A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 30.
Summary
How is chromatin architecture established and what role does it play in activation of transcription? We show that a regulatory locus in yeast (the UASg) bears, in addition to binding sites for the activator Gal4, sites bound by the protein RSC. RSC tightly positions a nucleosome, evidently partially unwound, in a structure that facilitates Gal4 binding to its sites. The complex comprises a barrier that suffices to impose characteristic features of chromatin architecture. Removal of RSC allows ordinary nucleosomes to form more broadly over the UASg, and these nucleosomes compete with (but do not exclude) Gal4 binding to its sites. Taken with our previous work, the results show that both prior to and following induction specific DNA binding proteins are the predominant determinants of chromatin architecture at the GAL1/10 genes. RSC/nucleosome complexes are found scattered throughout the yeast genome. We surmise, also, that Gal4 works in higher eukaryotes despite whatever obstacle broadly positioned nucleosomes present.
Introduction
“Chromatin architecture” refers, generally, to the disposition of nucleosomes along DNA molecules in a population of cells. The classical approach to determine nucleosome positioning is to digest chromatin with micrococcal nuclease (MNase) such that the primary product comprises mononucleosomes, and then to identify the protected DNA fragments. Only nucleosomes (and not, for example, the transcription complex (Bryant et al., 2008)) protect segments of DNA in this assay, and the recovered nucleosomal fragments usually span about 150 bp. When populations of yeast cells are analyzed in this way recurrent features of chromatin architecture are observed at regions in and around promoters (reviewed in (Jiang and Pugh, 2009; Rando and Chang, 2009)). These features include, in addition to nucleosomes positioned more or less at random, “phased” nucleosomes that occupy identical positions on DNA throughout the population; 100–300 bp segments that bear no nucleosomes (called nucleosome-free regions, NFRs); 10–20 bp segments that register as hypersensitive sites (HS s) in nuclease digestion experiments; and, often, nucleosomes containing H2A.Z, a variant of the more common nucleosome subunit H2A. What determines nucleosome identities and positions, and to what end?
A partial answer to these questions has come from studies of certain inducible genes in yeast. In these cases, one or another transcriptional activator effects removal of nucleosomes that form in adjacent promoter regions prior to induction. This reaction clears the way for subsequent recruitment by the activator of the transcriptional machinery, and absent this step, induction is delayed (Bryant et al., 2008; Korber and Horz, 2004). For example, upon induction of either the PHO5 or GAL1/10 genes, a DNA-bound transcriptional activator (Pho4, in one case and Gal4 in the other) recruits the ‘nucleosome remodeler’ Swi/Snf, which rapidly removes nucleosomes lying adjacent to the site of binding of each activator. NFRs of 100–300 bp are thus created upon induction of these genes. It has been suggested that another member of the Swi/Snf family, RSC, can also be recruited to DNA by specific DNA binding proteins (Badis et al., 2008; Hartley and Madhani, 2009), see also (Ng et al., 2002; Parnell et al., 2008). Unlike Swi/Snf, RSC bears specific zinc-cluster DNA binding determinants (Angus-Hill et al., 2001), and it has been suggested that RSC, either recruited to DNA by another protein, or binding DNA on its own, removes nucleosomes (Badis et al., 2008; Hartley and Madhani, 2009). RSC, as we shall describe, plays an important role at the GAL1/10 genes, but not by removing nucleosomes.
These findings left open the question of what role chromatin architecture might play prior to induction. For example, do nucleosomes compete with regulatory proteins (e.g. Gal4) for binding to DNA, and if so, how significant is that effect and how might it be overcome or avoided? One possibility is that DNA sequences in eukaryotes have evolved with differing nucleosome-forming propensities, and the sites of binding of regulatory proteins are maintained relatively nucleosome-free. Were this the case, one would expect that reconstitution experiments with purified histones and DNA would produce a distribution of nucleosomes that would leave critical sites unoccupied. Contrary to this expectation, it was reported (in contrast to an earlier claim (Terrell et al., 2002)), that reconstitution of such a nucleosome pattern at the yeast PHO5 gene requires, in addition to histones, one or more unidentified proteins in a cell extract (Korber and Horz, 2004). Genome-wide nucleosome reconstitution experiments with yeast DNA have not settled the problem; two recent such studies differ significantly in the reported degree to which nucleosomes reconstituted in vitro occupy positions similar to those found in vivo (Kaplan et al., 2009; Zhang et al., 2009). The problem is of general interest in view of the fact that Gal4 is a ‘universal’ activator. That is, when ectopically expressed, it can activate any of a wide array of genes in higher eukaryotes modified so as to bear Gal4 binding sites nearby.
We recently described a quantitative MNase protection assay that reveals not only nucleosome positioning, but also, especially for well positioned nucleosomes, the fraction of the population that bears a protecting nucleosome for any given position and instant (Bryant et al., 2008). In outline, we measure the nuclease sensitivity of each of a wide array of 60 bp segments (amplicons). The typical DNA fragment yields a biphasic curve, indicating the presence of two populations: one that is highly sensitive (as though it were naked) and the other that is highly protected (as though it bears a nucleosome). These curves differ in their inflection points and thus reveal the fraction of templates in the population, for any given segment, that bears a nucleosome. Rarely, curves are seen that indicate that every member of the population is naked (hypersensitive, HS) or, in contrast, is occupied.
We applied this method to analyzing the chromatin architecture prior to and following induction of the yeast GAL1/10 genes ((Bryant et al., 2008) and see Results, Figure 1A). One striking finding was that the UASg, which bears Gal4 binding sites, behaved differently than did any other DNA segment in the region. First, it was protected by some unknown factor in 100% of the population, and second, the protected segment was some 30 bp shorter than that protected by a typical nucleosome. Previous studies had variously suggested that the UASg is nucleosome-free, that it bears a nucleosome, and/or that it bears some unusual factor (Bryant et al., 2008; Cavalli and Thoma, 1993; Fedor and Kornberg, 1989; Fedor et al., 1988; Kaplan et al., 2009; Lee et al., 2007; Lohr, 1984, 1993).
