Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer and prognosis: cohort study and literature review (original) (raw)
. Author manuscript; available in PMC: 2011 Dec 1.
Published in final edited form as: J Pathol. 2010 Dec;222(4):350–366. doi: 10.1002/path.2774
Abstract
The abundance of tumour-infiltrating T-cells has been associated with microsatellite instability (MSI) and a favorable prognosis in colorectal cancer. Because molecular alterations in colon cancer including MSI, the CpG island methylator phenotype (CIMP), BRAF mutation and global DNA hypomethylation have been associated with clinical outcome, potential confounding by these molecular features needs to be controlled when assessing the prognostic significance of tumour-infiltrating T-cells. We utilized a database of clinically and molecularly-annotated colon and rectal carcinoma cases (N=768; stage I-IV) in two prospective cohort studies (the Nurses' Health Study and the Health Professionals Follow-up Study). Using tissue microarray and automated Ariol image analysis system, we quantified densities of CD3+, CD8+, CD45RO+ (PTPRC) and FOXP3+-cells within neoplastic epithelial areas. We used Cox proportional hazard models to compute mortality hazard ratio, adjusting for clinical and molecular features including KRAS, BRAF, and PIK3CA mutations, MSI, CIMP and LINE-1 hypomethylation. The densities of CD8+, CD45RO+ and FOXP3+-cells were significantly associated with patient survival in univariate analyses (Ptrend<0.007). In the multivariate model, tumour-infiltrating CD45RO+-cell density, but not that of CD3+, CD8+ or FOXP3+-cell, was significantly associated with survival (p=0.0032). In multivariate linear regression analysis, MSI-high (p<0.0001) and high-level tumour LINE-1 methylation (p=0.0013) were independently associated with higher CD45RO+-cell density. Nonetheless, the survival benefit associated with CD45RO+-cells was independent of MSI and LINE-1 status. In conclusion, tumour-infiltrating CD45RO+-cell density is a prognostic biomarker associated with longer survival of colorectal cancer patients, independent of clinical, pathological and molecular features. In addition, MSI-high and tumour LINE-1 methylation level are independent predictors of CD45RO+-cell density. Our data offer a possible mechanism by which MSI confers an improved clinical outcome, and support efforts to augment host immune response in the tumour microenvironment as a strategy of targeted immunotherapy.
Keywords: colon cancer, reaction, mismatch repair, epigenetics, immunology, therapeutic target, immunity
Introduction
The abundance of tumour-infiltrating T-cells has been associated with improved clinical outcome of colorectal cancer patients (Table 1) [1-23]. Although the exact mechanism remains uncertain, the adaptive immune system may play an important role in suppressing tumour progression [18,24]. Tumour-infiltrating T-cells may be an indicator of host immune response to tumour and an attractive target for immunotherapy [25-28].
Table 1. Studies on tumour-infiltrating T-cell subsets and colorectal cancer patient survival.
| Ref. | Authors (year) | No. of hospitals | Sample size | No. of events | Disease stage | Additional therapy | High density of T-cells (vs. low density) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OS | CS | Image analyzer | T-cell subset analyzed | 5-year CS, OS or DFS, log-rank p value | CS, OS or DFS univariate HR (95% CI)a, p value | CS, OS or DFS multivariate HR (95% CI)a, p value | Variables examined in multivariate analysis | ||||||
| [1] | Naito et al. (1998) | 2 | 131 | - | - | Duke's A-D | - | No | CD8+ | p=0.0003 (OS) | - | 0.61 (0.41-0.89)p=0.011 (OS) | inflammatory cells, invasive pattern, stage, tumour grade |
| [2] | Guidoboni et al. (2001) | 1 | 109 | 37 | - | II-III | No treatment or 5-FU adjuvant chemo therapy | No | CD3+ | p=0.004 (OS) | - | 0.40 (0.19-0.85) (OS) | age, sex, stage |
| CD8+ | p=0.0008 (OS) | - | 0.33 (0.15-0.73) (OS) | ||||||||||
| GZMB+ | p=0.0001 (OS) | - | 0.23 (0.10-0.50) (OS) | ||||||||||
| [3] | Oberg et al. (2002) | 1 | 93 | 59 | 47 | Duke's C | No treatment or chemo or radiation therapy | No | CD8+ | 77% (vs. 38%)p=0.011 (CS) | - | - | - |
| CD45RO+ | 66% (vs. 33%)p=0.002 (CS) | - | - | ||||||||||
| CD68+ | 60% (vs. 37%)p=0.033 (CS) | - | - | ||||||||||
| [4] | Petty et al. (2002) | 2 | 72 | - | - | I-IV | No treatment or chemo or radiation therapy | No | CD134+ | p=0.02 (CS) | - | - | - |
| [5] | Diederichsen et al. (2003) | 2 | 41 | 25 | - | Duke's A-D | No treatment | No | CD4+/CD8+ ratio | 22% (vs. 61%) (OS) | - | p=0.046 (OS) | age, stage |
| [6] | Funada et al. (2003) | - | 97 | - | - | - | - | No | CD8+ | - | p=0.01 (OS) | - | - |
| [7] | Prall et al. (2004) | 1 | 152 | - | - | III | 5FU/LV adjuvant chemo or radiation therapy | No | CD8+ | p<0.00l (CS) | - | - | - |
| [8] | Menon et al. (2004) | - | 93 | - | - | II-III | - | No | CD8+ | p=0.03 (DFS) | - | 0.56 (0.32-0.99)p=0.04 (DFS) | age, sex, location, stage, tumour grade |
| [9] | Chiba et al. (2004) | 3 | 371 | - | 74 | I-IV | - | No | CD8+ | p<0.0001 (CS) | - | 0.43 (0.23-0.83)p=0.01 (CS) | age, sex, tumour grade, invasive pattern, location, MMR protein, TIL, stage |
| [10] | Pages et al. (2005) | 1 | 336 | 158 | - | Duke's A-D | 5FU based adjuvant chemo therapy | Yes | CD45RO+ | 65%? (vs. 35%?) p<0.00l (OS)72%? (vs. 50%?) p<0.00l (DFS) | - | p=0.02 (OS) | stage(pT, pN, pM) |
| [11] | Baeten et al. (2006) | 1 | 117 | - | - | Duke's A-D | No treatment or radiation therapy | No | CD3+ | p<0.05 (CS) | - | - | |
| CD8+ | p<0.25 (CS) | - | - | - | |||||||||
| CD16+ | p<0.04 (CS) | - | - | ||||||||||
| [12] | Galon et al. (2006) | 1 | 406 | - | - | I-III | 5FU based adjuvant chemo therapy | Yes | CD3+ | 73% (vs. 40%) p<0.0001 (OS)81% (vs. 54%) p=0.0012 (DFS) | - | 0.53 (0.40-0.70) p<0.0001 (OS) 0.42 (0.29-0.60)p<0.0001 (DFS) | stage (pT, pN) |
| CD8+ | 72% (vs. 50%) p<0.0001 (OS)2% (vs. 56%) p=0.0002 (DFS) | - | - | - | |||||||||
