncogenesis. Neuron, 18, 847–855. [DOI] [PubMed] [Google Scholar]
- Cantor C.R. and Schimmel,P.R. (1980) Biophysical Chemistry: Techniques for the Study of Biological Structure and Function. W.H.Freeman and Co., New York. [Google Scholar]
- Carraway K.L., III and Cantley,L.C. (1994) A Neu acquaintance for erbB3 and erbB4: a role for receptor heterodimerization in growth signaling. Cell, 78, 5–8. [DOI] [PubMed] [Google Scholar]
- Cohen B.D., Green,J.M., Foy,L. and Fell,H.P. (1996) HER4-mediated biological and biochemical properties in NIH 3T3 cells. Evidence for HER1–HER4 heterodimers. J. Biol. Chem., 271, 4813–4818. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick V.D., Pisacane,P.I., Vandlen,R.L. and Sliwkowski,M.X. (1998) Formation of a high-affinity heregulin binding site using the soluble extracellular domains of ErbB2 with ErbB3 or ErbB4. FEBS Lett., 431, 102–106. [DOI] [PubMed] [Google Scholar]
- Gamett D.C., Pearson,G., Cerione,R.A. and Friedberg,I. (1997) Secondary dimerization between members of the epidermal growth factor receptor family. J. Biol. Chem., 272, 12052–12056. [DOI] [PubMed] [Google Scholar]
- Goldman R., Levy,R.B., Peles,E. and Yarden,Y. (1990) Heterodimerization of the erbB-1 and erbB-2 receptors in human breast carcinoma cells: a mechanism for receptor transregulation. Biochemistry, 29, 11024–11028. [DOI] [PubMed] [Google Scholar]
- Grasberger B., Minton,A.P., DeLisi,C. and Metzger,H. (1986) Interaction between proteins localized in membranes. Proc. Natl Acad. Sci. USA, 83, 6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graus-Porta D., Beerli,R.R., Daly,J.M. and Hynes,N.E. (1997) ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J., 16, 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield C., Hiles,I., Waterfield,M.D., Federwisch,M., Wollmer,A., Blundell,T.L. and McDonald,N. (1989) Epidermal growth factor binding induces a conformational change in the external domain of its receptor. EMBO J., 8, 4115–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther N., Betzel,C. and Weber,W. (1990) The secreted form of the epidermal growth factor receptor: characterization and crystallization of the receptor–ligand complex. J. Biol. Chem., 265, 22082–22085. [PubMed] [Google Scholar]
- Harari D., Tzahar,E., Romano,J., Shelly,M., Pierce,J.H., Andrews,G.C. and Yarden Y. (1999) Neuregulin-4: a novel growth factor that acts through the ErbB-4 receptor tyrosine kinase. Oncogene, 18, 2681–2689. [DOI] [PubMed] [Google Scholar]
- Heldin C.-H. (1995) Dimerization of cell surface receptors in signal transduction. Cell, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- Holmes W.E.S. et al. (1992) Identification of heregulin, a specific activator of p185erbB2. Science, 256, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Honegger A.M., Schmidt,A., Ullrich,A. and Schlessinger,J. (1990) Evidence for epidermal growth factor (EGF)-induced intermolecular autophosphorylation in living cells. Mol. Cell. Biol., 10, 4035–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan T., Wen,J., Arakawa,T., Liu,N., Brankow,D., Hu,S., Ratzkin,B. and Philo,J.S. (1995) Binding of Neu differentiation factor with the extracellular domain of Her2 and Her3. J. Biol. Chem., 270, 24604–24608. [DOI] [PubMed] [Google Scholar]
- Huang G.C., Ouyang,X. and Epstein,R.J. (1998) Proxy activation of protein erbB2 by heterologous ligands implies a heterotetrameric mode of receptor tyrosine kinase interaction. Biochem. J., 331, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S.R., Mohammadi,M. and Schlessinger,J. (1998) Autoregulatory mechanisms in protein-tyrosine kinases. J. Biol. Chem., 273, 11987–11990. [DOI] [PubMed] [Google Scholar]
- Hurwitz D.R., Emanuel,S.L., Nathan,M.H., Sarver,N., Ullrich,A., Felder,S., Lax,I. and Schlessinger,J. (1991) EGF induces increased ligand binding affinity and dimerization of soluble epidermal growth factor (EGF) receptor extracellular domain. J. Biol. Chem., 266, 22035–22043. [PubMed] [Google Scholar]
- Jones F.E. and Stern,D.F. (1999) Expression of dominant-negative ErbB2 in the mammary gland of transgenic mice reveals a role in lobuloalveolar development and lactation. Oncogene, 18, 3481–3490. [DOI] [PubMed] [Google Scholar]
- Jones F.E., Welte,T., Fu,X.Y. and Stern,D.F. (1999) ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol., 147, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.T., Akita,R.W. and Sliwkowski,M.X. (1999) Binding specificities of egf domains for ErbB receptors. FEBS Lett., 447, 227–231. [DOI] [PubMed] [Google Scholar]
- Karunagaran D., Tzahar,E., Beerli,R.R., Chen,X., Graus-Porta,D., Ratzkin,B.J., Seger,R., Hynes,N.E. and Yarden,Y. (1996) ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: implications for breast cancer. EMBO J., 15, 254–264. [PMC free article] [PubMed] [Google Scholar]
- King C.R., Borrello,I., Bellot,F., Comoglio,P. and Schlessinger,J. (1988) EGF binding to its receptor triggers a rapid tyrosine phosphorylation of the erbB2 protein in the mammary tumor cell line SKBR-3. EMBO J., 7, 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax I., Mitra,A.K., Ravera,C., Hurwitz,D.R., Rubinstein,M., Ullrich,A., Stroud,R.M. and Schlessinger,J. (1991) Epidermal growth factor (EGF) induces oligomerization of soluble, extracellular, ligand-binding domain of EGF receptor. J. Biol. Chem., 266, 13828–13833. [PubMed] [Google Scholar]
