15q13.3 microdeletions increase risk of idiopathic generalized epilepsy (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 25.
Published in final edited form as: Nat Genet. 2009 Jan 11;41(2):160–162. doi: 10.1038/ng.292
Abstract
We identified 15q13.3 microdeletions encompassing the CHRNA7 gene in 12 of 1,223 individuals with idiopathic generalized epilepsy (IGE), which were not detected in 3,699 controls (joint P = 5.32 × 10−8). Most deletion carriers showed common IGE syndromes without other features previously associated with 15q13.3 microdeletions, such as intellectual disability, autism or schizophrenia. Our results indicate that 15q13.3 microdeletions constitute the most prevalent risk factor for common epilepsies identified to date.
Idiopathic generalized epilepsies (IGE) are common seizure disorders accounting for up to one-third of all epilepsies1. The vast majority of individuals with IGE have a complex genetic etiology2, for which the underlying genetic alterations remain largely unknown. Recently, a 15q13.3 microdeletion syndrome has been identified in 0.2–0.3% of individuals with mental retardation and epilepsy3, schizophrenia4,5, autism and other neuropsychiatric features6. The critical region of the 1.5-Mb deletion on 15q13.3 contains at least seven genes, including the CHRNA7 gene coding for the α7 subunit of the nicotinergic acetylcholine receptor, which is considered a plausible candidate gene for the epilepsy phenotype.
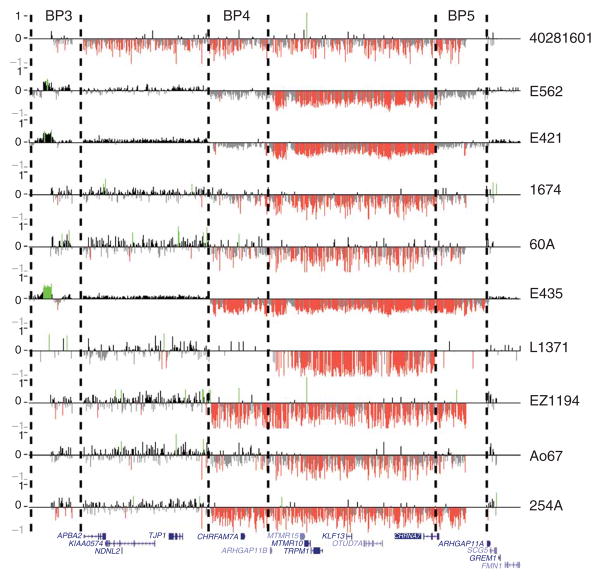
Susceptibility loci for common idiopathic epilepsies, comprising benign epilepsy of childhood with centrotemporal spikes7 and common IGE syndromes8,9, have also been mapped to the 15q13–q14 region. To test whether the 15q13.3 deletion increases risk of common epilepsies, we screened for structural variants within the 15q13.3 region in two independent samples of individuals with IGE and ancestrally matched controls. The first sample comprised 647 unrelated IGE cases of Western European ancestry (EPICURE sample) and 1,202 German controls (PopGen) genotyped using the Affymetrix Genome-Wide Human SNP array 6.0. We identified the 15q13.3 microdeletion in 7 of 647 IGE cases (Supplementary Fig. 1 online) with different IGE syndromes (Table 1). All but one of these showed segmental breakpoints BP4 and BP5, as observed in the 15q13.3 microdeletion syndrome4. The other showed a 3.8-Mb deletion defined by BP3 and BP5. The 15q13.3 deletion was not detected in any of 1,202 controls examined.
Table 1.