| CD45RO+ | 68% (vs. 33%) p<0.0003 (OS) 77% (vs. 37%) p=0.002 (DFS) | - | - | - | |||||||||
| GZMB+ | 72% (vs. 61%) p=0.15 (OS)84% (vs. 68%) p=0.39 (DFS) | - | - | - | |||||||||
| CD3+, CD45RO+ | p<0.0001 (DFS) | - | - | - | |||||||||
| [13] | Zlobec et al. (2008) | 3 | 587 | - | - | I-II (MSS only) | - | No | CD8+ | - | 0.47 (0.33-0.68)p<0.00l (CS) | 0.47 (0.30-0.73)p=0.001 (CS) | p27, TIL, uPA |
| [14] | Zlobec et al. (2008) | 3 | 392 | - | 226 | I-II (Rectum only) | - | No | CD8+ | - | 0.55 (0.41-0.74)p<0.00l (CS) | 0.63 (0.45-0.88)p=0.006 (CS) | age, size, stage (PT, pN) |
| [15] | Sinicrope et al. (2009) | 2 | 101 | - | - | II-III | 5FU based adjuvant chemo therapy | No | CD3+/FOXP3+ratio | 71% (vs. 62%) (OS)67% (vs. 46%) (DFS) | 0.57 (0.30-1.09)p=0.087 (OS) 0.46 (0.24-0.90) p=0.021 (DFS) | 0.47 (0.24-0.94)p=0.039 (DFS) | age, lymph node, MSI, stage, tumour grade |
| [16] | Laghi et al. (2009) | 1 | 286 | - | 136 | II-III | No treatment or adjuvant chemo therapy | No | CD3+ | node negative p=0.01 (CS)node positive p=0.66 (CS)node negative p=0.006 (DFS)node positive p=0.62 (DFS) | - | - | - |
| [17] | Frey et al. (2009) | - | 1232 | - | - | I-III | - | No | FOXP3+ | MSS group1 62% (vs. 46%) p=0.004 (CS) | - | 0.73 (0.60-0.90) p=0.019 (CS) | age, sex, stage (pT, pN), tumour grade, vascular invasion, tumour border configuration |
| MSS group2 (CS) 60% (vs. 44%) p<0.00l (CS) | - | 0.70 (0.60-0.90) p=0.007 (CS) | |||||||||||
| MSI 75%? (vs. 63%?) p=0.029 (CS) | - | 0.63 (0.3-1.2) p=0.13 (CS) | |||||||||||
| [18] | Salama et al. (2009) | 1 | 445 | - | - | II-III | 5FU/LV based adjuvant chemo therapy | Yes | CD8+ | - | 0.74 (0.67-0.82)p<0.0001 (CS) | NS (CS) | lymphocytic response, other T-cell subsets, MSI, stage, tumour grade, vascular, lymphatic and perineural invasion |
| CD45RO+ | - | 0.74 (0.65-0.84)p<0.0001 (CS) | NS (CS) | ||||||||||
| FOXP3+ | - | 0.78 (0.70-0.87)p<0.0001 (CS) | 0.54 (0.38-0.77)p=0.001 (CS) | ||||||||||
| [19] | Pages et al. (2009) | 1 | 411 | - | - | I-II | 5FU/LV based adjuvant chemo therapy | Yes | CD8+ | p<0.0001(OS and DFS) | - | - | stage, bowel perforation |
| CD45RO+ | p<0.000l(OS and DFS) | - | - | ||||||||||
| CD8+ plus CD45RO+ | p<0.0001(OS and DFS) | - | p<0.0001 (CS, OS and DFS) | ||||||||||
| [20] | Deschool meester et al. (2010) | - | 209 | 100 | 100 | I-IV | No treatment or adjuvant or neo-adjuvant chemo therapy | No | CD3+ | p=0.04 (OS) | - | 0.54 (0.18-1.59)p=0.26 (OS) | age, sex, location, stage tumour grade, adjuvant treatment, other T-cell subsets |
| CD8+ | p=0.04 (OS) | - | 2.06 (0.67-6.39)p=0.21 (OS) | ||||||||||
| GZMB+ | - | - | 1.18 (0.45-3.13)p=0.74 (OS) | ||||||||||
| [21] | Suzuki et al. (2010) | 1 | 94 | - | - | I-IV | - | No | CD8+/FOXP3+ratio | p=0.01 (OS) | 0.35 (0.15-0.81)p=0.014 (OS) | 0.40 (0.17-0.94)p=0.035 (OS) | stage, venous invasion |
| [22] | Correale et al. (2010) | - | 57 | - | - | - | FOLFOX chemo therapy | No | FOXP3+ | - | p=0.0009 (DFS)p=0.0005 (OS) | - | - |
| [23] | Lee et al. (2010) | 1 | 87 | - | - | II | - | Yes | CD3+ | p=0.010 (DFS)p=0.061 (OS) | p=0.003 (DFS)p=0.039 (OS) | 0.20 (0.02-2.60)p=0.22 (DFS) | vascular invasion, neural invasion, other T-cell subsets? |
| CD25+ | p=0.013 (DFS)p=0.15 (OS) | p=0.002 (DFS)p=0.017 (OS) | 0.22 (0.02-2.35)p=0.21 (DFS) | ||||||||||
| CD45RO+ | p=0.049 (DFS)p=0.16 (OS) | p=0.014 (DFS)p=0.037 (OS) | 0.24 (0.02-1.10)p=0.014 (DFS) | ||||||||||
| FOXP3+ | p=0.009 (DFS)p= 0.027 (OS) | p=0.005 (DFS)p=0.040 (OS) | 0.14 (0.07-0.85)p=0.027 (DFS) | ||||||||||
| Current study | Many | 768 | 366 | 229 | I-IV | - | Yes | CD3+ | 79% (vs. 75%) Q4 (vs. Q1)b p=0.19 (CS) | 0.73 (0.49-1.08) Q4 (vs. Q1)b p=0.070c(CS) | 1.30 (0.81-2.07) Q4 (vs. Q1)b p=0.16c (CS) | age, sex, BMI, family history, year of diagnosis, location, stage, tumour grade, LINE-1, CIMP, MSI, BRAF, KRAS,PIK3CA, other T-cell subsets | |
| CD8+ | 78% (vs. 66%) Q4 (vs. Q1)b p=0.026 (CS) | 0.61 (0.42-0.88) Q4 (vs. Q1)b p=0.007c(CS) | 0.81 (0.52-1.27) Q4 (vs. Q1)b p=0.34c (CS) | ||||||||||
| CD45RO+ | 83% (vs. 68%) Q4 (vs. Q1)b p<0.0001 (CS) | 0.40 (0.26-0.60) Q4 (vs. Q1)b P<0.0001c (CS) | 0.51 (0.32-0.80) Q4 (vs. Q1)b p=0.034c (CS) | ||||||||||
| FOXP3+ | 80% (vs. 64%) Q4 (vs. Q1)b p<0.0001 (CS) | 0.48 (0.32-0.70) Q4 (vs. Q1)b p<0.0001c (CS) | 0.89 (0.59-1.34) Q4 (vs. Q1)b p=0.76c (CS) |
Tumour-infiltrating lymphocytes may also reflect specific molecular alterations associated with indolent tumour behavior. Previous studies have shown that lymphocytic infiltration is associated with microsatellite instability (MSI) in colorectal cancer [29-32]. Truncated peptides produced by frameshift mutations due to MSI may be immunogenic and contribute to host immune response [24,26,33]. However, little is known on the interrelationship between tumour-infiltrating T-cells, MSI and other tumour molecular features, including the CpG island methylator phenotype (CIMP), global DNA hypomethylation and KRAS, BRAF and PIK3CA mutations.
Previous studies have reported that MSI [34], CIMP [35], BRAF mutation [35,36], PIK3CA mutation [37], and tumour LINE-1 hypomethylation [38] are associated with prognosis, and that lymphocytic infiltration is associated with many of these molecular variables [32]. As such, to define the prognostic effect of tumour-infiltrating T-cells independent of those potential confounders, large studies of colorectal cancers with extensive molecular characterization are needed.
We, therefore, examined the prognostic role of tumour-infiltrating T-cell subsets in a database of 768 colorectal cancers from two prospective cohort studies. Because we concurrently assessed the densities of CD3+, CD8+, CD45RO+ (PTPRC) and FOXP3+-cells as well as other relevant molecular and pathologic features, we could evaluate the independent effect of each T-cell subset density on patient survival. Our data have shown that the density of CD45RO+-cells, but not that of CD3+, CD8+, or FOXP3+-cells, is an independent prognostic biomarker associated with longer survival of colorectal cancer patients.
