Activation of the RAS/Cyclic AMP Pathway Suppresses a TOR Deficiency in Yeast (original) (raw)
Abstract
The TOR (target of rapamycin) and RAS/cyclic AMP (cAMP) signaling pathways are the two major pathways controlling cell growth in response to nutrients in yeast. In this study we examine the functional interaction between TOR and the RAS/cAMP pathway. First, activation of the RAS/cAMP signaling pathway confers pronounced resistance to rapamycin. Second, constitutive activation of the RAS/cAMP pathway prevents several rapamycin-induced responses, such as the nuclear translocation of the transcription factor MSN2 and induction of stress genes, the accumulation of glycogen, the induction of autophagy, the down-regulation of ribosome biogenesis (ribosomal protein gene transcription and RNA polymerase I and III activity), and the down-regulation of the glucose transporter HXT1. Third, many of these TOR-mediated responses are independent of the previously described TOR effectors TAP42 and the type 2A-related protein phosphatase SIT4. Conversely, TOR-controlled TAP42/SIT4-dependent events are not affected by the RAS/cAMP pathway. Finally, and importantly, TOR controls the subcellular localization of both the protein kinase A catalytic subunit TPK1 and the RAS/cAMP signaling-related kinase YAK1. Our findings suggest that TOR signals through the RAS/cAMP pathway, independently of TAP42/SIT4. Therefore, the RAS/cAMP pathway may be a novel TOR effector branch.
The Saccharomyces cerevisiae targets of rapamycin, TOR1 and TOR2, are functionally and structurally conserved protein kinases that control a large and diverse set of growth-related readouts in response to nutrient availability (53). In yeast, rapamycin treatment or TOR depletion results in several physiological changes characteristic of starved cells, including inhibition of translation initiation, inhibition of ribosome biogenesis, specific changes in transcription, sorting and turnover of nutrient permeases, accumulation of storage carbohydrates (such as glycogen), and induction of autophagy (reviewed in references 12 and 52). Nitrogen (in particular glutamine) and possibly carbon are important nutrients in the context of yeast TOR signaling (13, 56). Mammalian TOR (mTOR) controls translation and other growth-related processes in response to amino acids and a growth factor signal (e.g., insulin). The growth factor input into the mTOR pathway is via phosphatidylinositol 3-kinase (PI3K), PDK1, and protein kinase B (PKB; also known as Akt). The precise nature of the link between mTOR and PI3K signaling remains to be determined but may involve the PKB-mediated phosphorylation and inhibition of the tuberous sclerosis protein complex (TSC1-TSC2) upstream of mTOR. Control of translation by mTOR is via activation of S6 kinase (S6K) and inhibition of the eIF4E inhibitor 4E-BP1 (reviewed in reference 30).
The yeast TORs control several readouts via a phosphatase switch composed of the type 2A-related phosphatase SIT4, the PP2A/SIT4-associated protein TAP42, and the TAP42-interacting protein TIP41 (16, 29, 31). Under good nutrient conditions, TOR promotes the binding of SIT4 to TAP42, thereby maintaining SIT4 inactive. On rapamycin treatment or nutrient depletion (i.e., TOR-inactivating conditions), SIT4 dissociates from its inhibitor TAP42 and is active. Activated SIT4 dephosphorylates and activates several targets, such as the GATA-type transcription factor GLN3, the Ser/Thr kinase NPR1, and TIP41 (2, 29, 54).
Under favorable nutrient conditions, TOR globally represses starvation-specific transcription by sequestering several nutrient-responsive transcription factors, such as the GATA factors GLN3 and GAT1, the zinc finger transcription factors MSN2 and MSN4, and the bHLH/Zip factor RTG1/3, in the cytoplasm (12, 35). In the case of GLN3, TOR prevents the transcription of genes normally induced on nitrogen limitation by promoting the association of GLN3 with the cytoplasmic URE2 protein (2, 4, 9, 25). The phosphorylation of GLN3, which is antagonized by SIT4, is critical for its interaction with URE2 and thus for its cytoplasmic retention (2). Similarly, TOR and TAP42 maintain the protein kinase NPR1 in an inactive, phosphorylated state whereas TOR inactivation results in the SIT4-dependent dephosphorylation and activation of NPR1. The phosphorylation state of NPR1, in turn, impinges on the sorting and turnover of amino acid permeases such as the tryptophan permease TAT2 and possibly the general amino acid permease GAP1 (3, 15, 54). Ultimately, both GLN3 and NPR1 are involved in scavenging or synthesizing alternative nutrient (nitrogen) sources. TOR also signals to the translation machinery via TAP42, but the mechanism by which TAP42 and/or the PP2A phosphatases are involved in the control of protein synthesis is unknown. TOR also negatively regulates RTG1/3, MSN2, and MSN4 but does so independently of SIT4 (2, 37). For other TOR readouts such as ribosome biogenesis or autophagy, the effector pathways are unknown but may involve novel TOR effectors or cross talk with other growth-controlling signaling pathways.
In addition to its redundant function with TOR1 in a rapamycin-sensitive signaling pathway, TOR2 performs a rapamycin-insensitive function that mediates the polarization of the actin cytoskeleton (53). Recently, two large TOR complexes comprising the TOR proteins and several partner proteins (KOG1, LST8, and AVO1 to AVO3) have been identified in yeast (38, 64). At least one of the complexes is conserved in mammals (24, 33, 38). These structurally and functionally distinct complexes account for the diversity and specificity of TOR signaling in yeast.
The RAS/cyclic AMP (cAMP) pathway in S. cerevisiae plays a major role in the control of growth and metabolism in response to nutrients. The core of the pathway consists of the guanine nucleotide exchange factor CDC25, which, presumably in response to nutrients, activates the redundant small GTPases RAS1 and RAS2 (RAS). RAS, in turn, activates adenylate cyclase (encoded by the CDC35 gene), resulting in the production of cAMP and activation of protein kinase A (PKA) by dissociation of the PKA regulatory subunit BCY1 and the PKA catalytic subunit(s) (redundantly encoded by the TPK1, TPK2, and TPK3 genes) (7). The RAS/cAMP pathway negatively regulates cellular physiology characteristic of stationary-phase/nutrient starvation. Thus, cells deficient in RAS/cAMP signaling, similarly to TOR-depleted or rapamycin-treated cells, exhibit a G1 cell cycle arrest, accumulation of storage carbohydrates (e.g., glycogen and trehalose), and specific changes in transcription (7, 59). RAS/cAMP signaling has also been implicated in the nutrient-mediated control of ribosome biogenesis (reviewed in reference 62; see Discussion). Consistently, cells with constitutive RAS/cAMP signaling fail to adapt their growth program in response to nutrient starvation and rapidly lose viability. Recently, the complexity of RAS/cAMP signaling has increased as a result of the discovery of a G-protein-coupled receptor system, consisting of GPR1 and its Gα protein GPA2, that appears to act upstream of adenylate cyclase to stimulate cAMP production, probably in response to glucose (36, 40, 66). Moreover, cAMP production is a very fine-tuned process that is regulated at different levels involving, for instance, feedback loops and the action of phosphodiesterases (59). The way in which nutrients are sensed by the RAS/cAMP pathway remains unclear.
The precise mechanism(s) by which PKA controls cell growth is not known. It has long been known that the majority of downstream targets of PKA are enzymes involved in intermediary (carbon) metabolism (59). More recently, however, other PKA targets have emerged. For instance, PKA negatively regulates the transcription of a large number of stress-responsive genes by phosphorylating and inactivating MSN2 and probably MSN4 (20, 21, 58). The MSNs are redundant, stress element (STRE, AG4)-binding transcription activators (6, 41, 55). The MSNs are also negatively controlled by TOR, via the 14-3-3 proteins BMH1 and BMH2 (2, 5). Moreover, PKA negatively regulates RIM15, a kinase involved in the control of postdiauxic transcription (48, 51).
The RAS/cAMP pathway is connected to other nutrient-regulated signaling components, such as the kinase SCH9 (a yeast PKB homologue) and YAK1/SOK1 (see below) (59). A functional interaction between the RAS/cAMP pathway and TOR has also been suggested (2, 14), but this interaction has not been investigated directly and systematically.
Here we report that constitutive activation of the RAS/cAMP pathway suppresses a TOR deficiency. We also provide evidence for regulation of PKA and YAK1 localization by TOR. These findings suggest that the RAS/cAMP pathway is a TOR effector pathway distinct from the previously characterized TAP42/SIT4 pathway.
MATERIALS AND METHODS
Strains, plasmids, and media.