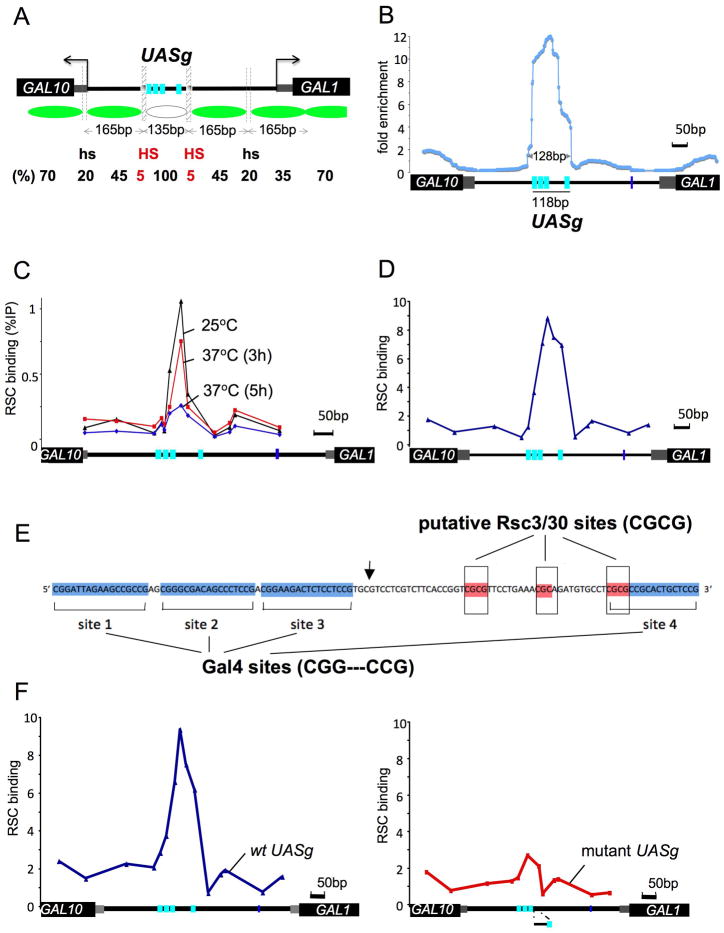
Figure 1. Disposition of nucleosomes and RSC at the GAL1/10 locus.
**(**A) Schematic of the chromatin architecture found at the GAL1/10 locus prior to induction as described in (Bryant et al., 2008). Nucleosomes are represented by green ovals, and two kinds of hypersensitive sites (HS and hs) are indicated by hatched bars. The white oval represents the unknown factor(s) that protect the UASg. The numbers between the arrows show the length of each segment protected from MNase digestion, and the numbers at the bottom show the percent of the population protected. The transcription start sites of the GAL1 and GAL10 genes are indicated by grey bars and the beginnings of the ORFs by black bars. The four Gal4 sites comprising the UASg are in cyan. The TATA box in the GAL1 promoter lies approximately coincident with the rightmost site marked hs.
(B) RSC binding to the GAL1/10 locus. Yeast bearing TAP-tagged RSC were cross-linked, sonicated, and the isolated chromatin digested with MNase to an extent that yielded primarily mononucleosomes. RSC bound DNA-fragments were isolated on IgG-beads. The purified DNA was then subjected to paired-end high throughput (Illumina) DNA sequencing. The resulting fragments were mapped to the S. cerevisiae genome to determine their sizes and positions. The number of fragments that cross any given base pair in the GAL1/10 locus is shown by the blue line (the fragment density). The data is represented as fold enrichment of RSC bound fragments over a random distribution. Cells were grown in glucose at 25°C, and the TAP-tag was added to the RSC subunit RSC8.
(C) Inactivation of the RSC DNA binding subunit Rsc3. Cells bearing the ts mutant RSC (rsc3-1), also TAP-tagged, were grown in glucose at 25°C and then shifted to 37°C for various times as indicated. Cells were treated as described in (B) and the recovered DNA analyzed by QPCR. RSC binding is presented as the percentage immunoprecipitated.
(D) RSC binding in the absence of Gal4. Cells deleted for gal4 and bearing TAP-tagged wt RSC were analyzed for RSC binding and MNase sensitivity as described in (C). RSC binding is shown as fold over a control locus in the PHO5 gene. Supplemental Figure S1 shows that the MNase protection pattern in and around the UASg is not altered by deletion of gal4.
(E) Gal4 and putative RSC binding sites in the UASg. The Gal4 binding sites are indicated in blue. Gal4 sites each bind a dimer of the protein, and each of the three strong binding sites has the sequence CGG-N11-CCG. Putative binding sites for Rsc3/30 (Badis et al., 2008) are indicated in red. The rightmost putative RSC site overlaps Gal4 binding site 4, which differs from the Gal4 consensus in one base pair as shown. The arrow indicates the site of truncation of the UASg in the mutant strain created for the experiments in (F). Sequences to the right of the arrow are deleted in the truncation mutant.
(F) RSC binding to a wt and a truncated UASg. Cells bearing TAP-tagged RSC and grown in raffinose were probed as described in (C).
Here we show that a complex comprising RSC and a nucleosome is bound constitutively, and independently of Gal4, to the UASg. The unusually small size (for a nucleosome) of the protected UASg DNA fragment(s), confirmed here by paired-end DNA sequencing (Illumina), reflects, we suggest, the presence of a partially unwound nucleosome. The complex, placed in an ectopic position, suffices to impose characteristic features of chromatin architecture found at the GAL1/10 locus - including phased nucleosomes and hypersensitive sites - on the flanking DNA. We attribute this effect to the tight positioning of the complex imposed by specific binding of RSC to sites in the UASg. Removal of RSC from the UASg (effected either by inactivation of RSC or by deletion of a segment of the UASg) allows general encroachment of nucleosomes over the locus. Absent RSC at the UASg, Gal4 binding is impeded (but not blocked), a result indicating that the RSC/nucleosome complex presents the Gal4 sites in the UASg for ready access. The loss of this complex at the UASg has biological consequences under at least one physiological condition. A preliminary survey reveals the presence of RSC/nucleosome complexes scattered throughout the yeast genome. The RSC found in higher eukaryotes, so far as we know, lacks the specific DNA binding determinants found on yeast RSC (Mohrmann and Verrijzer, 2005), and, consistent with nucleosome disposition on a UASg inserted into a mammalian cell as reported here, evidently Gal4 works in such cells against whatever obstacle broadly positioned nucleosomes might present.
Results
Figure 1A shows the chromatin architecture at the GAL1/10 locus as found in wild type cells determined using the method of (Bryant et al., 2008). Cells were grown in the absence of galactose, and so the GAL genes were silent. Rather precisely positioned nucleosomes (one to the left and two to the right in the figure) flank the UASg. The boundaries of the UASg are marked by short (ca. 10–20 bp) HS’s. As indicated in the figure, no more than 5% of these HS sequences in the population are occluded by nucleosomes. The regions separating flanking nucleosomes from each other are also unusually sensitive to the nuclease, but less so than the HS sites (20% vs. 5% of the population protected), and so are labeled hs. The repeat length of the nucleosomes, measured as the distance between the centers of hypersensitive sites, is about 165 bp. This is as expected if the core nucleosome includes the typical 147 bp with adjacent nucleosomes separated by about 18 bp of linker DNA. Each of these nucleosomes, however, fully protects less than 50% of the population at any given instant, a finding consistent with the observation that promoter nucleosomes in yeast tend to exchange more rapidly than do other nucleosomes (Dion et al., 2007; Linger and Tyler, 2006).