Materials and methods
Study group
We utilized a database of colorectal cancer cases in the Nurses' Health Study (N=121,701 women followed since 1976) [39], and the Health Professionals Follow-up Study (N=51,529 men followed since 1986) [39]. Based on availability of follow-up data and adequate tissue specimens for tissue microarray (TMA), 768 colorectal cancers (diagnosed up to 2004) were included (Table 2). Hospitals where patients underwent tumour resections were distributed throughout 48 States in the U.S. Patients were observed until death or June 30, 2009, whichever came first. Among our cohort studies, there was no significant difference in demographic features between cases with and without available tissue [39]. Tissue sections from all colorectal cancers were reviewed by a pathologist (S.O.) unaware of other data. Tumour grade was categorized as low vs. high (≥50% vs. <50% gland formation). Written informed consent was obtained from all study subjects. Tissue collection and analyses were approved by the Institutional Review Boards.
Table 2. Subset T-cell density in neoplastic epithelial areas, according to various features in colorectal cancer.
| Clinical, pathologic or molecular feature | Total N | CD3+ (cells/mm2) Median (25th -75th percentile) | P value | CD8+ (cells/mm2) Median (25th -75th percentile) | P value | CD45RO+ (cells/mm2) Median (25th -75th percentile) | P value | FOXP3+ (cells/mm2) Median (25th -75th percentile) | P value |
|---|---|---|---|---|---|---|---|---|---|
| All cases | 768 | 245 (86-581) | 237 (77-646) | 377 (159-727) | 26 (14-48) | ||||
| Age (years) | 0.097 | 0.07 | 0.98 | 0.74 | |||||
| <65 | 286 (37%) | 225 (77-532) | 270 (87-843) | 392 (154-694) | 26 (14-47) | ||||
| ≥65 | 482 (63%) | 257 (96-603) | 214 (72-565) | 373 (164-754) | 26 (13-49) | ||||
| Gender | 0.12 | 0.0025 | 0.0029 | 0.17 | |||||
| Men | 283 (37%) | 264 (85-735) | 304 (88-917) | 299 (153-611) | 25 (13-46) | ||||
| Women | 485 (63%) | 234 (92-517) | 193 (70-517) | 425 (169-839) | 27 (14-49) | ||||
| Body mass index (BMI, kg/m2) | 0.69 | 0.70 | 0.87 | 0.88 | |||||
| <30 | 632 (82%) | 244 (86-572) | 248 (75-685) | 376 (156-718) | 26 (14-48) | ||||
| ≥30 | 135 (18%) | 254 (94-590) | 200 (87-548) | 393 (181-783) | 25 (11-48) | ||||
| Family history of colorectal cancer in any first degree relative | 0.52 | 0.90 | 0.62 | 0.93 | |||||
| (-) | 589 (77%) | 244 (92-584) | 247 (75-615) | 372 (156-718) | 26 (14-49) | ||||
| (+) | 179 (23%) | 248 (81-548) | 215 (80-748) | 399 (163-805) | 27 (12-46) | ||||
| Year of diagnosis | <0.0001 | <0.0001 | 0.077 | <0.0001 | |||||
| Prior to 1995 | 291 (38%) | 159 (71-447) | 300 (127-892) | 402 (205-756) | 22 (11-37) | ||||
| 1995 to 2004 | 477 (62%) | 276 (121-640) | 188 (62-542) | 357 (142-699) | 30 (15-53) | ||||
| Tumour location | 0.72 | 0.74 | 0.030 | 0.30 | |||||
| Rectum | 153 (20%) | 255 (95-590) | 200 (76-655) | 284 (125-656) | 22 (12-48) | ||||
| Distal colon | 233 (30%) | 227 (87-551) | 255 (80-663) | 374 (175-681) | 27 (14-44) | ||||
| Proximal colon | 382 (50%) | 247 (84-570) | 239 (73-609) | 417 (174-857) | 27 (14-49) | ||||
| Tumour grade | 0.49 | 0.80 | 0.0051 | 0.89 | |||||
| Low grade | 690 (90%) | 241 (85-568) | 234 (77-647) | 373 (158-688) | 26 (14-48) | ||||
| High grade | 75 (10%) | 288 (143-717) | 245 (70-634) | 493 (183-1737) | 26 (15-48) | ||||
| Disease stage | 0.29 | 0.14 | 0.37 | <0.0001 | |||||
| I | 166 (21%) | 284 (103-588) | 219 (80-608) | 409 (156-847) | 34 (18-52) | ||||
| II | 236 (31%) | 239 (76-593) | 274 (69-836) | 397 (181-805) | 25 (14-47) | ||||
| III | 217 (28%) | 227 (102-520) | 250 (91-737) | 377 (145-680) | 25 (12-48) | ||||
| IV | 112 (15%) | 196 (81-468) | 141 (56-475) | 345 (135-548) | 20 (10-32) | ||||
| Unknown | 37 (5%) | 292 (79-620) | 300 (55-588) | 361 (220-1207) | 42 (17-103) | ||||
| CIMP status | 0.39 | 0.56 | <0.0001 | 0.19 | |||||
| CIMP-0/low | 630 (84%) | 240 (86-524) | 232 (77-636) | 348 (154-678) | 25 (13-46) | ||||
| CIMP-high | 124 (16%) | 252 (83-737) | 274 (70-710) | 538 (255-1314) | 28 (14-56) | ||||
| MSI status | 0.55 | 0.58 | <0.0001 | 0.0017 | |||||
| MSI-low/MSS | 630 (84%) | 233 (86-525) | 234 (73-647) | 339 (143-655) | 25 (13-46) | ||||
| MSI-high | 123 (16%) | 267 (73-714) | 270 (83-597) | 616 (316-1415) | 34 (18-57) | ||||
| BRAF mutation | 0.91 | 0.58 | 0.0006 | 0.059 | |||||
| (-) | 642 (85%) | 241 (87-558) | 232 (76-654) | 348 (154-684) | 25 (13-47) | ||||
| (+) | 114 (15%) | 256 (82-615) | 264 (77-592) | 529 (255-1023) | 33 (15-57) | ||||
| KRAS mutation | 0.13 | 0.066 | 0.0043 | 0.036 | |||||
| (-) | 481 (63%) | 245 (81-559) | 205 (71-559) | 412 (175-832) | 28 (14-50) | ||||
| (+) | 278 (37%) | 241 (123-568) | 265 (83-832) | 315 (146-627) | 23 (13-40) | ||||
| PIK3CA mutation | 0.94 | 0.033 | 0.12 | 0.45 | |||||
| (-) | 576 (84%) | 248 (86-586) | 226 (75-645) | 364 (146-692) | 27 (14-48) | ||||
| (+) | 112 (16%) | 229 (105-528) | 304 (122-864) | 425 (203-798) | 23 (13-42) | ||||
| LINE-1 methylation | 0.31 | 0.39 | 0.0035 | 0.25 | |||||
| ≥60% | 438 (59%) | 249 (94-588) | 246 (83-650) | 409 (191-797) | 28 (13-50) | ||||
| <60% | 306 (41%) | 216 (81-486) | 215 (66-636) | 290 (137-676) | 25 (14-42) |
KRAS, BRAF and PIK3CA sequencing, and MSI analysis
DNA was extracted from tumour, and PCR-Pyrosequencing targeted for KRAS (codons 12-13) [40], BRAF (codon 600) [41] and PIK3CA (exons 9 and 20) were performed [42]. MSI analysis was performed, using BAT25, BAT26, BAT40, D2S123, D5S346, D17S250, D18S55, D18S56, D18S67 and D18S487 [43]. MSI-high was defined as instability in ≥30% of the markers [43].
Analyses for CpG island methylation and LINE-1 methylation
Bisulfite DNA treatment and real-time PCR (MethyLight) were validated and performed [44]. We quantified DNA methylation in 8 CIMP-specific promoters [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and _SOCS1_] [45-47]. CIMP-high, CIMP-low and CIMP-0 were defined as ≥6/8, 1/8-5/8 and 0/8 methylated promoters, respectively, according to the previously-established criteria [47]. To accurately quantify LINE-1 methylation, we utilized Pyrosequencing [48,49].