The S. cerevisiae strains and plasmids used in this study are listed in Tables 1 and 2, respectively. All strains from our laboratory are isogenic derivatives of TB50a, TB50α, or JK9-3da. Strains from other laboratories, including the corresponding parental strains, are also listed in Table 1. Rich medium (YPD) or synthetic minimal medium (SD) complemented with the appropriate nutrients for plasmid maintenance were as described previously (3, 57). Nitrogen starvation experiments were performed with synthetic medium as previously described (54). Rapamycin was used at a final concentration of 200 ng/ml from a 1-mg/ml stock in 90% ethanol-10% Tween-20. Different incubation periods with rapamycin were used, depending on the readout being examined. Some readouts, due to their inherent delay in development, required longer incubation periods before they could be visualized.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| JK9-3da | MATaleu2-3,112 ura3-52 trp1 his4 rme1 HMLa | Hall laboratory |
| TB50a | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLa | Hall laboratory |
| TB50α | _MAT_α leu2-3,112 ura3-52 trp1 his3 rme1 HMLa | Hall laboratory |
| HM4-5D | TB50a_GLN3_-myc13-kanMX6 | 29 |
| JH11-1C | JK9-3daTOR1-1 | 27 |
| TB105-3B | TB50agln3::kanMX4 gat1::HIS3MX6 | 2 |
| TS65-2D | TB50asit4::kanMX4 | This study |
| TS83-6A | TB50α tap42::kanMX4/YCplac111::tap42-11 | This study |
| TS95-1D | TB50aras2::kanMX4 | This study |
| TS127-1A | TB50ayak1::kanMX4 | This study |
| TS129-8C | TB50a_YAK1_-myc13-kanMX6 | This study |
| TS135-2C | TB50ayak1::kanMX4 gln3::kanMX4 gat1::HIS3MX6 | This study |
| TS139 | TB50a_pho8_Δ60 | This study |
| TS140 | TB50aras2::_kanMX4 pho8_Δ60 | This study |
| TS141 | TB50abcy1::HIS3MX6 | This study |
| TS143 | TB50abcy1::_HIS3MX6 pho8_Δ60 | This study |
| TS145-1C | TB50asit4::_kanMX4 pho8_Δ60 | This study |
| TS160-6C | TB50α bcy1::HIS3MX6 gln3::kanMX4 gat1::HIS3MX6 | This study |
| TS177-7B | TB50aras2::_kanMX4 GLN3_-myc13-kanMX6 | This study |
| TS178-1C | TB50abcy1::_HIS3MX6 GLN3_-myc13-kanMX6 | This study |
| RH1602 | MATaleu2 ura3 his4 bar1 | Riezman laboratory |
| RH1597 | MATaend4-1 leu2 ura3 his4 bar1 | 50 |
| 23344C | _MAT_α ura3 | 26 |
| 27038a | MATanpi1 ura3 | 26 |
| SP1 | MATaleu2 ura3 trp1 his3 ade8 can1 | 8 |
| RS13-58A-1 | MATaleu2 ura3 trp1 his3 ade8 tpk1w1 tpk2::HIS3 tpk3::TRP1 bcy1::LEU2 | 8 |
| S18-1D | MATaleu2 ura3 trp1 his3 ade8 tpk1w1 tpk2::HIS3 tpk3::TRP1 | 8 |
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristics | Source or reference |
|---|---|---|
| pAS55 | YEplac195::HA-TAT2 | 3 |
| pAS103 | YEplac195::HA-NPR1 | 54 |
| pTB374 | pHAC195::HXT1 (2μm URA3) (C-terminally 3× HA tagged, CYC1 terminator; 2.2-kb _Xba_I fragment) | This study |
| pTB380 | pHAC111::HXT1 (CEN LEU2) (C-terminally 3× HA tagged, CYC1 terminator; 2.2-kb _Xba_I fragment) | This study |
| pTB419 | pHAC195::BMH2 (2μm URA3) (C-terminally 3× HA tagged, CYC1 terminator; 1.5-kb _Bam_HI-PstI fragment) | This study |
| pTB424 | YEplac195::CDC25ΔN (2μm URA3) (CDC25 promoter, 3× HA CDC25ΔN; 0.9-kb _Bam_HI-_Xba_I [promoter] and 1.6-kb _Xba_I [3× HA-CDC25 ORF C terminus including GEF domain/terminator] fragments) | This study |
| pTB429 | YEplac195::SOK1 (2μm URA3) (3.7-kb _Bam_HI-_Pst_I fragment) | This study |
| pTB439 | YEplac195::TPK1 (2μm URA3) (2.2-kb _Bam_HI-_Hin_dIII fragment) | This study |
| pTS118 | YCplac33::RAS2Val19 (CEN URA3) (2.1-kb _Eco_RI-_Hind_III fragment) | This study |
| pTS119 | YCplac33::RAS2Ala18Val19 (CEN URA3) (2.1-kb _Eco_RI-_Hind_III fragment) | This study |
| pTS120 | YCplac111::RAS2Val19 (CEN LEU2) (2.1-kb _Eco_RI-_Hind_III fragment) | This study |
| pTS134 | YCplac33::HA-TPK1 (CEN URA3) (N-terminally 2× HA tagged; 0.7-kb _Bam_HI-_Pst_I [promoter] and 1.5-kb _Pst_I-_Hin_dIII [2× HA TPK1 ORF/terminator] fragments/2.2-kb _Eco_RI-_Hin_dIII fragment) | This study |
| pTS137 | YCplac33::HA-BCY1 (CEN URA3) (N-terminally HA -tagged; 0.3-kb _Eco_RI-_Bam_HI [promoter + atg] and 1.6-kb _Bam_HI-_Hin_d III [HA-BCY1 ORF/terminator] fragments) | This study |
| pG2CT-112.2 | GPA2R273A expressed under control of GAPDH promoter (2μm TRP1) | 66 |
| pMSN2-GFP | YCplac111::ADH1 promoter-MSN2-GFP | 20 |
| pTN9 | TDH3 promoter-pho8Δ60 | 45 |
Genetic techniques and multicopy suppressor isolation.
Restriction enzyme digests, ligations, and isolation of plasmids were performed by standard methods. Yeast transformation was performed by the standard lithium acetate procedure (28). PCR cassettes were used to generate gene deletions and modifications as described previously (39). The PHO8 gene in strains used for autophagy assays (see below) was replaced with _pho8_Δ60 by inserting plasmid pTN9 into the PHO8 locus and excised as described previously (45).
Strain TB105-3B (gln3 gat1) was used to isolate multicopy suppressors that confer increased rapamycin resistance. Isolation of the multicopy suppressors was performed using a 2μm, URA3 (pSEY18 backbone)-based library derived from strain R1 (27). Cells were allowed to recover from the transformation in liquid YPD at 30°C for 5 h before being plated on YPD containing 200 ng of rapamycin per ml. Rapamycin-resistant transformants were isolated and passaged through 5-fluoroorotic acid plates to demonstrate that the increased resistance to rapamycin was plasmid linked. Plasmids were rescued and retransformed into the parental strain to recheck resistance. Deletions and subclones were made to ascertain which open reading frame (ORF) on each clone was responsible for rapamycin resistance in a gln3 gat1 mutant.
Glycogen staining.
Samples (7.5 optical density at 600 nm equivalents) from cultures treated with rapamycin or drug vehicle alone for 5 h were transferred to Millipore HA filters. The filters were subsequently placed on a solid agar matrix and exposed to iodine vapor as described previously (1).
Fluorescence microscopy. (i) Indirect immunofluorescence.
To localize HXT1-HA3, GLN3-myc13, HA2-TPK1, HA-BCY1, and YAK1-myc13, cells were grown at 30°C and treated with rapamycin or drug vehicle alone for the indicated times. Tagged proteins were visualized by indirect immunofluorescence on whole fixed cells, as described previously (3). High-affinity monoclonal antibodies (mouse anti-HA [HA.11, clone 16B12; BabCO] and anti-myc [clone 9E10; kindly provided by H.-P. Hauri, Biozentrum, Basel, Switzerland]) and Cy3-conjugated rabbit anti-mouse immunoglobulin G (Molecular Probes) were used to visualize tagged proteins. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) at a concentration of 1 μg/ml. The cells were visualized with a Zeiss Axiophot microscope (100× objective).
(ii) GFP microscopy.