In contrast, and as indicated in the figure, some unknown factor protects the UASg in virtually every member of the population. The protected region encompassing the UASg spans only some 135 bp measured as the distance between the centers of the flanking HS sites. We also found that the MNase protection pattern observed for wt cells is unaltered by deletion of gal4 (see Supplemental Figure S1). It was also noted that insertion of the UASg into a plasmid caused phased nucleosomes to form adjacent to the UASg (Fedor and Kornberg, 1989). A mutational analysis showed that Reb1, a protein thought to bind the UASg, was not responsible for this effect (Reagan and Majors, 1998). What then is the factor that binds to and strongly protects the UASg from nuclease digestion, and what is its physiological role?
RSC bound to the UASg
Figure 1B displays DNA fragments bearing both a nucleosome (as indicated by protection from MNase digestion) and RSC (as indicated by the presence of the TAP-tag). For this experiment, chromatin was digested under conditions that yielded primarily mononucleosomes, and RSC-bearing fragments were recovered on IgG-beads. Fragments (of size ca. 50–200 bp) were analyzed by paired-end high throughput DNA sequencing (Illumina). This technique determines the sequences found at both ends of each fragment, thus revealing the sizes and genomic origin of these fragments. Figure 1B shows the number of sequenced fragments that cross any given base pair (i.e. the “fragment density”) along the region between the GAL1 and GAL10 genes. The figure shows a strikingly well positioned peak over the UASg.
Figure 1C shows in a different way that RSC is bound at the UASg, and also shows that this binding depends upon the integrity of one of its DNA binding subunits. The mutation rsc3-1 renders heat sensitive the Rsc3 subunit, which bears one of RSC’s putative DNA-binding zinc clusters (Angus-Hill et al., 2001). For this experiment, cells bearing TAP-tagged RSC containing the rsc3-1 mutation were grown at various temperatures as indicated, chromatin subjected to MNase digestion and immunoprecipitated as in Figure 1B, and RSC-bearing fragments characterized by QPCR. The figure shows that the peak of RSC at the UASg is diminished as the cells are grown for longer times at the non-permissive temperature. Figure 1D shows the result of an experiment performed as that of Figure 1C, except that the cells were deleted for gal4, bore a wt RSC fused to the TAP-tag, and were grown at 30°C. The figure shows that the peak of RSC at the UASg forms independently of Gal4, and its MNase resistance indicates that a nucleosome is also present.
Figure 1E shows that the UASg bears, in addition to four Gal4 binding sites, at least two sites that match the proposed (weak) RSC consensus binding sequence (Badis et al., 2008), and a third that differs at one position. By deleting the sequences to the right of the arrow in the schematic, we generated a UASg bearing the three strongest Gal4 binding sites (sites 1–3) but lacking the putative RSC binding sites as well as the fourth, weak, Gal4 binding site. The experiment of Figure 1F, performed as that of Figure 1C, shows that binding of RSC was drastically reduced by this deletion.
A ‘small’, H2A.Z-containing nucleosome at the UASg
Our finding that the UASg was protected from nuclease (MNase) digestion indicates that this DNA, in addition to bearing RSC as just described, is also wrapped in a nucleosome. This surmise was confirmed in the experiments of Figures 2A and B. Chromatin digested as in the experiment of Figure 1C was precipitated with antibodies recognizing H2B, H3 and H4, with results shown in Figure 2A. Figure 2B shows that the H2A subunit was also present, but primarily in the form of the variant H2A.Z. The presence of all four histones indicates the presence of a complete nucleosome at the UASg. This corrects an earlier report of ours (Bryant et al., 2008), a matter discussed in Supplementary Materials. Figure 2B also shows that the nucleosomes occupying the site just to the right of the UASg comprise a mixed population, some of which contain H2A.Z and some of which contain H2A. Nucleosomes bearing H2A.Z at this site were reported by (Albert et al., 2007).
Figure 2. A nucleosome at the UASg.
(A) Histone H2B, H3 and H4 at and around the UASg. Crosslinked chromatin from wt cells (blue line), cells bearing myc-tagged histone H4 (red line) and cells expressing FLAG-tagged H2B (green line) was digested with MNase and then probed with the respective antibodies. DNA was analyzed by QPCR with results presented as fold over a control locus (in the PHO5 promoter). Cells were grown in media containing glucose and similar results were found for cells grown in raffinose (data not shown).
(B) Histone H2A and its variant H2A.Z at and around the UASg. An experiment as the one described in (A) was performed with wt cells or cells bearing HA-tagged H2A. MNase digested chromatin from wt cells was precipitated with an antibody against H2A.Z and that from cells bearing HA-tagged H2A was precipitated with an antibody recognizing the HA-epitope. The resulting DNA was analyzed as in (A). Supplemental Figure S2 shows that the MNase protection pattern in and around the UASg is not affected by deletion of htz1, the gene that encodes H2A.Z.
(C) Pattern of MNase protection at the GAL1/10 locus assayed by paired-end DNA sequencing. Cells bearing the rsc3-1ts mutation were grown at the permissive temperature in media containing raffinose, crosslinked, digested with MNase, and the resulting DNA subjected to paired-end sequencing (Illumina) as described in the legend of Figure 1B. The blue line indicates the density of the resulting MNase protected fragments over the GAL1/10 locus, and the fragments bound by RSC and protected from MNase digestion of Figure 1B are shown for comparison (red line). The distributions of the sizes of the mapped fragments at the UASg (left inset) and at the neighboring nucleosome are shown (right inset).
(D) Size distribution of MNase protected fragments over the GAL1/10 locus. MNase protected fragments were determined as described in (C) and the number of fragments of the sizes indicated is represented by the color saturation (see Experimental Procedures). The fragment density curve of (C) is superimposed for reference. The boxed areas indicate fragments of sizes corresponding to ordinary nucleosomes - in the ORFs and the one to the right of the UASg (labeled ‘normal’), - and fragments of smaller sizes that are associated with the RSC/nucleosome complex at the UASg (labeled ‘smaller’).