Immunohistochemistry and image analysis
Tissue microarrays (TMAs) were constructed [50]. For CD3, CD8 and FOXP3, antigen retrieval was performed, and deparaffinized tissue sections in EDTA Solution (pH 8.0) (Zymed, San Francisco, CA) were treated by a microwave for 30 sec in a pressure cooker. For CD45RO (the official gene symbol PTPRC), Citrate Buffer (pH 6.0) (Zymed) was used for antigen retrieval. Tissue sections were incubated with peroxidase block (5 min); protein block (20 min); and then, primary antibody against CD3 (rabbit polyclonal anti-human CD3 antibody, clone F7.2.38, 1:250 dilution, Dako Cytomation, Carpinteria, CA), CD8 (mouse monoclonal anti-human CD8 antibody, clone C8/144B, 1:100 dilution, Dako Cytomation), CD45RO (mouse monoclonal anti-human CD45RO antibody, clone UCHL1, 1:100 dilution, Dako Cytomation) or FOXP3 (mouse monoclonal anti-human FOXP3 antibody, clone 206D, 1:50 dilution; BioLegend, San Diego, CA) for 1 hr at room temperature. Next, we applied Envision System HRP-labeled polymer anti-rabbit (for CD3) or anti-mouse (for the other markers) (Dako Cytomation) for 30 min, diaminobenzidine (5 min) and hematoxylin counterstain (1 min).
After staining for each T-cell subset, TMA slides were scanned by an automated scanning microscope and Ariol image analysis system (Genetix, San Jose, CA) (Figure 1). The software was used to count the number of positive nuclei in each tissue core. We marked neoplastic epithelial areas, to exclude non-neoplastic areas (stroma, normal mucosa, and necrotic regions). We calculated the average density (cells/mm2) of each tumour-infiltrating T-cell subset. In addition, we analysed subset T-cell density in stromal areas as well as in whole TMA core (Supplementary Tables 1-2).
Figure 1.
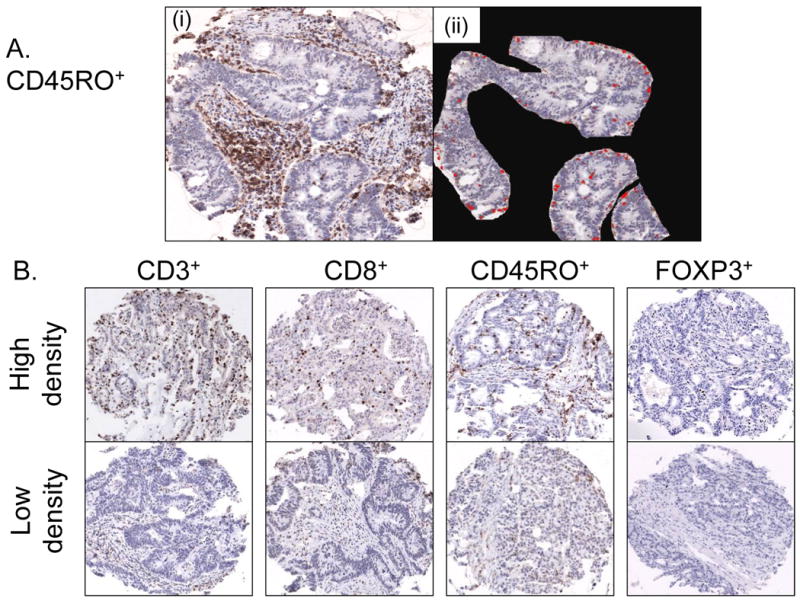
Image analysis of CD3+, CD8+, CD45RO+ and FOXP3+-cells in colorectal cancer.
A. CD45RO+-cells in a tissue microarray (TMA) core (i). Neoplastic epithelial areas are selected, and there are CD45RO+-cells (in red color) in neoplastic epithelial areas (ii).
B. TMA images for CD3+, CD8+, CD45RO+ and FOXP3+-cells in colorectal cancer. Upper panels show high cell density cases, and lower panels show low cell density cases. In these TMA cores, neoplastic epithelial areas are subsequently selected as shown in panel A(ii) for image analysis.
Immunohistochemical evaluation of T-cell subsets in cancer tissue has been a challenge, and there has been no standardized method. Pre-analytical variables such as tissue processing may have considerable impact on antigenicity of each T-cell subset, which may be substantially influenced by a subtle difference in conditions of immunohistochemical procedure. We tried to minimize such measurement errors in a number of ways. We constructed TMA and performed the immunohistochemical procedure in a very similar condition for all specimens. We obtained two to four tumour tissue cores from each case, considering within-tumour heterogeneity.
Statistical analyses
We used SAS program (Version 9.1, SAS Institute, Cary, NC). All p values were two-sided, and statistical significance was set at p=0.05. Nonetheless, when multiple hypothesis testing was performed, a p-value for significance was adjusted to p=0.0009 (=0.05/56) or p=0.0042 (=0.05/12) by Bonferroni correction. To assess unadjusted relationship with each T-cell subset density, we used the Wilcoxon rank-sum test for dichotomous variables or Kruskal-Wallis test for variables with more than two categories.
For survival analyses, Kaplan-Meier method and the log-rank test were used (Figure 2). For analyses of colorectal cancer-specific mortality, deaths as a result of causes other than colorectal cancer were censored. To control for confounding, we used stage-stratified proportional hazards model to compute hazard ratio (HR) of death according to each T-cell subset status. Disease stage [I, IIA, IIB (indicating invasion of visceral serosal surface or adjacent organ), IIIA, IIIB, IIIC, IV, unknown] was used as a stratifying variable (by the “strata” option in the SAS “proc phreg” command) to avoid residual confounding and overfitting. The model initially included age (continuous), sex, BMI (≥30 vs. <30 kg/m2), family history of colorectal cancer, year of diagnosis (continuous), tumour location (proximal colon vs. distal colon vs. rectum), tumour grade, CIMP, MSI, BRAF, KRAS, PIK3CA, LINE-1 methylation (continuous) and other T-cell subsets (quartiles). A backward stepwise elimination with a threshold of p=0.10 was used to select variables, but all of T-cell subsets were forced into the final model. We assigned a “missing” indicator variable for those cases with missing data on CD3+ (5.3%), CD8+ (7.7%), CD45RO+ (3.9%) or FOXP3+ (8.2%). For cases with missing information in any of the other categorical variables [BMI (0.1%), tumour grade (0.4%), CIMP (1.8%), MSI (1.8%), BRAF (1.5%), KRAS (1.1%), and PIK3CA (10%)], we included those cases in a majority category of that variable. An interaction was assessed by the Wald test on the cross product of the CD45RO+ variable and another variable of interest (without data-missing cases).
Figure 2.
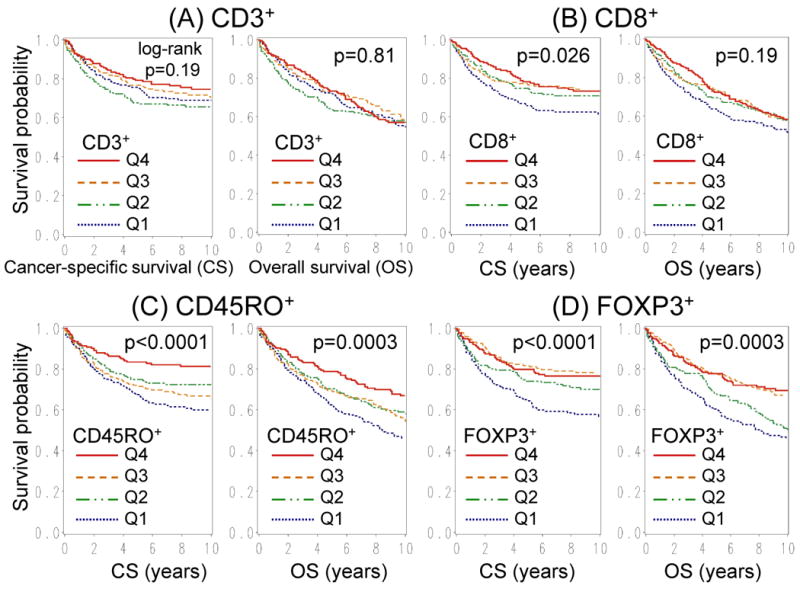
Kaplan-Meier survival curves with log-rank p values for colorectal cancer-specific survival (CS; left panel) and overall survival (OS; right panel) according to tumour-infiltrating T-cell subset density; CD3+-cells (A), CD8+-cells (B), CD45RO+-cells (C) and FOXP3+-cells (D). Q1-4, first to fourth quartile.