Cells expressing MSN2-green fluorescent protein (GFP) were grown to logarithmic phase, treated with rapamycin or drug vehicle alone or shifted to nitrogen-free medium for 25 min, and fixed for 2 h in phosphate-buffered saline-formaldehyde (3.7% final concentration). After three washes with PBS, samples were observed by using a Zeiss Axioplan microscope (XBO 75 W/2 xenon short-arc lamp).
Immunoblotting.
Whole-cell extracts for sodium dodecyl sulfate-polyacrylamide gel electrophoresis were prepared by glass bead lysis as described previously (3). Phosphatase (10 mM NaF, 10 mM NaN3, 10 mM _p_-nitrophenylphosphate, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate) and protease (1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail tablets Complete [Roche Diagnostics]) inhibitors were added to the extraction buffer (120 mM NaCl, 50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1% NP-40). The samples were denatured at 37°C for 15 min (HXT1 extracts) or 95°C for 5 min (NPR1 extracts). A total of 50 μg (HXT1 extracts) or 25 μg (NPR1 extracts) of protein was loaded per lane for standard SDS-PAGE and immunoblotting. For detection of HA-tagged proteins, a mouse anti-HA antibody (clone 12CA5) and horseradish peroxidase-conjugated secondary anti-mouse secondary antibodies and ECL reagents (Amersham Pharmacia Biotech) were used.
RNA isolation and Northern blot analysis.
RNA from cells treated with drug vehicle alone or rapamycin for the indicated times was analyzed. Total-RNA preparation and Northern blot analysis were performed by standard methods (11). Yeast DNA probes were generated by PCR using yeast genomic DNA as the template and oligonucleotide primers specific for the genes to be analyzed. PCR products were labeled with 32P-labeled phosphate by using a DecaLabel DNA labeling kit (MPI Fermentas).
Pol I and Pol III activity and [C3H3]methionine labeling.
To monitor RNA polymerase I (Pol I) and Pol III activity, l-[_methyl_-3H]methionine ([C3H3]Met) labeling was performed essentially as described previously (49, 63). Cells were grown in synthetic complete medium (lacking methionine) to logarithmic phase and then treated with rapamycin or drug vehicle alone for 45 min. Subsequently, 75 μCi of [C3H3]Met (70 to 85 Ci/mmol, 185 MBq/ml; Amersham Pharmacia Biotech) was added to 0.75 optical density at 600 nm equivalent of cells, and the samples were incubated for 5 min at 30°C (typically in a reaction volume of 1.8 ml) to allow the incorporation of label into nascent rRNA and tRNA transcripts. Then 10% of the cells were collected on Whatman glass microfiber filters to control for label uptake. The remaining 90% were harvested by centrifugation, washed with water, and subjected to total-RNA preparation by standard methods (11). Uptake counts on the filters and isolated total RNA were subsequently subjected to analysis in a scintillation counter. Incorporated RNA counts were normalized to the uptake of the label by the corresponding cells. We noticed that sit4 mutant cells reproducibly displayed reduced label uptake in comparison to wild-type cells.
Autophagy (alkaline phosphatase) assay.
Progression of autophagy was analyzed by the increase of alkaline phosphatase activity in cells expressing a cytosolic proform of the alkaline phosphatase protein (PHO8Δ60) with 55 mM α-naphthyl phosphate as the substrate. The protein concentration of samples after glass bead extraction in assay buffer (250 mM Tris-HCl [pH 9.0], 10 mM MgSO4, 10 μM ZnSO4) was determined by the Bio-Rad microassay. The assay was basically performed as described previously (46), with samples being incubated for 20 min at 30°C and the fluorescence intensity of emission at 472 nm being monitored after excitation at 345 nm. For each sample, the alkaline phosphatase activity determined was normalized to the corresponding protein concentration.
RESULTS
Activation of the RAS/cAMP pathway confers resistance to rapamycin.
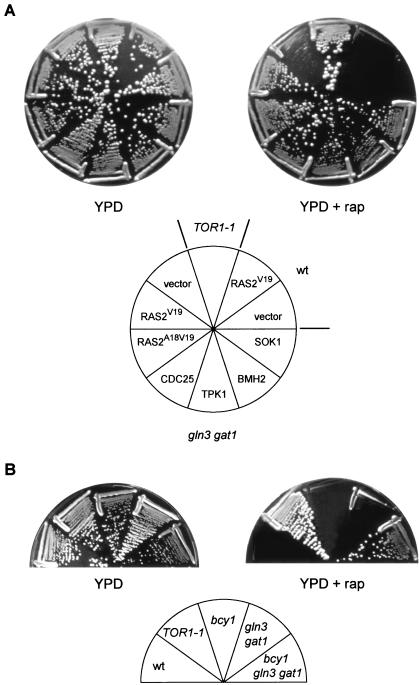
Yeast cells lacking the TOR-controlled GATA transcription factors GLN3 and GAT1 are only weakly resistant to the growth-inhibitory properties of rapamycin (2) (Fig. 1). Thus, rapamycin must still inhibit some growth-related processes in gln3 gat1 mutant cells. To identify such processes, we conducted a selection for multicopy suppressors that confer increased resistance to rapamycin in a gln3 gat1 mutant background. Essentially all the characterized multicopy suppressors encoded activators of the RAS/cAMP pathway or proteins functionally linked to the RAS/cAMP pathway. The suppressors included the RAS guanine nucleotide exchange factor CDC25, the PKA catalytic subunit TPK1, the previously isolated PKA suppressor of unknown function SOK1 (61), and the 14-3-3-protein BMH2 (Fig. 1A). We subsequently observed that activation of the RAS/cAMP pathway by expression of a constitutively active RAS2 (RAS2Val19 or RAS2Ala18Val19) or GPA2 (GPA2R273A) also confers increased rapamycin resistance in a gln3 gat1 mutant (Fig. 1A and data not shown for GPA2). None of the above suppressors conferred significant rapamycin resistance in our wild-type strain (shown for RAS2Val19 in Fig. 1A), indicating that activation of the RAS/cAMP pathway can confer resistance to rapamycin (200 ng/ml) only in the absence of the TOR-controlled factors GLN3 and GAT1.
FIG. 1.
Activation of the RAS/cAMP pathway confers resistance to rapamycin. (A) Wild-type (wt, TB50a) cells transformed with empty vector or pTS118 (RAS2V19), TOR1-1 (JH11-1C) cells, and gln3 gat1 (TB105-3B) cells transformed with empty vector, pTS118 (RAS2V19), pTS119 (RAS2A18V19), pTB424 (CDC25), pTB439 (TPK1), pTB419 (BMH2), or pTB429 (SOK1) were incubated at 30°C for 2.5 days on YPD medium with or without rapamycin (rap; 200 ng/ml). The mutant TOR1-1 allele encodes a protein that is insensitive to rapamycin; thus, TOR1-1 cells are fully resistant to rapamycin. (B) Wild-type (wt, TB50a), TOR1-1 (JH11-1C), bcy1 (TS141), gln3 gat1 (TB105-3B), and bcy1 gln3 gat1 (TS160-6C) cells were incubated at 30°C for 2.5 days on YPD medium with or without rapamycin (200 ng/ml).
Consistent with the above findings, deletion of components that antagonize the RAS/cAMP pathway also resulted in increased rapamycin resistance. In particular, deletion of BCY1, encoding the PKA regulatory subunit (60), or deletion of YAK1, encoding a DYRK family kinase and a SOK1 antagonist (19, 61), conferred pronounced rapamycin resistance in a gln3 gat1 mutant (Fig. 1B and data not shown for YAK1). BCY1 and YAK1 deletions by themselves did not confer detectable rapamycin resistance (Fig. 1B and data not shown for YAK1). Thus, constitutive activation of the RAS/cAMP pathway, but only in combination with the loss of GLN3 and GAT1, results in strong resistance to rapamycin. This strong rapamycin resistance is comparable to that conferred by the TOR1-1 allele.
Given the above, one might expect that a reduction in RAS/cAMP signaling would result in rapamycin hypersensitivity. However, contrary to this expectation, loss-of-function mutations in RAS2 (_ras2_Δ) or TPK (_tpk1w1 tpk2_Δ _tpk3_Δ) did not confer rapamycin (1 to 5 ng/ml) hypersensitivity (data not shown). The RAS/cAMP pathway is essential, and the above mutations might not reduce RAS/cAMP signaling sufficiently to uncover a rapamycin-hypersensitive phenotype. It is worth noting that the tpk (_tpk1w1 tpk2_Δ _tpk3_Δ) mutant was originally selected as a strain in which PKA activity is uncoupled from nutrients and has elevated cAMP levels (8).