Figure 2C displays MNase-protected fragments assayed using paired-end sequencing (Illumina). The density of sequenced fragments along the DNA is represented by the blue curve, and for comparison the density of sequenced fragments bearing RSC (taken from Figure 1B) is represented by the red curve. The two insets above the curves show the fragment size distributions of two regions of the blue curve: on the left are UASg fragments, and on the right are fragments from the region protected by the nucleosome just to the right of the UASg. The figure shows that the UASg fragments cluster around 120 bp, whereas those at the adjacent site cluster around 150 bp. The difference in size distribution is statistically significant to p < 0.001 as determined by a two-sample Kolmogorov-Smirnov test. Figure 2D shows the fragment size distribution along a large part of the GAL1/10 region. It is readily apparent that the weakly phased nucleosomes in the ORFs yield protected fragments of sizes similar to those in the promoters, whereas the UASg protected fragments are shorter. The important result is that, as inferred from our earlier studies (Bryant et al., 2008), the RSC/nucleosome complex at the UASg protects fragments that are, on average, some 30 bp smaller than those protected by a typical nucleosome. And the finding noted above, that 100% of the UASg’s in the population are fully protected from MNase digestion indicates that every UASg bears a nucleosome that protects the unusually small DNA fragments.
As a side note, we have observed a difference between the MNase protection patterns at the GAL1/10 locus depending on whether digested chromatin is assayed by QPCR or by Illumina sequencing. Thus the curve generated by Illumina sequencing seems to be missing two of the protected regions that appear in the other assays (see for example Figure 1A, and 2A and B). One of these regions lies just to the left of the UASg, and the other lies just downstream of the nucleosome flanking the UASg on the right. The explanation for this effect could be as follows: when QPCR is used to assay specific fragments, standard curves are used to correct for the rates at which individual fragments are amplified by PCR. In contrast, to obtain sufficient amounts of each fragment for Illumina sequencing all genomic fragments are amplified by PCR prior to sequencing. However, the rates at which individual fragments are amplified cannot be controlled for, and were fragments protected by a specific nucleosome to be under-amplified, those fragments (and hence the nucleosome) would be underrepresented.
RSC is required for imposition of chromatin architecture at the UASg
As RSC binding to the UASg is impaired (by inactivating RSC containing the rsc3-1ts mutant), the peak of H3 detected over the MNase-protected UASg broadens but does not diminish (Figure 3A). A similar result is observed if the cell bears a RSC with a mutation (sth1-3ts) that inactivates its catalytic domain (Du et al., 1998)(Figure 3B), and the cells are grown at the non-permissive temperature. Note in particular the loss of the HS sites as RSC is inactivated in both mutants. This loss is quantitated for the strain bearing the rsc3-1 mutation by our MNase protection assay as shown in Figure 3C. It can be seen that as RSC is inactivated, the region corresponding to the left HS site (in the figure) becomes increasingly less sensitive to nuclease, that is, it is increasingly occluded by nucleosomes. The experiment of Figure 3D examines the truncated UASg, which is severely deficient in RSC binding, but which bears a DNA sequence that is HS in the wild type. The figure shows that that hypersensitivity is essentially lost in the truncated mutant, consistent with the idea that RSC binding to the UASg is required to phase the nucleosomes and create the HS sites in the wt case.
Figure 3. The effect of RSC binding to the UASg on chromatin architecture.
(A) Histone H3 binding to the UASg in the presence and absence of bound RSC. Cells bearing rsc3-1ts (a mutation in a DNA binding subunit) were grown in glucose at 25°C (black) and then shifted to 37°C for the times indicated (red and blue). Cells were probed for histone H3 as described for Figure 2A.
(B) Histone H3 binding to the UASg in the absence of active RSC. Cells bearing RSC sth1-3ts (a mutation in RSC’s catalytic subunit) grown in glucose at the permissive (black) and non-permissive (red) temperature were probed for H3 as in (A).
(C) Quantitation of the loss of an HS site as RSC is inactivated. A MNase protection experiment was performed as described (Bryant et al., 2008). Cells bearing the rsc3-1ts mutation were grown in raffinose at 25°C and then shifted to 37°C, and MNase protection assayed at various times. The black bar above the schematic shows the position analyzed, and the dots show the increasing protection of the HS site as the cells were grown at 37°C. Similar results were found for the HS site to the right of the UASg in the schematic (not shown).
(D) Effect of truncating the UASg on an HS site. A MNase protection experiment was performed as in (C) with cells bearing the truncated UASg of Figure 2D.
(E) Nucleosome disposition at and around a UASg inserted at an ectopic position. A MNase protection experiment was performed using cells bearing a UASg inserted 551 bp downstream from the GAL1 translation start site. In this mutant the DNA spanning the GAL1-GAL10 promoters was deleted. Protection was analyzed after growth of cells in non-inducing medium (2% raffinose, blue curve) and 30 min following addition of 2% galactose (red curve). The numbers below the figure describe the percent protection of the hatched bars, indicating that HS sites flanking the UASg were created by the insertion.
(F) Gal4 binding to the UASg inserted at an ectopic position. Cells bearing the ectopically positioned UASg described in (E) were grown in raffinose and a ChIP experiment detecting Gal4 was performed as described (Floer et al., 2008) except that chromatin was digested with MNase prior to immunoprecipitation. Gal4 binding is shown as fold over a control location in the PHO5 gene.
(G) RSC binding to the UASg inserted at an ectopic position. Cells bearing the ectopically positioned UASg and TAP-tagged RSC were grown in raffinose, chromatin was treated with MNase and a ChIP experiment recognizing the TAP-tag was performed as in Figure 1C.
The complex at the UASg suffices to impose chromatin architecture
The experiment of Figure 3E shows the effect on chromatin architecture of the inserted UASg in the GAL1 ORF. The inserted UASg is flanked by HS sites and phased nucleosomes, just as is the UASg when located at its ordinary position in the GAL1/10 locus. The numbers below the curve show that HS sites were not present at the corresponding positions in the ORF prior to insertion of the UASg. Thus nucleosome phasing and the creation of HS sites occur independently of sequence context (for the two sequence contexts examined), and instead are imposed by the complex at the UASg. One difference between the architecture created in the ORF and that at the ordinary UASg location is that the flanking nucleosomes form more efficiently at the ectopic site. Nevertheless, as the figure shows, nucleosomes immediately adjacent to the ectopically positioned UASg were quickly removed upon addition of galactose, just as are the corresponding nucleosomes when the UASg is at its ordinary location. The result implies that Gal4 must bind to the ectopically positioned UASg, an inference confirmed by the ChIP experiment of Figure 3F. Figure 3G shows that, also as expected, RSC is bound to the ectopic UASg.