To assess relationship with CD45RO+-cell density, a multivariate linear regression model was constructed. Because CD45RO+-cell density showed skewed distribution, we used “loge(CD45RO+-cells/mm2)”, which distributed approximately normally. A backward stepwise elimination with a threshold of p=0.10 was used to select variables in the final model. After the final linear regression model was constructed, a distribution of residuals [observed minus predicted log(CD45RO+-cell density)] was visually inspected and confirmed that the assumption of residuals' normality and equal variance across predicted log(CD45RO+-cell density) was generally satisfied (data not shown). We assessed whether there was any influential case, by Cook's D statistics, a summary measure of influence, and found that there was no influential case (all Cook's D value <0.031).
Results
T-cell subsets in colorectal cancer
Utilizing tissue microarray (TMA), we quantified CD3+, CD8+, CD45RO+ and FOXP3+-cells in neoplastic epithelial areas by automated image analysis on 768 colorectal cancers. Density of each T-cell subset (cells/mm2) distributed as follows; CD3+ (mean 730; median 245; interquartile range 86-581), CD8+ (mean 806; median 237; interquartile rage 77-646), CD45RO+ (mean 670; median 377; interquartile range 159-727), and FOXP3+ (mean 38; median 26; interquartile range 14-48). Selected T-cell subset densities were positively associated with CIMP-high, MSI-high, BRAF mutation, and high-level global DNA methylation (as measured in LINE-1) (Table 2). Table 3 shows Spearman's correlation coefficients between T-cell subset densities in neoplastic epithelial areas.
Table 3. Correlation between tumour-infiltrating T-cell subset densities (r; Spearman's correlation coefficient).
| CD3+-cell | CD8+-cell | CD45RO+-cell | FOXP3+-cell |
|---|---|---|---|
| CD3+-cell | r = 0.47p<0.0001 | r = 0.33p<0.0001 | r = 0.061p=0.11 |
| CD8+-cell | r = 0.47p<0.0001 | r = 0.24p<0.0001 | r = -0.022p=0.58 |
| CD45RO+-cell | r = 0.33p<0.0001 | r = 0.24p<0.0001 | r = 0.23p<0.0001 |
| FOXP3+-cell | r = 0.061p=0.11 | r = -0.022p=0.58 | r = 0.23p<0.0001 |
Tumour-infiltrating T-cell subset density and patient survival
We examined density of each tumour-infiltrating T-cell subset and patient survival. There were 366 deaths including 229 colorectal cancer-specific deaths. Median follow-up for censored cases was 11.6 years. In Kaplan-Meier analysis (Figure 2), high CD45RO+ and FOXP3+ densities were associated with improved cancer-specific and overall survival (log-rank p<0.0004). We performed univariate and multivariate Cox regression analyses (Table 4). In univariate analysis, CD8+, CD45RO+ and FOXP3+-cells were associated with longer cancer-specific survival (Ptrend<0.007) and overall survival (Ptrend≤0.04). In multivariate analysis, however, only CD45RO+-cell density was significantly associated with overall survival (Ptrend=0.015), after adjusting for all known or suspected clinical and molecular features, including other T-cell subset densities. Compared to Q1 stratum, CD45RO+ Q4 stratum was associated with significantly longer cancer-specific survival [multivariate hazard ratio (HR), 0.51; 95% confidence interval (CI): 0.32–0.80: p=0.0032] and overall survival (multivariate HR, 0.61; 95% CI: 0.43–0.85: p=0.0036). In exploratory analyses, any of tumour-infiltrating CD3+/FOXP3+, CD8+/FOXP3+, and CD45RO+/FOXP3+ was not associated with survival (data not shown).
Table 4. Subset T-cell density in neoplastic epithelial areas and patient mortality in colorectal cancer.
| T-cell subset | Quartile | Colorectal cancer-specific survival | Overall survival | ||
|---|---|---|---|---|---|
| Univariate HR (95% CI) | Multivariate HR (95% CI) | Univariate HR (95% CI) | Multivariate HR (95% CI) | ||
| CD3+-cell (N=727) | Q1 (0-85/mm2) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| Q2 (86-244 /mm2) | 1.10 (0.77-1.58) | 1.29 (0.89-1.88) | 1.04 (0.78-1.39) | 1.04 (0.77-1.42) | |
| Q3 (245-579 /mm2) | 0.89 (0.62-1.30) | 1.40 (0.93-2.10) | 0.91 (0.67-1.22) | 1.19 (0.86-1.65) | |
| Q4 (≥580 /mm2) | 0.73 (0.49-1.08) | 1.30 (0.81-2.07) | 0.94 (0.70-1.26) | 1.37 (0.96-1.96) | |
| P for trenda | 0.070 | 0.16 | 0.50 | 0.051 | |
| CD8+-cell (N=709) | Q1 (0-75 /mm2) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| Q2 (76-235 /mm2) | 0.70 (0.49-1.01) | 0.75 (0.51-1.11) | 0.83 (0.62-1.12) | 0.78 (0.57-1.06) | |
| Q3 (236-645 /mm2) | 0.64 (0.44-0.92) | 0.79 (0.53-1.18) | 0.77 (0.57-1.04) | 0.83 (0.60-1.14) | |
| Q4 (≥646 /mm2) | 0.61 (0.42-0.88) | 0.81 (0.52-1.27) | 0.74 (0.54-0.99) | 0.85 (0.60-1.20) | |
| P for trenda | 0.0068 | 0.34 | 0.040 | 0.47 | |
| CD45RO+-cell (N=738) | Q1 (0-158 /mm2) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| Q2 (159-376 /mm2) | 0.64 (0.45-0.92) | 0.81 (0.56-1.18) | 0.74 (0.56-0.98) | 0.79 (0.59-1.06) | |
| Q3 (377-725 /mm2) | 0.80 (0.57-1.12) | 1.18 (0.81-1.70) | 0.73 (0.55-0.97) | 0.96 (0.71-1.30) | |
| Q4 (≥726 /mm2) | 0.40 (0.26-0.60) | 0.51 (0.32-0.80) | 0.52 (0.38-0.70) | 0.61 (0.43-0.85) | |
| P for trenda | <0.0001 | 0.034 | <0.0001 | 0.015 | |
| FOXP3+-cell (N=705) | Q1 (0-12 /mm2) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| Q2 (13-25 /mm2) | 0.65 (0.46-0.93) | 0.75 (0.52-1.09) | 0.85 (0.65-1.12) | 1.07 (0.81-1.43) | |
| Q3 (26-47 /mm2) | 0.44 (0.30-0.64) | 0.74 (0.49-1.11) | 0.52 (0.38-0.71) | 0.85 (0.62-1.17) | |
| Q4 (≥48 /mm2) | 0.48 (0.32-0.70) | 0.89 (0.59-1.34) | 0.48 (0.35-0.66) | 0.80 (0.57-1.14) | |
| P for trenda | <0.0001 | 0.76 | <0.0001 | 0.095 |
In addition, we performed survival analyses using each T-cell subset density in whole TMA tissue cores and in tumour stromal areas, which yielded results similar to Table 4 (Supplementary Tables 1-2).