Activation of the RAS/cAMP pathway prevents rapamycin-induced nuclear translocation of MSN2, STRE-dependent transcription, and accumulation of glycogen.
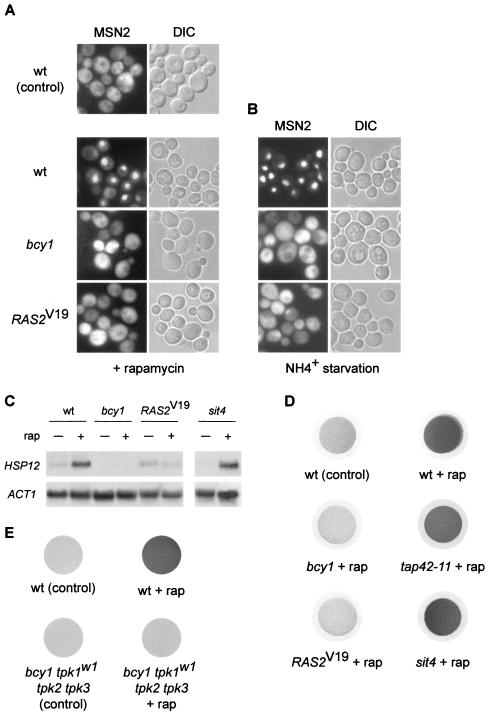
To understand how activation of the RAS/cAMP pathway contributes to increased rapamycin resistance, several TOR-controlled readouts were assayed in bcy1 or _RAS2_Val19 (_RAS2_V19) mutants, i.e., cells with an activated RAS/cAMP pathway. The readouts were examined in a wild-type GLN3 GAT1 strain to prevent indirect effects on the readouts due to rapamycin-resistant growth. We first investigated the localization of MSN2, using an MSN2-GFP fusion. As reported previously (2), MSN2 quickly translocated from the cytoplasm into the nucleus in wild-type cells treated with rapamycin. In cells with an activated RAS/cAMP pathway (bcy1 or _RAS2_Val19), however, the rapamycin-induced translocation of MSN2 to the nucleus was completely abolished (Fig. 2A). The nuclear accumulation of MSN2 typically seen on nitrogen (ammonium) starvation was also fully blocked by activation of the RAS/cAMP pathway (Fig. 2B). Thus, changes in MSN2 localization in response to TOR inactivation by rapamycin treatment or nitrogen starvation are prevented by activation of the RAS/cAMP pathway, suggesting that TOR and the RAS/cAMP pathway share downstream targets.
FIG. 2.
Activation of the RAS/cAMP pathway prevents rapamycin-induced stress response and accumulation of glycogen. (A) Wild-type (wt, TB50a), bcy1 (TS141), and _RAS2_Val19 (TS95-1D/pTS118) cells transformed with pMSN2-GFP were grown in synthetic medium at 30°C, treated with rapamycin or drug vehicle alone for 25 min, and subjected to GFP microscopy. bcy1 or _RAS2_Val19 cells treated with drug vehicle alone showed a cytoplasmic distribution of MSN2-GFP (not shown). The cellular distribution of MSN2-GFP is shown in the left column; the differential interference contrast (DIC) image of the same field of cells is shown in the right column. (B) The strains in panel A were subjected to nitrogen starvation by transferring the cells grown in synthetic medium in the presence of 10 mM (NH4)2SO4 to synthetic medium lacking (NH4)2SO4 for 25 min. Control cells were filtered into synthetic medium containing 10 mM (NH4)2SO4 and displayed a cytoplasmic distribution of MSN2-GFP [see wt (control) panel in panel A]. bcy1 and _RAS2_Val19 cells growing in synthetic medium containing 10 mM (NH4)2SO4 showed a cytoplasmic distribution of MSN2-GFP (not shown). The cellular distribution of MSN2-GFP is shown in the left column; the right column shows the differential interference contrast image of the same field of cells. (C) Wild-type (wt, TB50a), bcy1 (TS141), _RAS2_Val19 (TS95-1D/pTS118), and sit4 (TS65-2D) cells were pregrown in synthetic medium, shifted to YPD, grown for three or four generations, and treated with drug vehicle or rapamycin for 25 min. Total RNA was probed with 32P-labeled DNA probes specific for HSP12 and for actin (ACT1) as the loading control. (D) Wild-type (wt, TB50a), bcy1 (TS141), _RAS2_Val19 (TS95-1D/pTS118), tap42-11 (TS83-6A), and sit4 (TS65-2D) cells were grown in complete synthetic medium (at 30°C, except for TS83-6A, which was grown at 24°C) and treated with rapamycin or drug vehicle alone for 5 h. Cells collected on filters were subsequently exposed to iodine vapor to stain glycogen. (E) Wild-type (wt, SP1) and bcy1 tpk1w1 tpk2 tpk3 (RS13-58A-1) cells were grown in complete synthetic medium, treated with rapamycin or drug vehicle, and stained for glycogen as described for panel D.
MSN2 and its homologue MSN4 control the transcription of several genes whose products are involved in diverse stress responses. Binding of MSN2 and MSN4 to the promoter of these “stress genes” is mediated by a STRE promoter element (41, 55, 58). To confirm that an activated RAS/cAMP pathway blocks rapamycin-induced, MSN-dependent transcriptional activity, we analyzed the transcript levels of HSP12. The HSP12 gene harbors several STRE elements, and its induction upon diverse stresses is MSN dependent (41, 55). As expected from the above findings, HSP12 was induced in wild-type cells on rapamycin treatment but not in bcy1 or _RAS2_Val19 cells (Fig. 2C). Thus, activation of the RAS/cAMP pathway indeed prevents the rapamycin-induced transcription of stress genes.
Accumulation of the storage carbohydrate glycogen usually occurs on nutrient starvation and is an indicator of stationary phase. Barbet et al. have reported that cells treated with rapamycin strongly accumulate glycogen (1). As another TOR readout, we tested whether cells lacking BCY1 or expressing RAS2Val19 accumulate glycogen on rapamycin treatment. Wild-type, bcy1, and _RAS2_Val19 cells were treated with rapamycin and stained for glycogen by using iodine vapor. Whereas wild-type cells treated with rapamycin stained dark brown (indicating the accumulation of glycogen), bcy1 or _RAS2_Val19 cells showed no glycogen accumulation (Fig. 2D). The lack of rapamycin-induced glycogen accumulation by bcy1 or _RAS2_Val19 cells is contrary to a previous report (1), a discrepancy possibly due to strain variation.
To determine whether the repression of rapamycin-induced readouts by activation of RAS/cAMP signaling occurs only in strains with very elevated levels of PKA activity (bcy1 or _RAS2_Val19), we also investigated glycogen accumulation in bcy1 cells carrying a mutant TPK1 allele (tpk1w1) and lacking TPK2 and TPK3 (_bcy1 tpk1w1 tpk2_Δ _tpk3_Δ). These cells still display constitutive but not excessively elevated PKA activity, since they lack two of the three TPK genes and the tpk1w1 allele encodes a debilitated kinase (8). Similar to bcy1 or _RAS2_Val19 cells, _bcy1 tpk1w1 tpk2_Δ _tpk3_Δ cells did not accumulate glycogen following rapamycin treatment whereas the isogenic (parental) wild-type strain displayed strong rapamycin-induced glycogen accumulation (Fig. 2E). Thus, even moderate elevation of PKA activity is sufficient to prevent rapamycin-induced glycogen accumulation. This finding is in good agreement with our above observation that nonactivated alleles, such as wild-type TPK1 and SOK1, confer rapamycin resistance (Fig. 1).
In summary, activation of the RAS/cAMP pathway is sufficient to largely prevent the stress response and stationary-phase indicators associated with inactivation of TOR signaling. Cutler et al. have reported that activation of the RAS/cAMP pathway also prevents a rapamycin-induced block of pseudohyphal growth (14).
Activation of the RAS/cAMP pathway partly represses rapamycin-induced autophagy.