The effect of RSC on induction of the GAL genes
What effect does the RSC/nucleosome complex – with its attendant chromatin architecture - have on regulation of expression of the GAL genes? Figure 4A shows that addition of galactose to cells growing in raffinose induced expression of GAL1 equally quickly, and to the same extent, whether or not a ts RSC was inactivated by growth at the non-permissive temperature. Figure 4B shows that, consistent with this finding, induction of a strain growing in raffinose and bearing the truncated UASg, which does not bind RSC, followed the same time course as did induction of wt cells. In contrast, when cells were grown in glucose and then transferred to galactose, inactivation of RSC (or elimination of RSC binding sites) resulted in a marked delay in induction (Figure 4C and D). We explain these findings as follows.
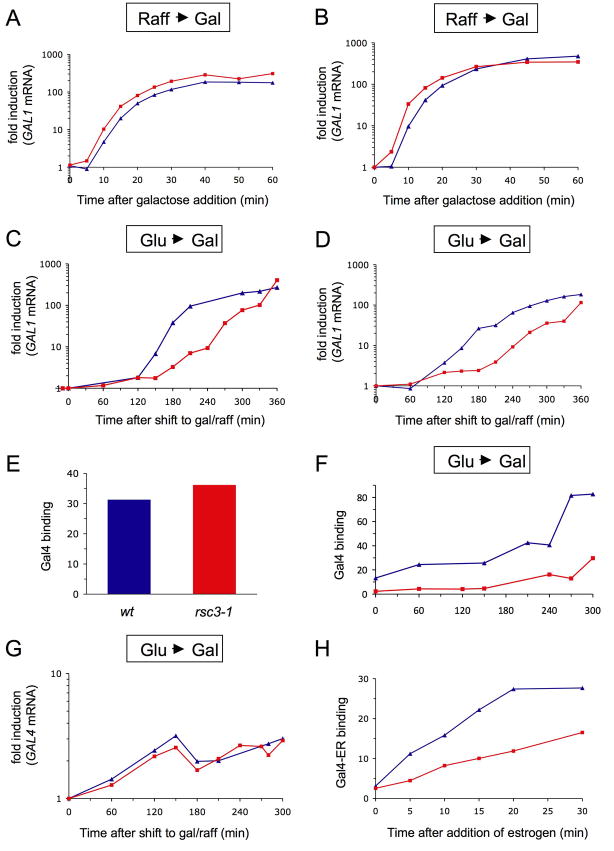
Figure 4. Effects of RSC on induction of GAL1 and on binding of Gal4 to the UASg.
(A) Effect of inactivating RSC on induction: Raff ⇒ Gal. Galactose was added to wt (blue) or rsc3-1ts mutant cells (red) growing in raffinose at 37°C for 3h. At the times indicated GAL1 mRNA levels were determined as described (Floer et al., 2008).
(B) Effect of deleting putative RSC binding sites on induction: Raff ⇒ Gal. Galactose was added to raffinose-grown cells that bore wt RSC and either the wt (blue) or the truncated UASg (red) of Figure 2D.
(C) Effect of inactivating RSC on induction: Glu ⇒ Gal. wt (blue) and rsc3-1ts cells (red) were grown in glucose for 3h at 37°C and then shifted to media lacking glucose but containing galactose and raffinose. GAL1 mRNA levels were measured as described in (A).
(D) Effect of removing RSC binding sites on induction: Glu ⇒ Gal. An experiment was performed as that described in (C) except that cells bearing wt RSC and either the wt (blue) or truncated UASg (red) were grown at 25°C.
(E) Effect of inactivating RSC on Gal4 bound to the UASg: Raff. wt cells (blue) and rsc3-1ts cells (red) were grown in raffinose at 25°C and then shifted for 3h to 37°C. Gal4 binding was analyzed as described for Figure 3F and is shown normalized to a control locus in the PHO5 gene.
(F) Effect of inactivating RSC on Gal4 binding to the UASg: Glu ⇒ Gal_. wt_ (blue) and rsc3-1ts cells (red) were grown in glucose for 3h at 37°C and shifted to medium containing galactose. Gal4 binding to the UASg was analyzed as described in (E).
(G) Effect of inactivating RSC on GAL4 mRNA production. GAL4 mRNA levels were measured for cells grown as described for the experiment of (C).
(H) Effect of deleting RSC binding sites on binding of a hormone regulated Gal4-fusion protein to the UASg. Cells were deleted for gal4 but contained a plasmid expressing a myc-tagged Gal4DBD-ER-VP16 fusion (Nalley et al., 2006), and either the wt UASg (blue) or its truncated derivative (red). Gal4 binding to the UASg was determined at the times indicated, following addition of estrogen to cells growing in glucose, by probing for myc. The data was normalized to a control locus in the PHO5 gene.
It is well known that in cells growing in raffinose, Gal4 is expressed and binds the UASg, but its activating region is covered by the inhibitor Gal80. Addition of galactose inactivates Gal80, thereby freeing Gal4’s activating region and triggering activation of transcription. Glucose, however, represses Gal4 expression (an aspect of the “glucose repression” effect at the GAL genes), and upon transfer to galactose, Gal4 levels must first be increased, and the newly synthesized Gal4 must bind the UASg before it can work. We imagine that in the absence of RSC, Gal4 must compete with broadly bound nucleosomes for binding to the UASg, whereas in its presence the UASg is held readily accessible to Gal4. According to this idea, for cells growing in raffinose and absent RSC at the UASg, Gal4 would slowly compete away the nucleosomes, bind its sites on the UASg, and thus be positioned to respond rapidly to galactose. In contrast, for identical cells growing in glucose and then transferred to galactose, some delay in induction would be incurred as Gal4 competes for binding to its sites.
These ideas make predictions realized in the experiments of Figures 4E–H. Figure 4E shows that cells grown overnight in raffinose indeed have Gal4 bound to the UASg whether or not RSC is present. Figure 4F shows that for cells growing in glucose and transferred to galactose, binding of Gal4 to the UASg is significantly delayed in the absence of RSC. Figure 4G shows that this delay is not accounted for by a delay in expression of Gal4 caused by RSC inactivation. That is, as measured by the levels of GAL4 mRNA, Gal4 is expressed equally well in this scenario whether or not RSC is inactivated. Figure 4H confirms a key aspect of this picture in another way. The fusion protein Gal4-ER-VP16 is held in the cytoplasm in the absence of estrogen, and upon addition of the hormone the fusion protein enters the nucleus and binds the UASg (Nalley et al., 2006). The figure shows that in such an experiment, the fusion protein binds the truncated UASg lacking RSC sites significantly more slowly than it binds a wt UASg.