Interaction between CD45RO+-cells and other variables in survival analysis
We examined whether the survival benefit associated with tumour-infiltrating CD45RO+-cells was modified by any of the clinical, pathologic and molecular variables (Figure 3). We did not observe a significant interaction between CD45RO+-cell density and any of the covariates (all Pinteraction>0.10), such as disease stage, MSI status, or CD3+, CD8+, or FOXP3+-cell density.
Figure 3.
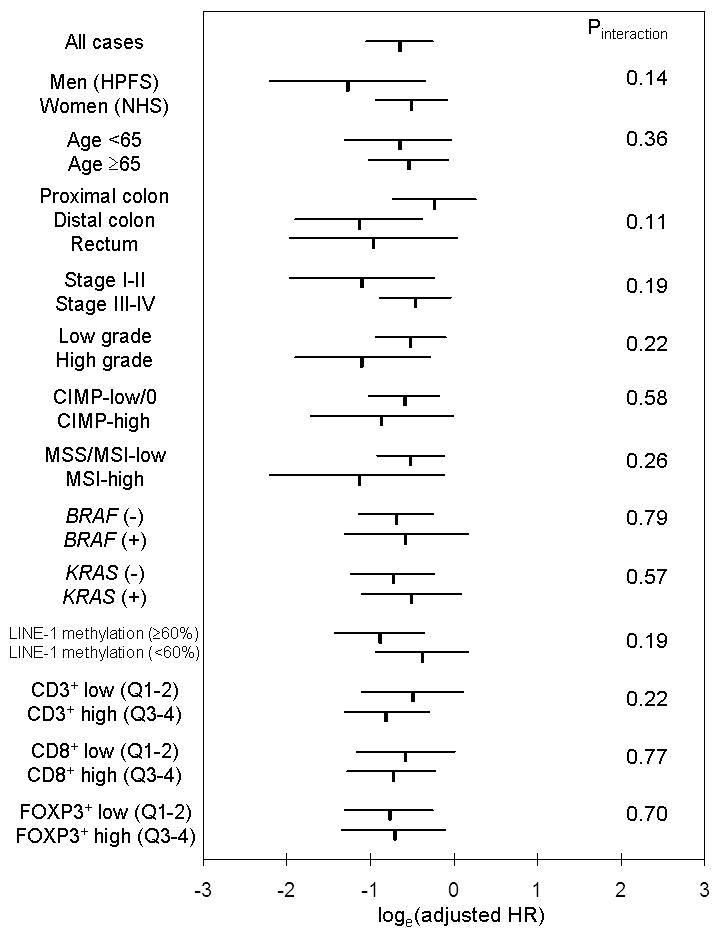
Stratified analysis of tumour-infiltrating CD45RO+-cell density and colorectal cancer-specific mortality. Loge(adjusted HR) with 95% confidence interval (CI) for CD45RO+-cell density Q4 (vs. Q1-3) in various strata is shown. There was no significant interaction (effect modification) by any of the variables including CD3+, CD8+, or FOXP3+-cell density.
CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; HR, hazard ratio; MSI, microsatellite instability; MSS, microsatellite stable; NHS, Nurses' Health Study; Q1-4, first to fourth quartile.
Multivariate linear regression analysis to assess relations with CD45RO+-cells
In view of the survival benefit specifically associated with tumour-infiltrating CD45RO+-cells, we constructed a multivariate linear regression model to define the clinical and molecular features that were independently associated with tumour-infiltrating CD45RO+-cells (Table 5). The presence of MSI-high was positively associated with CD45RO+-cell density (p<0.0001), whereas LINE-1 hypomethylation was inversely associated with CD45RO+-cell density (p=0.0013), adjusting for other potential factors. Nonetheless, as described above (Table 4), the influence of tumour-infiltrating CD45RO+-cells on patient survival was independent of tumour MSI and LINE-1 methylation status.
Table 5. Multivariate linear regression analysis to predict tumour-infiltrating CD45RO+-cell density [loge(CD45RO+ cells/mm2)] in colorectal cancers.
| Variables in the final model | Adjusted β coefficient [change in loge(CD45RO+-cells/mm2) by a given variable] | 95% confidence limits | P value (T-test) |
|---|---|---|---|
| MSI-high (vs. low/MSS) | 0.53 | 0.31, 0.76 | <0.0001 |
| LINE-1 hypomethylation (for a unit of 30% decrease in methylation level) | -0.43 | -0.69, -0.17 | 0.0013 |
Discussion
We conducted this study to examine the independent prognostic effect of tumour-infiltrating T-cell subsets (CD3+, CD8+, CD45RO+ and FOXP3+) in a large population of colorectal cancer patients. We found that tumour-infiltrating CD45RO+-cell density was significantly associated with improved prognosis in colorectal cancers, independent of clinical, pathologic and molecular features, including densities of the other T-cell subsets. Thus, our findings suggest tumour-infiltrating CD45RO+-cell density as a prognostic biomarker in colorectal cancer. Furthermore, microsatellite instability (MSI)-high and high-level LINE-1 methylation were independently associated with tumour-infiltrating CD45RO+-cell density, raising potential roles of these molecular features in stimulating host immune response to tumour. Nonetheless, the survival benefit associated with tumour-infiltrating CD45RO+-cells was independent of tumour MSI and LINE-1 methylation status.
Examining tumour and host factors and patient outcome is important in cancer research [51-56]. Accumulating evidence suggests that effector/cytotoxic (CD3+ [2,12,15,16,33] and CD8+ [1-3,6-9,11,12,18-21]) and memory (CD45RO+ [3,12,18,19,23]) T-cells play important roles in antitumour immune response. With regard to regulatory T-cells, one specific marker is nuclear transcription factor FOXP3 [25,57,58]. FOXP3+ regulatory T-cell has been shown to modulate antitumour immune response [17-19,21-23,27,59-62] and suppress the activity of cytotoxic T-cells [18]. Thus, these specific subsets of effector/cytotoxic (CD3+ and CD8+), memory (CD45RO+) and regulatory (FOXP3+) T-cells are thought to be indicators of host immune response to tumour cells and might be a target for immunotherapy [25-27].
To examine T-cell subsets, we utilized a digitized, high-resolution image analysis, which could count the number of lymphocytes and minimise observer bias. The median densities of tumour-infiltrating T-cell subsets in the current study were compatible with data in the previous studies (CD3+ [12], CD8+ [21], CD45RO+ [18], and FOXP3+ [17,21,59]).
Previous studies have reported the relationship between MSI-high and density of CD3+ [2,16,33,59], CD8+ [2,7,8,18,59], CD45RO+ [18], or FOXP3+-cells [15,59]. Our current study has shown that densities of CD45RO+ and FOXP3+-cells, but not that of CD3+ or CD8+-cells, are significantly associated with MSI-high. The strong association between MSI and CD45RO+-cell density supports the hypothesis that truncated peptides produced by MSI and frameshift mutations may elicit host immune response and recruit CD45RO+-cells [24,33]. In most studies, MSI in colon cancer has been associated with improved survival [18,24,34,63-67], although the mechanism underlying this association is largely unknown. Our results suggest that stimulation of a host immune response to MSI-high cancer may offer one explanation. In addition, recent studies have reported a detrimental effect of adjuvant chemotherapy on MSI-high colon cancer patients [63,68], although there have been conflicting data [69]. It is possible that cytotoxic chemotherapy may attenuate the host immune response associated with MSI-high colon cancer. Further studies are needed to elucidate the mechanism of host immune response and modifying effect of chemotherapy.
In addition, we were able to detect an independent relationship between CD45RO+-cell density and high-level LINE-1 methylation, which has not been examined before. Previous studies have reported a link between LINE-1 hypomethylation, global DNA hypomethylation and chromosomal instability (CIN) [38,48,70]. Global DNA hypomethylation and CIN affect epigenetic status and gene copy numbers, respectively, in a genome-wide scale. Thus, it is not surprising that global DNA hypomethylation and/or CIN may affect expression levels of genes which contribute to interactions with host cells and regulation of host immune response to tumour. Additional studies are necessary to elucidate the exact mechanism of the relationship between LINE-1 methylation level and CD45RO+-cells.