- Lemmon M.A. and Schlessinger,J. (1994) Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci., 19, 459–463. [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., Bu,Z., Ladbury,J.E., Zhou,M., Pinchasi,D., Lax,I., Engelman,D.M. and Schlessinger,J. (1997) Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J., 16, 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach H., Middaugh,C.R. and Lewis,R.V. (1992) Statistical determination of the average values of the extinction coefficient of tryptophan and tyrosine in native proteins. Anal. Biochem., 200, 74–80. [DOI] [PubMed] [Google Scholar]
- Qian X., LeVea,C.M., Freeman,J.K., Dougall,W.C. and Greene,M.I. (1994) Heterodimerization of epidermal growth factor receptor and wild-type or kinase-deficient Neu: a mechanism of interreceptor kinase activation and transphosphorylation. Proc. Natl Acad. Sci. USA, 91, 1500–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese D.J. II and Stern,D.F. (1998) Specificity within the EGF family/ErbB receptor family signaling network. BioEssays, 20, 41–48. [DOI] [PubMed] [Google Scholar]
- Riese D.J. II, van Raaij,T.M., Plowman,G.D., Andrews,G.C. and Stern,D.F. (1995) The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol. Cell. Biol., 15, 5770–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese D.J. II, Kim,E.D., Elenius,K., Buckley,S., Klagsbrun,M., Plowman,G.D. and Stern,D.F. (1996) The epidermal growth factor receptor couples transforming growth factor-α, heparin-binding epidermal growth factor-like factor and amphiregulin to Neu, ErbB-3 and ErbB-4. J. Biol. Chem., 271, 20047–20052. [DOI] [PubMed] [Google Scholar]
- Schechter Y., Hernaez,L., Schlessinger,J. and Cuatrecasas,P. (1979) Local aggregation of hormone-receptor complexes is required for activation by epidermal growth factor. Nature, 278, 835–838. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. (1979) Receptor aggregation as a mechanism for transmembrane signaling: Models for hormone action. In De Lisi,C. and Blumenthal,R. (eds), Physical Chemistry of Cell Surface Events in Cellular Regulation. Elsevier, pp. 89–118. [Google Scholar]
- Schlessinger J. (1994) SH2/SH3 signaling proteins. Curr. Opin. Genet. Dev., 4, 25–30. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. and Ullrich,A. (1992) Growth factor signaling by receptor tyrosine kinases. Neuron, 9, 383–391. [DOI] [PubMed] [Google Scholar]
- Sliwkowski M.X. et al. (1994) Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J. Biol. Chem., 269, 14661–14665. [PubMed] [Google Scholar]
- Sliwkowski M.X., Lofgren,J.A., Lewis,G.D., Hotaling,T.E., Fendly,B.M. and Fox,J.A. (1999). Nonclinical studies addressing the mechanism of action of trastuzumab (Herceptin). Semin. Oncol., 26 (Suppl. 12), 60–70. [PubMed] [Google Scholar]
- Songyang Z. et al. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell, 72, 767–778. [DOI] [PubMed] [Google Scholar]
- Spivak-Kroizman T., Rotin,D., Pinchasi,D., Ullrich,A., Schlessinger,J. and Lax,I. (1992) Heterodimerization of c-erbB2 with different epidermal growth factor receptor mutants elicits stimulatory or inhibitory responses. J. Biol. Chem., 267, 8056–8063. [PubMed] [Google Scholar]
- Stern D.F. and Kamps,M.P. (1988) EGF-stimulated tyrosine phosphorylation of p185neu: a potential model for receptor interactions. EMBO J., 7, 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahar E. et al. (1997) Bivalence of EGF-like ligands drives the ErbB signaling network. EMBO J., 16, 4938–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Qian,X. and Greene,M.I. (1990) Intermolecular association of the p185neu protein and EGF modulates EGF receptor function. Cell, 61, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Wright J.D., Reuter,C.W.M. and Weber,M.J. (1995) An incomplete program of cellular tyrosine phosphorylations induced by kinase-defective epidermal growth factor receptors. J. Biol. Chem., 270, 12085–12093. [DOI] [PubMed] [Google Scholar]
- Wyatt P.J. (1993) Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta, 272, 1–40. [Google Scholar]
- Yarden Y. and Schlessinger,J. (1987a) Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry, 26, 1434–1442. [DOI] [PubMed] [Google Scholar]
- Yarden Y. and Schlessinger,J. (1987b) Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry, 26, 1443–1451. [DOI] [PubMed] [Google Scholar]
- Zhang K., Sun,J., Liu,N., Wen,D., Chang,D., Thomason,A. and Yoshinaga,S.K. (1996) Transformation of NIH3T3 cells by HER3 or HER4 receptors requires the presence of HER1 or HER2. J. Biol. Chem., 271, 3884–3890. [PubMed] [Google Scholar]
- Zhou M., Felder,S., Rubinstein,M., Hurwitz,D.R., Ullrich,A., Lax,I. and Schlessinger,J. (1993) Real-time measurements of kinetics of EGF binding to soluble EGF receptor monomers and dimers support the dimerization model for receptor activation. Biochemistry, 32, 8193–8198. [DOI] [PubMed] [Google Scholar]