RSC/nucleosome complexes elsewhere in the genome
Having surveyed the entire genome we have found some 4,100 RSC/nucleosome peaks listed in Supplementary Table S1. These peaks were identified by comparing genome-wide Illumina sequencing data from the MNase protection experiment of Figure 2C with that of the MNase-protection/RSC ChIP experiment of Figure 1B. Where a peak of RSC was greater in the latter data compared with the former, we assigned a location for a RSC/nucleosome complex. Preliminary analysis suggests that the majority of these RSC/nucleosome complexes protect fragments similar in sizes to those protected by an ordinary nucleosome (not shown). But a significant fraction of the complexes protect fragments similar in sizes to those protected by the complex at the UASg. Figure 5 shows four examples from chromosome II (which contains the GAL1/10 locus) of RSC/nucleosome complexes found in or near promoters, all of which yield protected fragments of sizes similar to those seen for the UASg. We do not know the function of any of these RSC/nucleosome complexes found outside the GAL1/10 locus.
Figure 5. ‘Small’ nucleosomes associated with RSC at various locations in the genome.
The distribution of fragments protected from MNase digestion at four promoters found on chromosome II. The data is displayed as in Figure 2D, except that the disposition of RSC (taken from data of the experiment of Figure 1B) is overlayed in red. The names of the genes, and the coordinates along the genome are shown in the figure.
A UASg placed in a mammalian cell
The specific DNA binding determinants of RSC are found in the two subunits Rsc3 and Rsc30 (Angus-Hill et al., 2001). The mammalian homologue of yeast RSC lacks the Rsc3 and Rsc30 subunits (Mohrmann and Verrijzer, 2005; Wilson et al., 2006). We expected, therefore, that a UASg transferred to mammalian cells would lack the characteristic structure found at the UASg in wt yeast, and would more closely resemble that seen in a yeast rsc3-1 mutant. This expectation is borne out in the experiment of Figure 6. HeLa cells were transfected with an integrating plasmid bearing a 700 bp region spanning the GAL1/10 promoters and the UASg. MNase-protected chromatin from selected integrants was then probed for histone H2B as in the experiment performed with yeast in Figure 2A. A signal was broadly distributed over the region, similar to that observed for the UASg in yeast in the absence of RSC (data not shown). Consistent with these findings, the MNase protection experiment of Figure 6 shows that HS sites are not found associated with the UASg inserted into a mammalian genome. Thus both the ChIP and MNase protection assays indicate a broad distribution of nucleosomes over the UASg in a mammalian cell, similar to that seen in yeast lacking RSC activity.
Figure 6. Chromatin architecture at a UASg inserted into a mammalian cell.

HeLa cells were transfected with an integrating plasmid bearing a 700 bp DNA segment spanning the UASg and the flanking GAL1/10 promoters, and integrant were selected. A MNase protection experiment was performed with yeast wt for RSC or bearing the rsc3-1ts mutation, and with HeLa cells bearing the inserted UASg segment. The blue bars indicate the protection of the DNA segment in the UASg as indicated in the schematic above the figure. The red bars indicate protection of the sequence just to the left of the UASg that is HS in wt yeast.
Discussion
Determinants of chromatin architecture at the GAL1/10 genes
We show that two DNA binding proteins, each of which recognizes specific sites in the UASg, determine chromatin architecture independent of sequence context. Thus, RSC, which traps an unusual nucleosome on the UASg, establishes chromatin architecture prior to induction, and Gal4, bound to the UASg, directs removal of promoter nucleosomes upon induction. This conclusion implies that different intrinsic nucleosome-forming potentials of different DNA sequences play little role in this architecture. This conclusion might be tempered by the following considerations. First, the regions flanking the UASg are relatively depleted of nucleosomes. That is, although the nucleosomes form at more or less specified (phased) sites, they tend to form less frequently than do typical nucleosomes - those found in ORFs, for example (see Figure 1A and (Bryant et al., 2008)). It is possible that this relatively low occupancy of predetermined sites reflects the inherent nucleosome-forming propensities of these DNA sequences, a notion consistent with our finding that, when the UASg is positioned at an ectopic site, the flanking phased nucleosomes form more readily than when the UASg is at its wild type location. Whether DNA sequence plays this role, and what might be its biological significance, remains for further investigation. Second, perhaps the UASg spontaneously forms a nucleosome with high frequency in vivo, and perhaps that property helps stabilize the final RSC/nucleosome complex. Experiments performed in vitro indicate that the UASg readily wraps into nucleosomes (Kaplan et al., 2009; Rainbow et al., 1989).
The RSC/nucleosome complex and chromatin architecture
How does the RSC/’small’ nucleosome complex, at the UASg, cause phasing of flanking nucleosomes and the creation of HS sites? The complex is held in a tight position by the specific DNA binding determinants on RSC. That tight positioning, we imagine, presents a barrier that excludes nucleosomal encroachment (Kornberg, 1981). Such a barrier would tend to cause flanking nucleosomes to be “phased”, an effect that would diminish as we move away from the barrier. The hypersensitive sites would also be explained by the barrier effect. Thus every UASg in the population would present an identical barrier, and the inability of nucleosomes to encroach on that barrier, would render a short bit of DNA sensitive to MNase in every cell in the population. Such a short sequence would appear as hypersensitive in the MNase protection assay. Nucleosome phasing and HS sites are thus determined not by the identities of the sequences adjacent to the UASg, but rather are a consequence of the barrier effect. A typical nucleosome would not present a well-defined barrier because, unlike a specific DNA binding protein, a nucleosome, even if bound to a favorable site, will tend to occupy a distribution of sites, which differ modulo 10 bp. This is because DNA, in wrapping around the histone octamer, makes many contacts with the protein (at intervals of some 10 bp), and so ratcheting the nucleosome by 10 bp will have little effect on its stability (see (Ioshikhes et al., 2006) for a fuller discussion).
Our experiments raise the possibility that for many genes the presumed role of RSC – i.e. to remove promoter nucleosomes – might usefully be reconsidered. It is reported that mutation of RSC causes an increase in nucleosome density and decreased gene activation at various loci in the genome (Badis et al., 2008; Hartley and Madhani, 2009). A similar finding would apply to the GAL genes, but our analysis shows that at that locus RSC plays no direct role in nucleosome removal, but rather facilitates activator binding. Furthermore, nucleosome depletion at the GAL locus (low nucleosome occupancy prior to induction and absence of nucleosomes following induction) does not play the role often ascribed to NFRs, i.e. to facilitate activator binding. Rather, the GAL1 and GAL10 NFRs are created by the activator (Gal4 in this case), and there is no obvious way the depletion of nucleosomes prior to induction could influence Gal4 binding.