Phenotypic features of individuals with 15q13.3 microdeletions
| Individual | Descent | Diagnosis | Seizure types | Age of onset | EEG | Cognition |
|---|---|---|---|---|---|---|
| EPICURE sample | ||||||
| Ao67 | German | JME | MyoclonusGTCS | 16 y19 y | GSW | Normal |
| 1674 | German | CAE | Absence | 5 y | GSW | ID |
| 254A | German | JAE | AbsenceGTCS | 9 y13 y | GSW | Normal |
| 60A | German | JAE | AbsenceGTCS | 12 y14 y | GSW | Normal |
| EZ1194 | Austrian | JME | AbsenceMyoclonusGTCS | 6 y10 y20 y | GSW | Normal |
| 40281601 | German | CAE | AbsenceGTCS | Uncertain3 y | GSW | Normal |
| D04u0213 | Dutch | JME | AbsenceMyoclonusGTCS | 4 y13 y | Irreg. GSWPPR | Normal |
| Mixed IGE sample | ||||||
| E421 | Northern African | JME | MyoclonusGTCS | <26 y26 y | Irreg. GSW | Normal |
| E435 | French | JME | MyoclonusGTCS | 12 y12 y | GSW | Mild deficits |
| L1371 | German | CAE | Absence | 4 y | Irreg. GSW | Normal |
| D07u0771 | Dutch | JAE | Absence | 14 y | GSW | Normal |
| E562 | French | JME | MyoclonusGTCS | 12 y13 y | GSW | Mild ID (IQ = 73) |
| Affected first-degree family members | ||||||
| 2376 (brother of 1674) | German | None | Severe ID | |||
| 254B (sister of 254A) | German | JME | AbsenceMyoclonus | 15 y15 y | GSW | Normal |
| E562M (mother of E562) | French | None | Panic disorder | |||
| E562B (brother of E562) | French | EGTCS | GTCS | Uncertain | Unknown | Normal |
We next examined the frequency of 15q13.3 microdeletions in a second independent sample of 576 IGE cases from Switzerland (n = 205), North America (n = 133) and Northern Europe (n = 238) as well as 2,497 controls from North America of predominantly European ancestry genotyped with various methods (Supplementary Methods online). In this sample, we found 15q13.3 microdeletions in 5 of 576 cases and 0 of 2,497 controls. Altogether, we identified 15q13.3 deletions in 12 of 1,223 IGE cases and in 0 of 3,699 controls (P = 5.32 × 10−8, Fisher’s exact test). We used custom array CGH (10 of 12 cases; Fig. 1) or SNP and CNV arrays (2 of 12 cases) to verify the deletions in all IGE probands and their available first-degree family members. We identified duplications involving CHRNA7 in 12 of 1,223 cases and 23 of 3,699 controls (P = 0.27, Pearson’s _χ_2 test; Supplementary Methods and Supplementary Fig. 2 online). Thus, our results suggest that the 15q13.3 deletion only, and not the reciprocal duplication, represents a major risk factor for IGE.
Figure 1.
Confirmation of 15q13.3 microdeletions using custom array CGH. High-resolution oligonucleotide array mapping of the 15q12–q13.3 region in 10 of 12 IGE probands with 15q13.3 microdeletions. Probes with log2 ratios above or below a threshold of 1.5 s.d. are colored green (duplications) or red (deletions). Hashed lines indicate the breakpoint regions BP3–BP5.
In our study, parental DNA was available for 5 of 12 probands with the deletion (Supplementary Fig. 3 online). In one individual (L1371), the deletion was apparently de novo, as we found that both clinically unaffected parents did not carry the deletion using custom array CGH. For the other four individuals, we observed parental transmission from one father and three mothers. One mother (E562M) suffered from panic disorder, a phenotype previously associated with 15q13.3 deletions6. The other three transmitting parents were apparently clinically unaffected, although we cannot exclude that subclinical manifestations such as age-dependent paroxysmal EEG discharges or undetected mild and remitting IGE phenotypes might have been missed. In one family (E562), both siblings carried the deletion and were affected by IGE. In family 254, the proband’s brother (254B) also carried the deletion and was affected by IGE. In the family of IGE proband 1674, the 15q13.3 microdeletion was present in a brother (2376) with severe intellectual disability but without a history of seizures.
The phenotypes of most of the individuals reported here with the 15q13.3 microdeletion differ notably from those reported originally for the 15q13.3 microdeletion syndrome, in which individuals showed marked intellectual disability, seizures, growth retardation and dysmorphic features3. Although we observed severe intellectual disability in 1 of 12 and mild intellectual disability in 2 of 12 probands, 9 of 12 of the probands in our sample had characteristic features of IGE without dysmorphic features or intellectual disability (Table 1). Seizure types in these individuals included typical absence seizures, myoclonic seizures and primary generalized tonic-clonic seizures, all of them occurring at the typical age of onset. Electroencephalographic (EEG) records were available for all individuals and showed normal background activity with paroxysmal generalized spike-wave discharges, which represent the EEG hallmark of IGE.
Two studies have reported association of the 15q13.3 microdeletion with schizophrenia and related psychoses4,5. Consistent with our study, those studies observed intellectual disability in only a small fraction of individuals carrying the deletion. In the current study, none of the individuals with IGE carrying a 15q13.3 deletion had a history of a psychotic episode. Thus, our findings extend the phenotypic spectrum related to the 15q13.3 deletion to common IGE syndromes without the previous reported neuropsychiatric features. Taken together, the current studies reveal extensive variability in the phenotypic manifestation associated with the 15q13.3 deletion, ranging from apparently healthy individuals to severely affected individuals with a broad spectrum of neuropsychiatric disorders3–6. These findings imply that shared mechanisms are involved in the pathogenesis of a spectrum of seemingly unrelated neuropsychiatric disorders, and argue for a new framework for understanding complex genetic diseases.