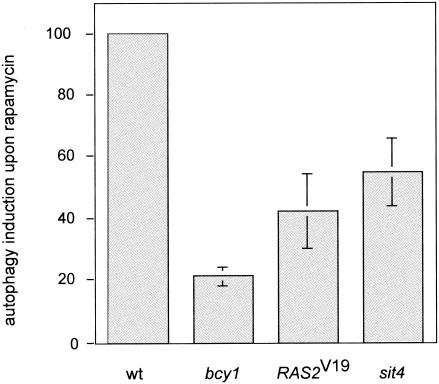
Autophagy, the bulk degradation of ribosomes and other cytoplasmic constituents on nutrient starvation, is negatively controlled by TOR via the APG1-APG13 complex (32, 46). We investigated whether activation of the RAS/cAMP pathway prevents rapamycin-induced autophagy. To monitor autophagy, we took advantage of a cytoplasmic proform of alkaline phosphatase (PHO8Δ60) (45). As reported previously (46), treatment of wild-type yeast cells with rapamycin resulted in a significant induction (approximately 10-fold, considered 100%) of autophagy, as assayed by alkaline phosphatase activity. Autophagy was induced only 20 or 40% in rapamycin-treated bcy1 or _RAS2_Val19 cells, respectively, indicating that activation of the RAS/cAMP pathway partly prevents autophagic processes induced on inactivation of TOR (Fig. 3). These results are consistent with previous reports demonstrating that exogenous cAMP prevents starvation- or rapamycin-mediated induction of autophagy (46).
FIG. 3.
Activation of the RAS/cAMP pathway prevents rapamycin-induced autophagy. Wild-type (wt, TS139), bcy1 (TS143), _RAS2_Val19 (TS140/pTS118), and sit4 (TS145-1C) cells (all expressing the modified alkaline phosphatase form PHO8Δ60) were grown in synthetic complete medium and treated with rapamycin or drug vehicle for 3 h. They were then subjected to alkaline phosphatase (autophagy) assay as described in Materials and Methods. Shown is the induction of autophagic activity on rapamycin treatment, set to 100 (%) for wild-type cells. Means and standard deviations of the means of at least three assays were plotted.
Activation of the RAS/cAMP pathway prevents rapamycin-induced down-regulation of ribosome biogenesis.
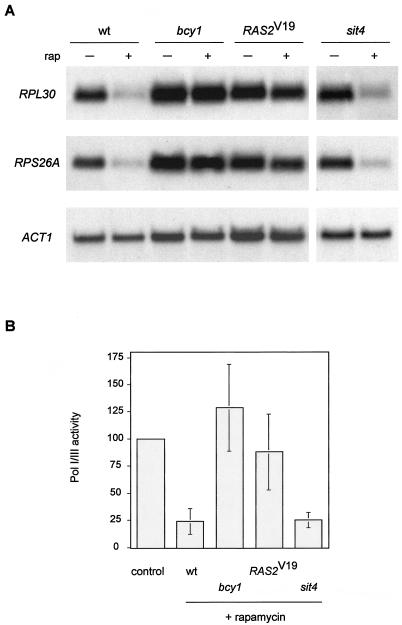
Rapamycin treatment, similar to nutrient depletion, leads to a rapid and pronounced down-regulation of Pol II-dependent ribosomal protein (RP) genes, a down-regulation of Pol I- and Pol III-dependent rRNA and tRNA genes (9, 25, 49, 67), and a down-regulation in rRNA processing (49). Thus, TOR signaling broadly controls ribosome biogenesis. The RAS/cAMP pathway also controls ribosome biogenesis, at least at the level of RP gene transcription (62). We examined if activation of the RAS/cAMP pathway blocks the rapamycin-induced down-regulation of RP gene transcription. Wild-type, bcy1, and _RAS2_Val19 cells were treated with rapamycin, and total RNA was extracted and probed for the two mRNAs encoding the large and small ribosomal subunit proteins RPL30 and RPS26A, respectively. Whereas wild-type cells displayed a pronounced down-regulation of both RP gene transcripts after rapamycin treatment, a similar down-regulation was absent or only very weakly detected in bcy1 or _RAS2_Val19 cells (Fig. 4A).
FIG. 4.
Activation of the RAS/cAMP pathway prevents rapamycin-induced down-regulation of ribosome biogenesis. (A) Wild-type (wt, TB50a), bcy1 (TS141), _RAS2_Val19 (TS95-1D/pTS118), and sit4 (TS65-2D) cells were grown in synthetic complete medium and treated with drug vehicle or rapamycin for 45 min. Total RNA was probed with 32P-labeled DNA probes specific for the RP genes RPL30, and RPS26A and also for actin (ACT1) as the loading control. (B) The strains described in panel A were grown in synthetic complete medium and treated with drug vehicle or rapamycin for 45 min. They were then labeled for 5 min with l-[_methyl_-3H]methionine, and extracted total RNA was subjected to scintillation counting. Values obtained for nucleic acid-incorporated radioactivity were normalized to the values for label uptake in each reaction. The normalized value for drug vehicle-treated cells (control), which was similar for all strains used in this experiment, was defined as 100(%) of Pol I and Pol III activity. Means and standard deviations of the mean of at least three assays were plotted.
To monitor Pol I- and Pol III-dependent synthesis of rRNA and tRNA, we assayed biosynthetic methylation of precursor rRNA and tRNA. The assay consisted of selective in vivo labeling of rRNA and tRNA with [C3H3]Met. Label incorporation into nascent Pol I and III transcripts was calculated by subjecting extracted total RNA to scintillation counting and normalizing the obtained values to the corresponding values for label uptake. As reported previously, uptake of the label was only slightly affected by rapamycin treatment (reference 49 and data not shown). Also as shown previously, label incorporation into nascent rRNA and tRNA decreased approximately fourfold in rapamycin-treated wild-type cells (49) (Fig. 4B). In contrast, rapamycin-treated bcy1 and _RAS2_Val19 cells displayed little to no decrease in label incorporation into nascent rRNA and tRNA. These results suggest that Pol I and Pol III activity in bcy1 or _RAS2_Val19 cells, in contrast to that in wild-type cells, persists despite TOR inactivation. Thus, activation of the RAS/cAMP pathway contributes to growth of cells on rapamycin-containing medium by preventing the down-regulation of ribosome biogenesis via the maintenance of RP gene, rRNA and tRNA transcription.
Activation of the RAS/cAMP pathway prevents rapamycin-induced down-regulation of the glucose transporter HXT1.
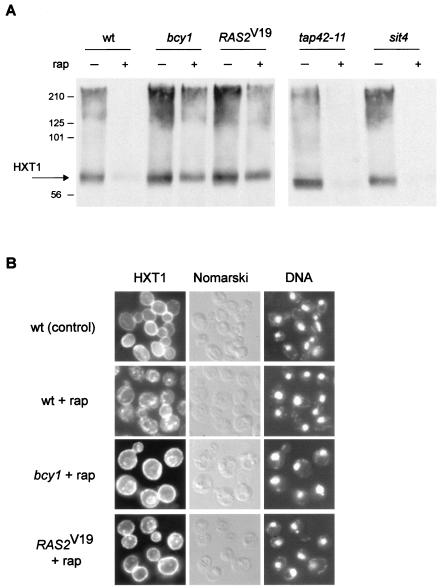
Yeast cells possess a large number of functionally redundant HXT genes that encode hexose (glucose) transporters belonging to the major facilitator superfamily of transporters. Via the combined action of different regulatory mechanisms, only the glucose transporters appropriate for the amount of extracellular glucose are expressed and/or maintained at any given time (reviewed in reference 47). The HXT1 gene encodes a major, low-affinity, high-capacity glucose transporter that is present when glucose is abundant, i.e., under favorable growth conditions. TOR controls, via the kinase NPR1, the sorting and thereby the stability of the high-affinity tryptophan permease, TAT2. On TOR inactivation, TAT2 is targeted to the vacuole and degraded (3). To investigate if TOR signaling also regulates the major hexose transporter, HXT1, we examined the expression of C-terminally HA-tagged HXT1 in rapamycin-treated cells by immunoblotting. Rapamycin treatment resulted in loss of the HXT1 signal, indicating that TOR maintains HXT1 (Fig. 5A). This finding suggests that HXT1 is a (heretofore unknown) target of TOR signaling. We also examined the subcellular localization of HXT1 in rapamycin-treated cells by indirect immunofluorescence of whole fixed cells. Whereas HXT1 was located mainly at the plasma membrane (rim staining) in control cells, rapamycin treatment resulted in a significant reduction in the amount of plasma membrane-localized HXT1 and the appearance of a signal in intracellular compartments, possibly the vacuole or other “degradation” compartments (Fig. 5B). Interestingly, the rapamycin-induced down-regulation of HXT1 was blocked in sla2 or rsp5 mutant cells, as observed by immunoblotting (data not shown). SLA2 (also called END4) is required for the endocytosis of several plasma membrane proteins (50), and RSP5 (also called NPI1) encodes a ubiquitin protein ligase (26), indicating that rapamycin-induced HXT1 down-regulation requires ubiquitination and endocytosis. Therefore, TOR appears to broadly control the stability of nutrient transporters, possibly to adjust the import rate of diverse nutrients to the growth rate. Interestingly, mTOR also controls several mammalian nutrient transporters, possibly including the principal glucose transporter Glut1 (18).