A “small” nucleosome
The ‘paired end” DNA sequencing technique (Illumina), which determines sequences from both ends of each sequenced fragment, confirms our earlier, surprising, finding that the size of the UASg fragment protected from MNase digestion is about 30 bp smaller than that protected by the typical nucleosome. Moreover, as we show here, the protecting factor includes RSC plus all four histones found in a nucleosome. These results indicate that the UASg is wrapped in a nucleosome complexed with RSC in such a manner that the DNA is partially unwound from the histone octamer. MNase digestion experiments can readily miss the presence of “smaller” nucleosomes such as that found at the UASg. Thus if one assumes that nucleosomes protect DNA of size 150 bp, and first isolates such sized fragments prior to further analysis, the shorter protected regions would be missed. Another way that shorter fragments might be overlooked would be by analyzing protected fragments by any method that sequences just one end of any individual fragment and assuming the location of the other end based on the usual size of sequences protected by nucleosomes.
The predominant form of the H2A subunit at the UASg is the minor variant H2A.Z. Perhaps this subunit interacts more efficiently with RSC than does the major H2A species. If so, the preference is not absolute, as indicated by the fact that in a strain deleted for H2A.Z, the RSC nucleosome complex at the UASg forms (Supplemental Figure S2). Other experiments have suggested a relation between H2A.Z incorporation into chromatin and RSC (Hartley and Madhani, 2009).
RSC/nucleosome complexes elsewhere in the genome
We have detected some 4,100 RSC/nucleosome peaks along the S. cerevisiae genome using the criteria described in the Supplemental Experimental Procedures. Preliminary analysis indicates that some 5–20% of these complexes protect fragments shorter than those associated with ordinary nucleosomes. These RSC/’small’ nucleosome complexes are overrepresented in or near promoters (not shown), four examples of which we show in Figure 5. Other RSC/nucleosome complexes, found more commonly in ORFs, protect fragments of sizes expected to be protected by ordinary nucleosomes (not shown). It is possible that different positioning of RSC binding sites produces these different structures. These and related matters remain for further investigation.
Role of chromatin architecture in gene regulation
Our results indicate that the RSC/partially unwound nucleosome complex facilitates Gal4 binding to its sites in the UASg, and this slower binding has a physiological consequence when cells grown in glucose are transferred to galactose. In this scenario newly made Gal4 more rapidly binds the UASg, and induces transcription, if RSC is present at the UASg. Thus the RSC/partially unwound nucleosome complex, bound at the UASg, would confer a significant growth advantage to yeast on the assumption that a rapid response to the environmental change (glucose to galactose) can be important.
The fact that the RSC found in higher eukaryotes lacks the DNA binding determinants of yeast RSC, and our finding that (as therefore expected) a UASg placed in a mammalian cell bears only broadly positioned nucleosomes (Figure 6), suggests the possibility that in such organisms there will be a necessary delay between the introduction of Gal4 and the activation of transcription of the target gene. We imagine that the speed with which this will happen could well depend upon the concentration of Gal4, the number of binding sites present in synthetic UASg’s, and so on. How such considerations might apply to other activators and genes in yeast and higher eukaryotes remains to be seen.
A Model for The RSC/nucleosome complex at the UASg
Figure 7A shows a model for a RSC/nucleosome complex (Chaban et al., 2008), based on cryo-EM and biochemical studies of a RSC/mononucleosome complex (Asturias et al., 2002; Lorch et al., 2001). In this model, RSC engulfs the nucleosome and partially unwinds it, contacting but not entirely covering the DNA remaining on the histone octamer surface. Inspired by this model, we projected a 118 bp UASg onto a sphere the size of a nucleosome (Figure 7B). In the projection_,_ the center of the UASg is aligned with the dyad axis of the nucleosome, and 80 bp of the UASg wraps around the histone octamer. Comparison of Figure 7A and B suggests that the putative RSC binding sites in the UASg (red segments) could contact RSC; the Gal4 binding sites (blue) 1 and 4 would lie in the unwrapped portion of the DNA; and Gal4 binding sites 2 and 3 would lie on the surface of the nucleosome not covered by RSC. Figure 7C (modeled for us by Francisco Asturias) shows that Gal4 (here represented by two dimeric Gal4 DNA binding domains) could bind sites 2 and 3 without destroying the structure. This model remains speculative at this point because, among other uncertainties, the path of the DNA in the structure of (Chaban et al., 2008) is not well defined. Nevertheless the model is strikingly consistent with our results.
Figure 7. A model for the RSC/nucleosome complex.
(A) Cryo-EM structure of RSC bound to a nucleosome (taken from (Chaban et al., 2008)). RSC interacts closely with nucleosomal DNA at three different positions (grey-colored RSC density labeled 1-3). Some DNA density (solid black line) is apparent in the structure, but large portions of the DNA (hatched black lines) were not detected, suggesting that they were highly mobile/disordered as a result of interaction of the nucleosome with RSC.
(B) Projection of the UASg onto a nucleosome. The UASg was modeled onto a single turn of a nucleosome (corresponding to 80 bp) with the dyad axis placed in the center of the UASg. Gal4 sites are shown in blue and putative RSC binding sites in red. Alignment of this projection with the structure in (A) places Gal4 sites 1 and 4 on the unwrapped ends of the nucleosome and sites 2 and 3 on a part of the nucleosome that is largely accessible in the structure. The positions of the putative RSC binding sites in the UASg correspond closely to the three RSC densities shown to contact nucleosomal DNA.
(C) Model of Gal4 binding to sites 2 and 3 in a UASg bound by a RSC/nucleosome complex. The UASg was positioned in the RSC/nucleosome structure of (A) as described in (B). Two Gal4 dimers (as represented by their DNA binding and dimerization domains, shown in red and purple) were positioned on Gal4 binding sites 2 and 3. The orientation of Gal4 dimers on DNA was taken from the Gal4/DNA structure of (Hong et al., 2008). The model shows that sites 2 and 3 are exposed along a surface of the RSC/UASg/nucleosome complex and that Gal4 can bind these sites without disrupting the structure.