The critical region affected by the BP4-BP5 deletions harbors at least seven genes (ARHGAP11B, MTMR15, MTMR10, TRPM1, KLF13, OTUD7A and CHRNA7) that might contribute to the seizure phenotype, including CHRNA7 as the prime candidate gene. Compelling evidence suggests that the impairment of neuronal ion channel function may be pivotal to the pathology of IGE10. Cholinergic pathways have several important functions in the brain, and nicotinergic acetylcholine receptors containing the α7 subunit are widely expressed throughout the central nervous system11. These receptors are localized both pre- and postsynaptically and are thought to modulate excitatory and inhibitory pathways. In particular, CHRNA7 is highly expressed in the reticular thalamus12, indicating a role in modulating thalamo-cortical pathways, which are central to the generation of primarily generalized seizures seen in IGE13. CHRNA2, CHRNA4 and CHRNB2, which code for the α2, α4 and β2 subunits of the nicotinergic acetylcholine receptor, respectively, have a causative role in autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE)11. Together, these lines of evidence strongly support an involvement of CHRNA7 in epileptogenesis.
The genetic architecture of common seizure disorders is thought to show a biological continuum ranging from rare monogenic forms to common epilepsies with complex inheritance. Positional cloning of epilepsy genes has been successful in large families with monogenic inheritance, and multiple rare gene variants, including mutations in CACNA1H and EFHC1, have been found in a small proportion of individuals with IGE10. However, common susceptibility alleles have not been identified so far. Here we describe a structural variant that is virtually absent in the general population (<0.02%)4,5 and roughly 50 times more frequent in individuals with IGE in the present study (1%). Furthermore, the frequency of 15q13.3 deletions in IGE seems to be higher than that reported in intellectual disability or schizophrenia (Supplementary Table 1 online). Given the strong epileptogenic effect, the 15q13.3 microdeletion represents the most prevalent major risk factor for IGE identified to date.
Supplementary Material
Supplementary Material
Acknowledgments
We thank all the subjects and their families for participating in this study. We are also grateful to H. Bode for referral of subjects and A. Ackerhans and K. Moldenhauer for database management. This study used samples from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), as well as clinical data. NINDS Repository sample numbers corresponding to the samples used are available upon request. This study was supported by grants from the German Research Foundation (SA434/4-1, T.S., P.N.), the German Federal Ministry of Education and Research (National Genome Research Network, NGFN-2: NeuroNet, NGFNplus: EMINet), the European Community (FP6 Integrated Project EPICURE, LSHM-CT-2006-037315; grant agreement 219250, A.J.S.), the PopGen biobank, the University of Kiel (I.H.), the Danish National Research Foundation (R.S.M.), in part by grants from the NIH (HD043569, E.E.E.), the National Epilepsy Funds (NEF, grant no. 04-08, B.P.C.K., D. Lindhout), the Netherlands Organization for Scientific Research (NOW, grant no. 917.66.315, B.P.C.K., C.d.K.) and the German Research Foundation/German Federal Ministry of Education and Research (DFG/BMBF) excellence cluster “Inflammation at Interfaces” (A.F., M.W., S.S.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Participating centers in the EPICURE Integrated Project are as follows
Department of Neuropediatrics, University Medical Center Schleswig-Holstein (Kiel Campus), Schwanenweg 20, 24105 Kiel, Germany (I.H., H.M., S.v.S., K.L.K., I.S., U.S.). Institute for Clinical Molecular Biology, University Medical Center Schleswig-Holstein (Kiel Campus), Arnold-Heller-Strasse 3, 24105 Kiel, Germany (A.F., M.W., S.S.). Institute of Medical Informatics and Statistics, University Medical Center Schleswig-Holstein (Kiel Campus), Brunswiker Strasse 10, 24105 Kiel, Germany (M.N.). Cologne Center for Genomics, University of Cologne, Zülpicher Strasse 47, 50674 Cologne, Germany (T.S., C.L., P.N.). Max-Delbrück-Center for Molecular Medicine, Robert-Rössle-Strasse 10, 13125 Berlin, Germany (C.L., V.G., T.S.). Department of Epileptology, University of Bonn, Sigmund Freud Strasse 1, 53105 Bonn, Germany (A.A.K.