FIG. 5.
Activation of the RAS/cAMP pathway prevents down-regulation of the glucose transporter HXT1. (A) Wild-type (wt, TB50a), bcy1 (TS141), _RAS2_Val19 (TS95-1D/pTS120), tap42-11 (TS83-6A), and sit4 (TS65-2D) cells transformed with pTB374 (HXT1-HA3, 2μm) were grown in synthetic complete medium (tap42-11 at 24°C, other strains at 30°C) and treated with drug vehicle or rapamycin for 150 min. They were then harvested and processed for immunoblotting. The predicted molecular mass of HXT1-HA3 is approximately 65 kDa; the molecular reason for the broad HXT1 signal is currently unknown. The molecular mass standard (in kilodaltons) is indicated on the left. (B) Wild-type (wt, TB50a), bcy1 (TS141), and _RAS2_Val19 (TS95-1D/pTS118) cells transformed with pTB380 (HXT1-HA3, CEN) were grown in synthetic complete medium and treated with drug vehicle or rapamycin for 75 min. They were then subjected to indirect immunofluorescence analysis to visualize HXT1. bcy1 and _RAS2_Val19 cells treated with drug vehicle alone showed a plasma membrane distribution of HXT1 similar to the one in wild-type control cells (not shown). The cellular distribution of HXT1-HA3 is shown in the left column; the same field of cells is shown in the middle (Nomarski) and right (DNA; DAPI staining) columns.
We investigated whether activation of the RAS/cAMP pathway counters the rapamycin-induced down-regulation of HXT1. As assayed by immunoblotting, bcy1 or _RAS2_Val19 prevented the down-regulation of HXT1 (Fig. 5A). Also, as detected by immunofluorescence on whole cells, bcy1 or _RAS2_Val19 largely, but not completely, prevented the internalization of HXT1 (Fig. 5B). Taken together, these results suggest that activation of the RAS/cAMP prevents the down-regulation of HXT1 following TOR inactivation, thus providing yet another possible reason for the growth of bcy1 or _RAS2_Val19 cells on rapamycin-containing medium.
TOR-controlled, TAP42/SITA-mediated events and TOR-controlled, RAS/cAMP-related readouts define distinct rapamycin-sensitive signaling pathways.
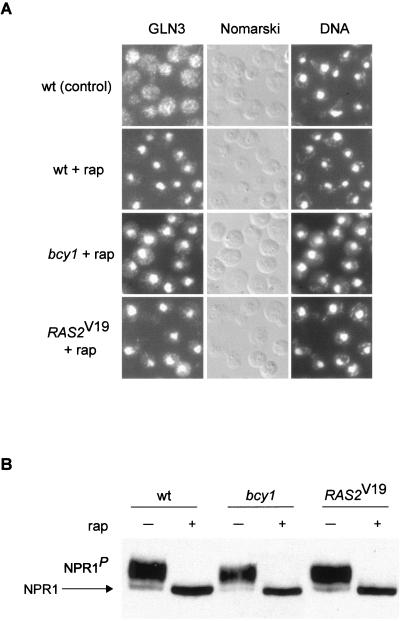
On TOR inactivation by nutrient depletion or rapamycin treatment, the TAP42 inhibitor protein dissociates from the phosphatase SIT4, resulting in SIT4 activation and, subsequently, SIT4-mediated dephosphorylation of the GATA factor GLN3 and the kinase NPR1 (2, 29). To further understand the specificity of the effects of RAS/cAMP signaling on TOR-controlled readouts, we investigated whether activation of the RAS/cAMP pathway affects the SIT4-dependent readouts GLN3 and NPR1. As described previously (2), GLN3 efficiently translocated from the cytoplasm to the nucleus in rapamycin-treated wild-type cells (Fig. 6A). In bcy1 or _RAS2_Val19 cells, the nuclear translocation of GLN3 observed upon rapamycin treatment was similar to that observed in wild-type cells (Fig. 6A). Thus, activation of the RAS/cAMP pathway does not affect TOR signaling through TAP42 and SIT4 to GLN3. This accounts for the earlier observation that an activated RAS/cAMP pathway confers rapamycin resistance only in a gln3 (and gat1) mutant background.
FIG. 6.
Rapamycin-induced TAP42/SIT4-regulated events are not affected by activation of the RAS/cAMP pathway. (A) Wild-type (wt, HM4-5D), bcy1 (TS178-1C), and _RAS2_Val19 (TS177-7B/pTS118) cells (all expressing GLN3-myc13) were grown in synthetic complete medium and treated with drug vehicle or rapamycin for 20 min. They were then subjected to indirect immunofluorescence analysis to visualize GLN3. bcy1 and _RAS2_Val19 cells treated with drug vehicle alone showed a cytoplasmic distribution of GLN3 similar to the one in wild-type control cells (not shown). The cellular distribution of GLN3-myc13 is shown in the left column; the same field of cells is shown in the middle (Nomarski) and right (DNA; DAPI staining) columns. (B) Wild-type (wt, TB50a), bcy1 (TS141), and _RAS2_Val19 (TS95-1D/pTS119) cells transformed with pAS103 (HA-NPR1) were grown in synthetic complete medium to logarithmic phase and treated with drug vehicle or rapamycin for 15 min. They were then harvested and processed for immunoblotting. The more rapidly migrating form of NPR1 represents the dephosphorylated species (54). P indicates phosphorylation.
To test whether the rapamycin-induced, SIT4-controlled dephosphorylation of NPR1 is affected by the RAS/cAMP pathway, the phosphorylation state of NPR1 in rapamycin-treated wild-type, bcy1, and _RAS2_Val19 cells was monitored. Dephosphorylation of NPR1, assayed by detection of altered electrophoretic mobility of NPR1 (54), occurred to a similar extent in wild-type, bcy1, and _RAS2_Val19 cells (Fig. 6B).
Moreover, and in good agreement with the results obtained for NPR1, an activated RAS/cAMP pathway (bcy1 and _RAS2_Val19) did not prevent the rapamycin-induced degradation of the tryptophan permease TAT2 (data not shown). This again indicates that rapamycin-induced, TAP42/SIT4-regulated events are not affected by activation of the RAS/cAMP pathway. Given the apparent difference in TOR signaling to HXT1 (RAS/cAMP) and TAT2 (TAP42) stability, the common involvement of ubiquitination (RSP5) and endocytosis (SLA2) in the degradation of HXT1 (see above) and TAT2 (3) may reflect a general rather than a TOR-specific requirement.
In summary, activation of the RAS/cAMP pathway prevents some rapamycin-induced readouts, such as MSN2 translocation, ribosome biogenesis, and HXT1 down-regulation, but does not overcome other rapamycin-induced readouts, such as GLN3 translocation, NPR1 dephosphorylation, and TAT2 degradation. The readouts that are not prevented by RAS/cAMP correspond to those controlled by TAP42/SIT4. The different signaling components for different TOR readouts suggest a signaling bifurcation downstream of TOR. To examine this suggestion, we investigated whether the absence of SIT4 (or, in some cases, the presence of a rapamycin-resistance allele of TAP42, tap42-11 [16]) affects the TOR-controlled readouts that were demonstrated above to be prevented by activation of the RAS/cAMP pathway. Rapamycin-induced translocation of MSN2, in contrast to GLN3 relocalization or NPR1 dephosphorylation, was not blocked in a sit4 or tap42-11 mutant (reference 2 and data not shown). In agreement with this finding, sit4 and tap42-11 mutants displayed wild-type-like glycogen accumulation and (for sit4) HSP12 induction on rapamycin treatment (Fig. 2C and D). Furthermore, transcription of the RP genes RPL30 and RPS26A was reduced in rapamycin-treated sit4 cells, as in wild-type cells (Fig. 4A). Moreover, Pol I and Pol III activity, assayed by biosynthetic methylation of rRNA and tRNA, was reduced approximately fourfold in sit4 mutant cells on rapamycin treatment, a reduction similar to that obtained with wild-type cells (Fig. 4B). Finally, rapamycin-induced down-regulation of the hexose transporter HXT1 occurred in sit4 and tap42-11 cells (Fig. 5A). Therefore, a sit4 mutation did not affect the readouts shown above to be affected by the RAS/cAMP pathway. Taken together, these results suggest that TAP42/SIT4 and RAS/cAMP signaling do indeed function in separate rapamycin-sensitive signaling pathways downstream of TOR.