The ubiquity of inhibitors and small effects
In a broader context, our findings illustrate principles that apply to many biological regulatory processes, especially in eukaryotes. First, where those systems are regulated by binding reactions, transcription being a salient example, inhibitors are required to suppress basal reactions. Those inhibitors must be readily overcome when the system is activated. Nucleosomes are widely believed to suppress basal transcription, and we and others have previously shown how promoter nucleosomes can be removed upon command (Bryant et al., 2008; Reinke and Horz, 2003). Second, many regulatory features may be regarded as add-ons that facilitate, but are not absolutely required, for any particular case. The GAL genes show us two examples: recruitment of Swi/Snf and its subsequent action, which facilitates the initiation of transcription as described, is not absolutely required – that reaction occurs in the absence of Swi/Snf, but more slowly (Bryant et al., 2008). A similar description of the RSC/partially unwound nucleosome complex would seem to apply here as well – its presence facilitates Gal4 binding to the UASg upon induction, but is not absolutely required. And, it seems likely that in the many cases where ectopically expressed Gal4 is used to express heterologous genes in higher eukaryotes, it does so in the absence of the facilitating RSC/partially unwound nucleosome complex. Add-ons, such as the RSC/partially unwound nucleosome complex, are found widely in biology, and they make systems that work, work better. Put another way, the prevalence of machinery with small effects illustrates the power of natural selection (Ptashne, 2009).
Experimental Procedures
Yeast strains and growth conditions
The strains and plasmids used in the experiments as well as the growth conditions are listed in the Supplemental Data. In general yeast cells were grown exponentially in SC media containing 2% of the sugars indicated.
Mammalian plasmid construction and cell transfection
The plasmid used for integrating the yeast UASg into mammalian chromatin was generated by inserting a 790 bp fragment containing the GAL1/10 regulatory region and the initial base pairs of the GAL1 and GAL10 ORFs into the pAcGFP1-1 vector (Clontech Laboratories, INC). The plasmid was integrated into HeLA cells using Fugene 6 (Roche Applied Science, Indianapolis, IN) and integrants were selected.
ChIP experiments and mRNA determination
ChIP experiments probing for RSC, histones or Gal4 were performed essentially as described (Floer et al., 2008) except that were indicated crosslinked DNA was digested with limiting amounts of MNase prior to immunoprecipitation. The immunoprecipitated DNA was analyzed either by quantitative PCR as described (Bryant and Ptashne, 2003) or by paired-end high throughput sequencing (Illumina) as described below. GAL1 and GAL4 mRNA was assayed as described (Floer et al., 2008).
MNase protection experiments
MNase protection experiments of yeast chromatin were performed as described (Bryant et al., 2008). For experiments with mammalian chromatin, clones bearing the integrated UASg were selected and the MNase digestion experiments were performed with six individual clones with essentially identical results. The exact sequences of the primers used can be given upon request.
Paired-end DNA sequencing (Illumina)
For these experiments cells were crosslinked with formaldehyde, sonicated and treated with MNase as described for the ChIP experiments (see above). The resulting MNase protected fragments were purified (Qiagen) without further size-separation. (Inspection of the resulting fragments by agarose gel electrophoresis revealed fragments of sizes 50–200 bp). For the analysis of fragments bound by RSC MNase protected DNA (from cells bearing TAP-tagged RSC) was precipitated on IgG-beads, followed by Qiagen purification. A detailed description of the paired-end sequencing (Illumina) method and of the analysis to determine RSC bound peaks can be found in the Supplemental Experimental Procedures. In brief, a DNA library for paired-end sequencing was created and the sequencing data was processed on a Illumina GA analysis pipeline. Reads passing Illumina quality filters were mapped to the S. cerevisiae genome obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/) on July 24, 2009. The 36 base paired end reads were mapped using MAQ alignment software with default settings (Li et al., 2008). The fragment density maps of Figures 1B and 2C and D were generated by calculating the number of fragments that cross any given base pair along the genome. The fragment size distributions (histograms of the insets in Figure 2C) were generated by calculating the number of fragments at a given size that cross the range of DNA indicated by the gray box. The fragment size distribution along the GAL1/10 locus shown in Figure 2D and at the four loci shown in Figure 5 was generated as follows. At each genomic position (in modules of 8 bp), the number of fragments of each size (in modules of 4 bp) were plotted with the y-axis representing the fragment size and the color saturation representing the average fragment count. The GEO accession number for the genome-wide datasets is GSE20078.
Supplementary Material
01
02
Acknowledgments
We thank Francisco Asturias for preparing the space filling model of the RSC/nucleosome complex bound to the UASg together with two Gal4 DNA binding domain dimers. We would also like to thank Timothy Hughes, Brad Cairns, Tom Kodadek, Francesc Posas and David Allis for yeast strains and plasmids, Hiten Madhani for the antibody against H2A.Z, and Alex Gann for helpful comments on the manuscript. This work was supported by a grant (R01GM032308) from the NIH to Mark Ptashne.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Structural analysis of the RSC chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2002;99:13477–13480. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS biology. 2008;6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. Embo J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genet. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ, Kornberg RD. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol Cell Biol. 1989;9:1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ, Lue NF, Kornberg RD. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. Journal of molecular biology. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci U S A. 2008;105:2975–2980. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Fitzgerald MX, Harper S, Luo C, Speicher DW, Marmorstein R. Structural basis for dimerization in DNA recognition by Gal4. Structure. 2008;16:1019–1026. doi: 10.1016/j.str.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nature genetics. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P, Horz W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J Biol Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981;292:579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nature genetics. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome research. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger J, Tyler JK. Global replication-independent histone H4 exchange in budding yeast. Eukaryot Cell. 2006;5:1780–1787. doi: 10.1128/EC.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic acids research. 1984;12:8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. Chromatin structure and regulation of the eukaryotic regulatory gene GAL80. Proc Natl Acad Sci U S A. 1993;90:10628–10632. doi: 10.1073/pnas.90.22.10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. RSC unravels the nucleosome. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Binding reactions: epigenetic switches, signal transduction and cancer. Curr Biol. 2009;19:R234–241. doi: 10.1016/j.cub.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Rainbow M, Lopez J, Lohr D. The yeast GAL1-10 UAS region readily accepts nucleosomes in vitro. Biochemistry. 1989;28:7486–7490. doi: 10.1021/bi00444a048. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-Wide Views of Chromatin Structure. Annu Rev Biochem. 2009 doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan MS, Majors JE. The chromatin structure of the GAL1 promoter forms independently of Reb1p in Saccharomyces cerevisiae. Mol Gen Genet. 1998;259:142–149. doi: 10.1007/s004380050799. [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Terrell AR, Wongwisansri S, Pilon JL, Laybourn PJ. Reconstitution of nucleosome positioning, remodeling, histone acetylation, and transcriptional activation on the PHO5 promoter. J Biol Chem. 2002;277:31038–31047. doi: 10.1074/jbc.M204662200. [DOI] [PubMed] [Google Scholar]
- Wilson B, Erdjument-Bromage H, Tempst P, Cairns BR. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genet. 2006;172:795–809. doi: 10.1534/genetics.105.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02