-L., C.E.E.). Department of Neurology, Charité University Medicine, Campus Virchow Clinic, Humboldt University of Berlin, Augustenburger Platz 1, 13353 Berlin, Germany (V.G., B.S., T.S.). Interdisciplinary Epilepsy-Center, Department of Neurology, Philipps University Marburg, 35033 Marburg, Germany (K.M.K., P.S.R., F.R.). Department of Neurology, University of Ulm, Helmholtzstrasse 8/1, 89081 Ulm, Germany (Y.W., H.L.). Department of Clinical Neurology (F.Z.) and Department of Pediatrics and Neonatology (L.U., M.F.), Medical University of Vienna, Währinger Gürtel 18–20, A-1090 Vienna, Austria. Center of Brain Research, Department of Biochemistry and Molecular Biology, Medical University of Vienna, Spitalgasse 4, A-1090 Vienna, Austria (K.F.). Department of Neurology, Danish Epilepsy Centre, Dianalund Kolonivej 1, 4293 Dianalund, Denmark (R.S.M., H.H.). Wilhelm Johannsen Centre for Functional Genome Research, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen N, Denmark (R.S.M.). Netherlands Section Complex Genetics, Department of Medical Genetics, University Medical Center Utrecht, Str. 2.112 Universiteitsweg 100, 3584 CG Utrecht, The Netherlands (B.P.C.K., C.d.K., D. Lindhout). Department of Neurology, Oosterschelde Hospital, P.O. Box 106, 4460 BB Goes, The Netherlands (F.V.). SEIN Epilepsy Institute in The Netherlands, P.O. Box 540, 2130AM Hoofddorp, The Netherlands (G.-J.d.H., D. Lindhout).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
T.S. and E.E.E. initiated and designed the study; I.H., H.M., S.v.S., I.S., A.A.K.-L., V.G., B.S., K.M.K., P.S.R., F.R., Y.W., H.L., F.Z., L.U., K.F., M. Feucht, F.V., G.-J.d.H., R.S.M., H.H., D. Luciano, C.R., D. Lindhout, C.E.E., U.S. and T.S. recruited and phenotyped the EPICURE sample; H.C.M., A.J.S., M.G., M. Fichera, C.B., P.G., P.T., A.M. and E.E.E. recruited and phenotyped the mixed IGE sample; A.F., M.W., M.N. and S.S. recruited and phenotyped the PopGen control sample; I.H., A.F., C.L., K.L.K., I.S., M.W., M.N., P.N. and T.S. performed the CNV analysis on SNP arrays; H.C.M., A.J.S., M. Fichera, C.B. and D. Luciano performed the qPCR screening; H.C.M., M. Fichera, C.B. and D. Luciano performed the screening using Illumina Genotyping BeadChips; H.C.M., A.J.S. and C.B. performed the confirmation using NimbleGen arrays; C.d.K., B.P.C.K. and D. Lindhout performed the confirmation using Illumina CNV BeadChips; I.H., H.C.M., A.J.S., M.G., M. Fichera, A.F., C.d.K., K.L.K., C.R., B.P.C.K., D. Lindhout, E.E.E. and T.S. coordinated the work and prepared the manuscript.
References
- 1.Jallon P, Latour P. Epilepsia. 2005;46(Suppl 9):10–14. doi: 10.1111/j.1528-1167.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 2.Ottman R. Epilepsia. 2005;46(Suppl 10):7–14. doi: 10.1111/j.1528-1167.2005.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp AJ, et al. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Schizophrenia Consortium. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefansson H, et al. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller DT, et al. J Med Genet advance online publication. 2008 September 19; doi: 10.1136/jmg.2008.059907. [DOI] [Google Scholar]
- 7.Neubauer BA, et al. Neurology. 1998;51:1608–1612. doi: 10.1212/wnl.51.6.1608. [DOI] [PubMed] [Google Scholar]
- 8.Elmslie FV, et al. Hum Mol Genet. 1997;6:1329–1334. doi: 10.1093/hmg/6.8.1329. [DOI] [PubMed] [Google Scholar]
- 9.Sander T, et al. Hum Mol Genet. 2000;9:1465–1472. doi: 10.1093/hmg/9.10.1465. [DOI] [PubMed] [Google Scholar]
- 10.Helbig I, Scheffer IE, Mulley JC, Berkovic SF. Lancet Neurol. 2008;7:231–245. doi: 10.1016/S1474-4422(08)70039-5. [DOI] [PubMed] [Google Scholar]
- 11.Steinlein OK, Bertrand D. Biochem Pharmacol. 2008;76:1175–1183. doi: 10.1016/j.bcp.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Breese CR, et al. J Comp Neurol. 1997;387:385–398. doi: 10.1002/(sici)1096-9861(19971027)387:3<385::aid-cne5>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Blumenfeld H. Epilepsia. 2005;46(Suppl 9):21–33. doi: 10.1111/j.1528-1167.2005.00311.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material