Interestingly, rapamycin-induced autophagy was reduced in sit4, bcy1, and _RAS2_Val19 cells. In the absence of SIT4, rapamycin treatment induced autophagy to about 55% of wild-type levels (Fig. 3), indicating that a sit4 mutation blocks autophagy but does so less effectively than does activation of the RAS/cAMP pathway. Therefore, contrary to the seemingly separate control of different TOR readouts by TAP42/SIT4 and RAS/cAMP described above, SIT4 and RAS/cAMP may jointly control autophagy. Given the complexity of the autophagic process (formation and trafficking of large membrane structures), a contribution of both pathways to the control of autophagy is plausible (10, 32, 46).
TOR controls the localization of TPK1 and YAK1.
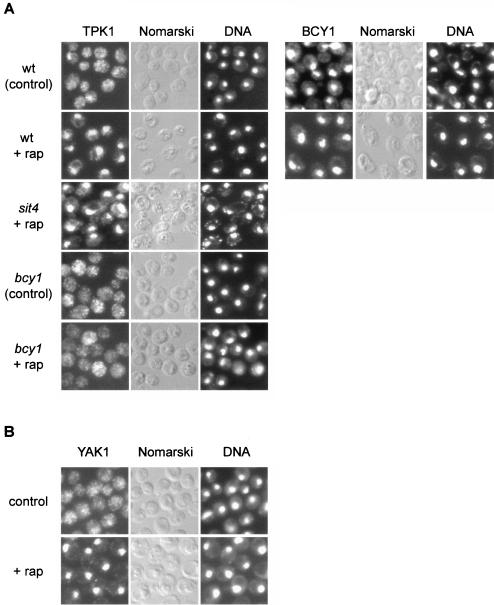
Recent studies have established that PKA localization in S. cerevisiae is regulated by cAMP levels. Deprivation of cAMP in rapidly growing yeast cells results in a pronounced nuclear accumulation of the PKA catalytic subunit TPK1. cAMP addition, which activates PKA via dissociation of the TPK-BCY1 complex, leads to rapid relocalization of TPK1 to the cytoplasm. The regulatory (kinase-inhibitory) BCY1 subunit remains localized in the nucleus in the presence and absence of cAMP. Hence, catalytically inactive PKA appears to reside in the nucleus (22, 23). To investigate how TOR and the RAS/cAMP pathway cooperate in signaling to cell growth readouts, we examined TPK1 and BCY1 localization in rapamycin-treated cells. Single-copy, HA-tagged versions of TPK1 and BCY1 expressed under the control of their own promoters were constructed and used to determine the subcellular localization of TPK1 and BCY1. In control cells, TPK1 was generally distributed throughout the cell; on rapamycin treatment, TPK1 accumulated in the nucleus, as observed previously in cAMP-deprived cells (Fig. 7A). The localization of BCY1 was not significantly altered on rapamycin treatment; i.e., BCY1 remained nuclear (Fig. 7A). Thus, similar to cAMP deprivation, rapamycin treatment induces the nuclear accumulation and presumably BCY1 binding of TPK1, suggesting that TOR maintains PKA activity. That TOR maintains PKA activity also explains the suppression of a TOR deficiency by activation of the RAS/cAMP pathway.
FIG. 7.
TOR controls the localization of the kinases TPK1 and YAK1. (A) Wild-type (wt, TB50a), bcy1 (TS141), and sit4 (TS65-2D) cells transformed with pTS134 (HA2-TPK1) or wild-type cells transformed with pTS137 (HA-BCY1) were pregrown in selective synthetic medium, shifted to YPD, grown for three or four generations, and treated with drug vehicle or rapamycin for 45 min. They were then subjected to indirect immunofluorescence analysis to visualize TPK1 or BCY1, respectively. The exposure time used to visualize TPK1 in bcy1 cells was approximately fourfold longer than for wild-type cells. The cellular distribution of HA2-TPK1 or HA-BCY1, respectively, is shown in the left column; the same field of cells is shown in the middle (Nomarski) and right (DNA; DAPI staining) columns. (B) Wild-type cells expressing YAK1-myc13 (TS129-8C) were grown to logarithmic phase in YPD medium and treated with drug vehicle or rapamycin for 30 min. They were then subjected to indirect immunofluorescence analysis to visualize YAK1. The cellular distribution of YAK1-myc13 is shown in the left column; the same field of cells is shown in the middle (Nomarski) and right (DNA; DAPI staining) columns.
To characterize the rapamycin-induced nuclear accumulation of TPK1, we investigated the localization of TPK1 in rapamycin-treated cells lacking SIT4 or BCY1. In rapamycin-treated sit4 cells, as in wild-type cells, TPK1 accumulated in the nucleus, indicating that the nuclear translocation of TPK1 in response to TOR inactivation occurs independently of SIT4 (Fig. 7A). In rapamycin-treated bcy1 cells, TPK1 remained largely cytoplasmic on rapamycin treatment (Fig. 7A). This finding is in good agreement with a previous report suggesting that nuclear accumulation of TPK1 depends on an interaction with nuclear BCY1 (22).
We also observed that HA-TPK1 expression is reproducibly and significantly reduced in bcy1 cells. The TPK1 promoter contains several putative MSN-binding STRE elements (43). In a bcy1 mutant, MSN2 and MSN4 remain cytoplasmic (see above) and thus presumably unable to activate TPK1 expression.
As mentioned above, deletion of YAK1 in a gln3 gat1 mutant background also confers resistance to rapamycin. Moreover, YAK1 accumulates in the nucleus in response to nutrient starvation (42). To investigate if YAK1, like TPK1, is controlled by TOR, we examined YAK1 localization in rapamycin-treated cells. Whereas YAK1 was generally distributed throughout the cell in control cells, we observed a pronounced nuclear YAK1 signal in rapamycin-treated cells, indicating that YAK1 localization is controlled by TOR (Fig. 7B). In summary, TOR controls the localization, and therefore possibly the activity, of both TPK1 and YAK1, suggesting that TOR signals through the RAS/cAMP pathway.
DISCUSSION
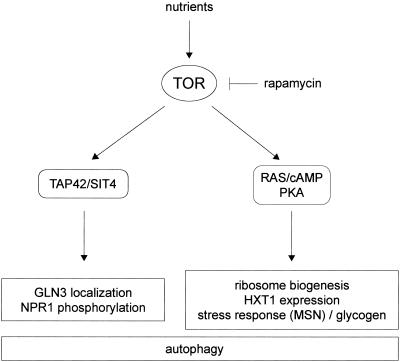
We report that constitutive activation of the RAS/cAMP pathway blocks several rapamycin-induced responses and thus confers rapamycin resistant growth. The rapamycin-induced responses include down-regulation of ribosome biogenesis, decreased expression of the glucose transporter HXT1, activation of stress (STRE) transcription, and induction of autophagy. Interestingly, the TOR effectors TAP42 and the PP2A-related phosphatase SIT4 are not involved in the control of most of these RAS/cAMP-regulated TOR readouts. Conversely, TAP42/SIT4-dependent events, such as GLN3 translocation or NPR1 dephosphorylation, are still rapamycin sensitive in cells with activated RAS/cAMP signaling. These findings suggest that rapamycin-sensitive TOR signaling may bifurcate into (at least) two separate effector pathways—TAP42/SIT4 and RAS/cAMP—each controlling a specific subset of TOR readouts (Fig. 8). The notion of two separate rapamycin-sensitive pathways downstream of TOR is also supported by the finding that activation of the RAS/cAMP pathway confers rapamycin-resistant growth only in a gln3 gat1 background (Fig. 1).
FIG. 8.
Model of rapamycin-sensitive TOR signaling in the control of cell growth. Rapamycin-sensitive TOR signaling bifurcates into two separate effector pathways, TAP42/SIT4 and RAS/cAMP/PKA. Each effector pathway controls a specific subset of TOR readouts. For details, see Discussion.
How does TOR impinge on RAS/cAMP signaling? The simplest model is that TOR signals through the RAS/cAMP pathway to control a TAP42/SIT4-independent set of readouts, including ribosome biogenesis and HXT1 expression. This model is supported by the key and novel finding that TOR controls the localization and presumably the activity of TPK1 (Fig. 7A). It remains to be determined how TOR controls TPK1 localization. It is also worth nothing that the RAS/cAMP pathway may not be controlled exclusively by TOR. Similar to the intersection of the glucose-responsive GPR1/GPA2 system with the RAS/cAMP pathway (59), TOR may interact with the RAS/cAMP pathway as a nitrogen-responsive input. The mechanism by which TOR interacts with the RAS/cAMP pathway at or upstream of TPK1 is unknown.