In early-stage colorectal cancer, infiltration of CD45RO+-cells has been associated with an gene expression pattern reflective of T-helper 1 cells and cytotoxic T-cells [19]. Moreover, a recent study has reported that infiltration of CD45RO+-cells is associated with high expression of specific chemokines (CXCL9 and CXCL10), which may play roles in favorable prognosis of colorectal cancer patients [71]. Previous studies reported that tumour-infiltrating CD45RO+-cell density was associated with improved survival in colorectal cancer [3,10,12,19,23]; however, none of those studies [3,10,12,19,23] examined the prognostic effect of CD45RO+-cells independent of established molecular alterations, including MSI, CIMP, BRAF mutation and LINE-1 methylation, all of which have been associated with colon cancer prognosis [34,35,38]. Because these molecular features are also associated with tumour-infiltrating T-cells, the independent effect of specific T-cell subsets on patient outcome can only be ascertained through a comprehensive, multivariate analysis model where data on clinical, pathologic, and molecular features are available simultaneously as in our current analyses.
A recent study by Salama et al. [18] reported that tumour-infiltrating FOXP3+-cell density was a prognostic factor in 445 stage II-III colorectal cancers in multivariate analysis which included clinical and pathological features, MSI, and CD8+ and CD45RO+ T-cell subsets. In contrast, our database of 768 stage I-IV colorectal cancers suggests that the survival benefit associated with tumour-infiltrating FOXP3+-cell density is absent after adjusting for CD45RO+ status and other clinical and molecular features. Of note, in contrast to the previous studies [15,59] and ours, Salama et al. [18] did not observe a significant relation between MSI and FOXP3+-cell density, and Le Gouvello et al. [61] found lower mRNA expression level of FOXP3 in MSI-high tumour tissue, perhaps reflecting differences in study populations and analysis methods on FOXP3 [15,59].
CD8+ and CD3+-cell densities have been associated with favorable prognosis in colorectal cancers [2,15,16,20]. Our analyses have shown that neither CD3+ nor CD8+-cell density is associated with improved survival in multivariate analysis. In particular, the attenuation in the effect of CD8+-cells in multivariate analysis is the result of adjusting for disease stage and CD45RO+-cells, suggesting that the improved outcome associated with CD8+-cells in previous studies [1,2,6-9,11,13,14,20] might be due, at least in part, to the confounding effect of CD45RO+-cells.
Recent studies have reported that the ratio of tumour-infiltrating CD3+/FOXP3+ [15] or CD8+/FOXP3+ [21] is associated with clinical outcome of colorectal cancers. However, in our current study, any of ratios of CD3+/FOXP3+, CD8+/FOXP3+, and CD45RO+/FOXP3+ was not associated with patient outcome (data not shown). Those previous studies had small sample sizes (N=94 [21], and N=101 [15]) and performed multiple hypothesis testing without validation cohort, and hence, generalisability of their findings may be limited.
Considering within-tumour heterogeneity, we evaluated two to four tumour tissue cores from each case in TMA. We admit that this approach might still fall short of capturing heterogeneity within tumour. Nonetheless, a misclassification, if any, in TMA analysis due to tumour heterogeneity would be non-differential and unrelated to patient outcome or molecular variables. Despite a possibility of such misclassified cases, we were able to demonstrate the prognostic significance of tumour-infiltrating CD45RO+-cells, because of our well-powered study design.
In summary, tumour-infiltrating CD45RO+-cell density is significantly associated with longer survival of colorectal cancer patients, independent of clinical, pathological and molecular features. In addition, MSI-high and LINE-1 methylation level are independent predictors of CD45RO+-cell density. Our findings suggest that tumour-infiltrating CD45RO+-cells may materially improve patient outcome, offer a possible mechanism by which MSI confers an improved patient outcome, and supports efforts to augment host immune response as a therapeutic strategy.
Supplementary Material
3
List of online supporting information
Supplementary Table 1. Subset T-cell density in whole TMA tumour tissue core and patient mortality in colorectal cancer
4
Supplementary Table 2. Subset T-cell density in tumour stromal areas and patient mortality in colorectal cancer
Acknowledgments
We deeply thank the Nurses' Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires; hospitals and pathology departments throughout the U.S. for providing us with tumour tissue materials. We thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
This work was supported by U.S. National Institute of Health [P01 CA87969 (to S. Hankinson), P01 CA55075 (to W. Willett), K07 CA122826 (to S.O.), R01 CA151993 (to S.O.), P50 CA127003 (GI SPORE to C.S.F.), and GI SPORE Developmental Project Award (to S.O.)]; the Bennett Family Fund for Targeted Therapies Research; and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. K.N. was supported by a fellowship grant from the Japan Society for Promotion of Science. Y.B. was supported by a fellowship grant from the Uehara Memorial Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
Abbreviations
CI
confidence interval
CIMP
CpG island methylator phenotype
HR
hazard ratio
MSI
microsatellite instability
MSS
microsatellite stable
OR
odds ratio
SD
standard deviation
TMA
tissue microarray
Footnotes
No conflicts of interest exist.
Statement of author contributions: study conception and design (CSF, SO); acquisition of data (KN, YB, NT, KS, MH, EG, CSF, SO); statistical analysis and interpretation of data (KN, YB, NT, JAM, GD, EG, CSF, SO); manuscript writing and critical revision (KN, YB, NT, JAM, GD, EG, CSF, SO); funding support (CSF, SO); final approval of manuscript (all authors).