An alternative model for how TOR impinges on RAS/cAMP signaling is that the RAS/cAMP pathway and an unknown, rapamycin-sensitive TAP42/SIT4-independent pathway converge on several common targets. This model seems unlikely because it requires that TOR and the RAS/cAMP pathway converge simultaneously on several targets and because of the finding that TOR controls PKA localization (Fig. 7A). However, TOR and the RAS/cAMP pathway may converge at least on the stress transcription factors MSN2 and MSN4. Others have reported that TOR and the RAS/cAMP pathway appear to signal in parallel to MSN2 (20, 21).
The two models described above are not mutually exclusive. TOR may intersect with the RAS/cAMP pathway at more than one level. Cross talk between signaling pathways is a common theme in growth control. Of particular relevance is the link between mTOR signaling and the PI3K/PDK1/PKB pathway in mammalian cells. This link involves both convergence on common targets, such as S6K or 4E-BP, and a regulator-effector relationship in which PKB controls mTOR via the TSC complex (see the introduction) (30). Interestingly, the mTOR and PKA signaling pathways in mammalian cells have also recently been linked as mediators of the cardiomyocyte response to glutamine (65).
Ribosome biogenesis is a major consumer of the cell's energy resources, and its regulation is therefore of particular importance in the context of cell growth control. Growth-controlling transcription of RP genes involves PKA-dependent regulation of the transcription factor RAP1 (34). Depletion of cAMP or nutrient starvation leads to repression of RP mRNA levels. However, it has also been proposed that nutrient availability may induce the expression of RP genes by a mechanism independent of the RAS/cAMP pathway (44). We found that activation of the RAS/cAMP pathway prevents the down-regulation of ribosome biogenesis (RP gene, rRNA and tRNA transcription) on TOR inhibition (Fig. 4). Also, we found that TOR-regulated ribosome biogenesis is independent of SIT4 (Fig. 4), in agreement with the recent report by Duvel et al. that TAP42 inactivation does not affect the expression of RP genes and their rapamycin-induced repression (17). Definition of the critical upstream nutrient signals and dissection of the transcriptional effectors involved in the TOR-mediated control of ribosome biogenesis should provide further insight into how TOR signals in conjunction with the RAS/cAMP pathway to regulate ribosome biogenesis and cell growth.
In summary, our results suggest that TOR, via different effector pathways, signals to different growth-related readouts. Furthermore, our findings link the TOR and RAS/cAMP pathways, the two major nutrient-responsive, growth-controlling pathways in yeast. Elucidation of the precise nature of the relationship between these two pathways and the identification of linking components will provide further insight into how TOR and RAS/cAMP jointly contribute to the control of yeast cell growth.
Acknowledgments
We thank the André, Hirsch, Ohsumi, Riezman, Schüller, Thevelein, and Wigler laboratories for strains and plasmids; members of our laboratory, in particular José L. Crespo and Robbie Loewith, for helpful discussion; and Kelly Tatchell for comments on the manuscript.
This work was supported by grants from the Canton of Basel and the Swiss National Science Foundation to M.N.H.
REFERENCES
- 1.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7**:**25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402**:**689-692. [DOI] [PubMed] [Google Scholar]
- 3.Beck, T., A. Schmidt, and M. N. Hall. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146**:**1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275**:**35727-35733. [DOI] [PubMed] [Google Scholar]
- 5.Bertram, P. G., C. Zeng, J. Thorson, A. S. Shaw, and X. F. Zheng. 1998. The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Curr. Biol. 8**:**1259-1267. [DOI] [PubMed] [Google Scholar]
- 6.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180**:**1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broach, J. R. 1991. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7**:**28-33. [DOI] [PubMed] [Google Scholar]
- 8.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53**:**555-566. [DOI] [PubMed] [Google Scholar]
- 9.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13**:**3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, T. F., P. G. Bertram, W. Ai, and X. F. Zheng. 2001. Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J. Biol. Chem. 276**:**6463-6467. [DOI] [PubMed] [Google Scholar]
- 11.Collart, M. A., and S. Oliviero. 1993. Preparation of yeast RNA, p. 13.12.1-13.12.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 12.Crespo, J. L., and M. N. Hall. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66**:**579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99**:**6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12**:**4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Craene, J. O., O. Soetens, and B. Andre. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276**:**43939-43948. [DOI] [PubMed] [Google Scholar]
- 16.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10**:**1904-1916. [DOI] [PubMed] [Google Scholar]
- 17.Duvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11**:**1467-1478. [DOI] [PubMed] [Google Scholar]
- 18.Edinger, A. L., and C. B. Thompson. 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell 13**:**2276-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrett, S., and J. Broach. 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3**:**1336-1348. [DOI] [PubMed] [Google Scholar]
- 20.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12**:**586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schuller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21**:**135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffioen, G., P. Anghileri, E. Imre, M. D. Baroni, and H. Ruis. 2000. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J. Biol. Chem. 275**:**1449-1456. [DOI] [PubMed] [Google Scholar]
- 23.Griffioen, G., and J. M. Thevelein. 2002. Molecular mechanisms controlling the localisation of protein kinase A. Curr. Genet. 41**:**199-207. [DOI] [PubMed] [Google Scholar]
- 24.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110**:**177-189. [DOI] [PubMed] [Google Scholar]
- 25.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96**:**14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hein, C., J. Y. Springael, C. Volland, R. Haguenauer-Tsapis, and B. Andre. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18**:**77-87. [DOI] [PubMed] [Google Scholar]
- 27.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5**:**105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153**:**163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8**:**1017-1026. [DOI] [PubMed] [Google Scholar]
- 30.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell. Biol. 4**:**117-126. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18**:**2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150**:**1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110**:**163-175. [DOI] [PubMed] [Google Scholar]
- 34.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14**:**1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151**:**863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32**:**1002-1012. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Z., T. Sekito, C. B. Epstein, and R. A. Butow. 2001. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 20**:**7209-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10**:**457-468. [DOI] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14**:**953-961. [DOI] [PubMed] [Google Scholar]
- 40.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154**:**609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15**:**2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 42.Moriya, H., Y. Shimizu-Yoshida, A. Omori, S. Iwashita, M. Katoh, and A. Sakai. 2001. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 15**:**1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moskvina, E., C. Schuller, C. T. Maurer, W. H. Mager, and H. Ruis. 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14**:**1041-1050. [DOI] [PubMed] [Google Scholar]
- 44.Neuman-Silberberg, F. S., S. Bhattacharya, and J. R. Broach. 1995. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell. Biol. 15**:**3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noda, T., A. Matsuura, Y. Wada, and Y. Ohsumi. 1995. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210**:**126-132. [DOI] [PubMed] [Google Scholar]
- 46.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273**:**3963-3969. [DOI] [PubMed] [Google Scholar]
- 47.Ozcan, S., and M. Johnston. 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63**:**554-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedruzzi, I., N. Burckert, P. Egger, and C. De Virgilio. 2000. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 19**:**2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10**:**987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120**:**55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinders, A., N. Burckert, T. Boller, A. Wiemken, and C. De Virgilio. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12**:**2943-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276**:**9583-9586. [DOI] [PubMed] [Google Scholar]
- 53.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103**:**253-262. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17**:**6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93**:**5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10**:**1574-1581. [DOI] [PubMed] [Google Scholar]
- 57.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194**:**3-21. [DOI] [PubMed] [Google Scholar]
- 58.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17**:**3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33**:**904-918. [DOI] [PubMed] [Google Scholar]
- 60.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7**:**1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward, M. P., and S. Garrett. 1994. Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene. Mol. Cell. Biol. 14**:**5619-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24**:**437-440. [DOI] [PubMed] [Google Scholar]
- 63.Warner, J. R. 1991. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 194**:**423-428. [DOI] [PubMed] [Google Scholar]
- 64.Wedaman, K. P., A. Reinke, S. Anderson, J. Yates, III, J. M. McCaffery, and T. Powers. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14**:**1204-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia, Y., H. Y. Wen, M. E. Young, P. H. Guthrie, H. Taegtmeyer, and R. E. Kellems. 2003. Mammalian target of rapamycin and protein kinase A signaling mediate the cardiac transcriptional response to glutamine. J. Biol. Chem. 278**:**13143-13150. [DOI] [PubMed] [Google Scholar]
- 66.Xue, Y., M. Battle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17**:**1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18**:**4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]