References
- 1.Naito Y, Saito K, Shiiba K, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 2.Guidoboni M, Gafa R, Viel A, et al. Microsatellite instability and high content of activated cytotoxic lymphocytes identify colon cancer patients with a favorable prognosis. Am J Pathol. 2001;159:297–304. doi: 10.1016/S0002-9440(10)61695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberg A, Samii S, Stenling R, et al. Different occurrence of CD8+, CD45R0+, and CD68+ immune cells in regional lymph node metastases from colorectal cancer as potential prognostic predictors. Int J Colorectal Dis. 2002;17:25–29. doi: 10.1007/s003840100337. [DOI] [PubMed] [Google Scholar]
- 4.Petty JK, He K, Corless CL, et al. Survival in human colorectal cancer correlates with expression of the T-cell costimulatory molecule OX-40 (CD134) Am J Surg. 2002;183:512–518. doi: 10.1016/s0002-9610(02)00831-0. [DOI] [PubMed] [Google Scholar]
- 5.Diederichsen AC, Hjelmborg JB, Christensen PB, et al. Prognostic value of the CD4+/CD8+ ratio of tumour infiltrating lymphocytes in colorectal cancer and HLA-DR expression on tumour cells. Cancer Immunol Immunother. 2003;52:423–428. doi: 10.1007/s00262-003-0388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funada Y, Noguchi T, Kikuchi R, et al. Prognostic significance of CD8+ T cell and macrophage peritumoral infiltration in colorectal cancer. Oncol Rep. 2003;10:309–313. [PubMed] [Google Scholar]
- 7.Prall F, Duhrkop T, Weirich V, et al. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Hum Pathol. 2004;35:808–816. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Menon AG, Janssen-van Rhijn CM, Morreau H, et al. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Lab Invest. 2004;84:493–501. doi: 10.1038/labinvest.3700055. [DOI] [PubMed] [Google Scholar]
- 9.Chiba T, Ohtani H, Mizoi T, et al. Intraepithelial CD8+ T-cell-count becomes a prognostic factor after a longer follow-up period in human colorectal carcinoma: possible association with suppression of micrometastasis. Br J Cancer. 2004;91:1711–1717. doi: 10.1038/sj.bjc.6602201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pages F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 11.Baeten CI, Castermans K, Hillen HF, et al. Proliferating endothelial cells and leukocyte infiltration as prognostic markers in colorectal cancer. Clin Gastroenterol Hepatol. 2006;4:1351–1357. doi: 10.1016/j.cgh.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 13.Zlobec I, Minoo P, Baumhoer D, et al. Multimarker phenotype predicts adverse survival in patients with lymph node-negative colorectal cancer. Cancer. 2008;112:495–502. doi: 10.1002/cncr.23208. [DOI] [PubMed] [Google Scholar]
- 14.Zlobec I, Baker K, Terracciano L, et al. Two-marker protein profile predicts poor prognosis in patients with early rectal cancer. Br J Cancer. 2008;99:1712–1717. doi: 10.1038/sj.bjc.6604729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinicrope FA, Rego RL, Ansell SM, et al. Intraepithelial effector (CD3+)/regulatory (FoxP3+) T-cell ratio predicts a clinical outcome of human colon carcinoma. Gastroenterology. 2009;137:1270–1279. doi: 10.1053/j.gastro.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laghi L, Bianchi P, Miranda E, et al. CD3+ cells at the invasive margin of deeply invading (pT3-T4) colorectal cancer and risk of post-surgical metastasis: a longitudinal study. Lancet Oncol. 2009;10:877–884. doi: 10.1016/S1470-2045(09)70186-X. [DOI] [PubMed] [Google Scholar]
- 17.Frey DM, Droeser RA, Viehl CT, et al. High frequency of tumor-infiltrating FOXP3(+) regulatory T cells predicts improved survival in mismatch repair-proficient colorectal cancer patients. Int J Cancer. 2009;126:2635–2643. doi: 10.1002/ijc.24989. [DOI] [PubMed] [Google Scholar]
- 18.Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 19.Pages F, Kirilovsky A, Mlecnik B, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27:5944–5951. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 20.Deschoolmeester V, Baay M, Van Marck E, et al. Tumor infiltrating lymphocytes: an intriguing player in the survival of colorectal cancer patients. BMC Immunol. 2010;11:19. doi: 10.1186/1471-2172-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki H, Chikazawa N, Tasaka T, et al. Intratumoral CD8(+) T/FOXP3 (+) cell ratio is a predictive marker for survival in patients with colorectal cancer. Cancer Immunol Immunother. 2010;59:653–661. doi: 10.1007/s00262-009-0781-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Correale P, Rotundo MS, Del Vecchio MT, et al. Regulatory (FoxP3+) T-cell tumor infiltration is a favorable prognostic factor in advanced colon cancer patients undergoing chemo or chemoimmunotherapy. J Immunother. 2010;33:435–441. doi: 10.1097/CJI.0b013e3181d32f01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee WS, Park S, Lee WY, et al. Clinical impact of tumor-infiltrating lymphocytes for survival in stage II colon cancer. Cancer. 2010 doi: 10.1002/cncr.25293. [DOI] [PubMed] [Google Scholar]
- 24.Schwitalle Y, Kloor M, Eiermann S, et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 26.Speetjens FM, Lauwen MM, Franken KL, et al. Prediction of the immunogenic potential of frameshift-mutated antigens in microsatellite instable cancer. Int J Cancer. 2008;123:838–845. doi: 10.1002/ijc.23570. [DOI] [PubMed] [Google Scholar]
- 27.Curiel TJ. Tregs and rethinking cancer immunotherapy. J Clin Invest. 2007;117:1167–1174. doi: 10.1172/JCI31202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Disis ML, Bernhard H, Jaffee EM. Use of tumour-responsive T cells as cancer treatment. Lancet. 2009;373:673–683. doi: 10.1016/S0140-6736(09)60404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander J, Watanabe T, Wu TT, et al. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shia J, Ellis NA, Paty PB, et al. Value of histopathology in predicting microsatellite instability in hereditary nonpolyposis colorectal cancer and sporadic colorectal cancer. Am J Surg Pathol. 2003;27:1407–1417. doi: 10.1097/00000478-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins MA, Hayashi S, O'Shea AM, et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133:48–56. doi: 10.1053/j.gastro.2007.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tougeron D, Fauquembergue E, Rouquette A, et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod Pathol. 2009;22:1186–1195. doi: 10.1038/modpathol.2009.80. [DOI] [PubMed] [Google Scholar]
- 34.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 35.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 37.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 40.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino S, Kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogino S, Brahmandam M, Cantor M, et al. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 44.Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 47.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogino S, Brahmandam M, Kawasaki T, et al. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–464. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jehan Z, Bavi P, Sultana M, et al. Frequent PIK3CA gene amplification and its clinical significance in colorectal cancer. J Pathol. 2009;219:337–346. doi: 10.1002/path.2601. [DOI] [PubMed] [Google Scholar]
- 52.Abubaker J, Bavi P, Al-Haqawi W, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–445. doi: 10.1002/path.2625. [DOI] [PubMed] [Google Scholar]
- 53.Poulogiannis G, Ichimura K, Hamoudi RA, et al. Prognostic relevance of DNA copy number changes in colorectal cancer. J Pathol. 2010;220:338–347. doi: 10.1002/path.2640. [DOI] [PubMed] [Google Scholar]
- 54.Webster JA, Beck AH, Sharma M, et al. Variations in stromal signatures in breast and colorectal cancer metastases. J Pathol. 2010 doi: 10.1002/path.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 56.Horst D, Scheel SK, Liebmann S, et al. The cancer stem cell marker CD133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J Pathol. 2009;219:427–434. doi: 10.1002/path.2597. [DOI] [PubMed] [Google Scholar]
- 57.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 58.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 59.Michel S, Benner A, Tariverdian M, et al. High density of FOXP3-positive T cells infiltrating colorectal cancers with microsatellite instability. Br J Cancer. 2008;99:1867–1873. doi: 10.1038/sj.bjc.6604756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaput N, Louafi S, Bardier A, et al. Identification of CD8+CD25+Foxp3+ suppressive T cells in colorectal cancer tissue. Gut. 2009;58:520–529. doi: 10.1136/gut.2008.158824. [DOI] [PubMed] [Google Scholar]
- 61.Le Gouvello S, Bastuji-Garin S, Aloulou N, et al. High prevalence of Foxp3 and IL17 in MMR-proficient colorectal carcinomas. Gut. 2008;57:772–779. doi: 10.1136/gut.2007.123794. [DOI] [PubMed] [Google Scholar]
- 62.Blatner NR, Bonertz A, Beckhove P, et al. In colorectal cancer mast cells contribute to systemic regulatory T-cell dysfunction. Proc Natl Acad Sci U S A. 2010;107:6430–6435. doi: 10.1073/pnas.0913683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinicrope FA, Sargent DJ. Clinical implications of microsatellite instability in sporadic colon cancers. Curr Opin Oncol. 2009;21:369–373. doi: 10.1097/CCO.0b013e32832c94bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. e2073. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin SA, Hewish M, Lord CJ, et al. Genomic instability and the selection of treatments for cancer. J Pathol. 2010;220:281–289. doi: 10.1002/path.2631. [DOI] [PubMed] [Google Scholar]
- 66.Ferreira AM, Westers H, Sousa S, et al. Mononucleotide precedes dinucleotide repeat instability during colorectal tumour development in Lynch syndrome patients. J Pathol. 2009;219:96–102. doi: 10.1002/path.2573. [DOI] [PubMed] [Google Scholar]
- 67.Kang MR, Kim MS, Oh JE, et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol. 2009;217:702–706. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- 68.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 70.Matsuzaki K, Deng G, Tanaka H, et al. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–8569. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 71.Mlecnik B, Tosolini M, Charoentong P, et al. Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology. 2010;138:1429–1440. doi: 10.1053/j.gastro.2009.10.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3
List of online supporting information
Supplementary Table 1. Subset T-cell density in whole TMA tumour tissue core and patient mortality in colorectal cancer
4
Supplementary Table 2. Subset T-cell density in tumour stromal areas and patient mortality in colorectal cancer