Complete genome sequence of Sphaerobacter thermophilus type strain (S 6022T) (original) (raw)
Abstract
Sphaerobacter thermophilus Demharter et al. 1989 is the sole and type species of the genus Sphaerobacter, which is the type genus of the family Sphaerobacteraceae, the order Sphaerobacterales and the subclass Sphaerobacteridae. Phylogenetically, it belongs to the genomically little studied class of the Thermomicrobia in the bacterial phylum Chloroflexi. Here, the genome of strain S 6022T is described which is an obligate aerobe that was originally isolated from an aerated laboratory-scale fermentor that was pulse fed with municipal sewage sludge. We describe the features of this organism, together with the complete genome and annotation. This is the first complete genome sequence of the thermomicrobial subclass Sphaerobacteridae, and the second sequence from the chloroflexal class Thermomicrobia. The 3,993,764 bp genome with its 3,525 protein-coding and 57 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Sphaerobacteridae, Thermomicrobia, thermophile, obligate aerobic, sewage sludge isolate, pleomorphic, non-motile, non-sporeforming, GEBA
Introduction
Strain S 6022T (DSM 20745 = ATCC 49802 = NCIMB 13125) is the type strain of the species Sphaerobacter thermophilus, representing the type species of the genus Sphaerobacter. S. thermophilus was described by Demharter et al. in 1989 [1]. It is Gram-positive, non-motile and non-sporeforming. It was originally isolated from thermal treated municipal sewage sludge from München-Grosslappen, Germany [2]. Cells of S. thermophilus were also identified in three other municipal sludge stabilization plants spread across Germany (Isenbüttel, Nettetal, and Gemmingen) using an immunolabelling procedure. From the operating parameters of these plants a minimum temperature growth range of 40-65°C can be predicted [2]. Here we present a summary classification and a set of features for S. thermophilus strain S 6022T, together with the description of the complete genomic sequencing and annotation.
Classification and features
The closest related cultivated organism with a 16S rRNA sequence recorded in Genbank is Thermomicrobium roseum (DSM 5159) [3,4], which shares a mere 87% sequence similarity with strain S 6022T, indicating that S. thermophilus is phylogenetically one of the most isolated bacterial species. Only some uncultivated bacterial clones show a slightly closer relationship, e.g. clone Amb_16S_1237 (EF018775) isolated from Populus tremula (trembling aspen, 92%), EU035785 and EF643378 from soil in a radish-rich area in Jaunpur (India), clone AKYG1722 from farm soil adjacent to a silage storage bunker in Minnesota (89%), and AM935838 from a pilot-scale bioremediation process of hydrocarbon-contaminated soil in France (88%). None of the phylotypes sequenced during environmental screenings or genomic surveys surpassed 82% sequence similarity with strain S 6022T, expressly underlining the phylogenetically isolated and rare occurrence of S. thermophilus (status May 2009).
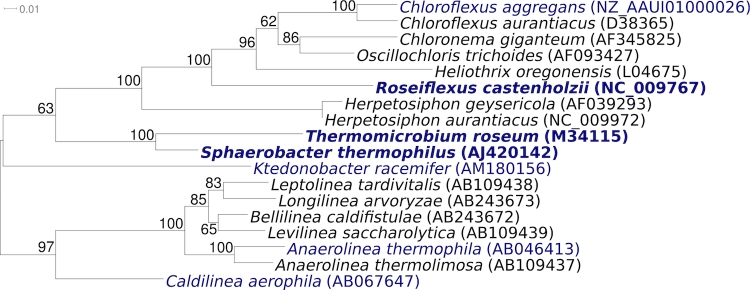
Figure 1 shows the phylogenetic neighborhood of S. thermophilus strain S 6022T in a 16S rRNA based tree. The sequence of the sole 16S rRNA gene in the genome of strain S 6022T differs by six nucleotides (0.4%) from the previously published 16S rRNA sequence generated from DSM 20745 (AJ420142). The difference between the genome data and the previously reported 16S rRNA in GenBank gene sequence is most likely due to sequencing errors in the latter.
Figure 1.
Phylogenetic tree of S. thermophilus strain S 6022T and all type strains of the phylum Chloroflexi, inferred from 1,304 aligned characters [5,6] of the 16S rRNA gene sequence under the maximum likelihood criterion [7]. The tree was rooted with the members of Anaerolineae and Caldilineae within the Chloroflexi. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [8] are shown in blue, published genomes in bold.
S. thermophilus S 6022T cells are coccoid (Figure 2), but are also described as coccoid rods, 1-1.5 by 1.5-3 µm, in older cultures or in glucose-free medium irregular club- or dumb-bell shaped forms [1]. Branched cells are not observed. Colonies on Ottow Medium (DSMZ Medium No. 467) [9] are opaque, circular with entire margin and reach a diameter of 1-2 mm after 3 days of incubation at 60°C. The strain grows strictly aerobically with optimal growth at 55°C and pH 8.5 (Table 1). There is no acid production from glucose. Strain S 6022T possesses catalase and oxidase and hydrolyzes starch but not gelatin, casein or cellulose [1]. Strain S 6022T shares many features such as thermophilia, optimal pH for growth, and lack of motility with its closest relative, T. roseum (DSM 5159) [3,4]. The genome sequence as presented here might contribute to the solution of the question if S. thermophilus, like T. roseum encodes a complete flagellar system [4], although neither strain is motile. Interestingly, none of the other species in the Chloroflexi for which a genome sequence currently exists encode for any flagellar structural components [4].
Figure 2.
Scanning electron micrograph of S. thermophilus S strain 6022T
Table 1. Classification and general features of S. thermophilus S 6022T in accordance to the MIGS recommendations [10].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [11] | |
| Phylum Chloroflexi | TAS [12] | ||
| Class Thermomicrobia | TAS [11-14] | ||
| Order Sphaerobacterales | TAS [15] | ||
| Suborder Sphaerobacterineae | TAS [16] | ||
| Family Sphaerobacteraceae | TAS [15] | ||
| Genus Sphaerobacter | TAS [1] | ||
| Species Sphaerobacter thermophilus | TAS [1] | ||
| Type strain S 6022 | |||
| Gram stain | positive | TAS [1] | |
| Cell shape | coccoid rods, irregular club- or dump-bell shaped | TAS [1] | |
| Motility | nonmotile | TAS [1] | |
| Sporulation | nonsporulating | TAS [1] | |
| Temperature range | thermophile, minimum 40-65°C | TAS [1,2] | |
| Optimum temperature | 55°C, pH 8.5 | TAS [1] | |
| Salinity | not reported | ||
| MIGS-22 | Oxygen requirement | obligate aerobic | TAS [1] |
| Carbon source | starch | NAS | |
| Energy source | unknown | ||
| MIGS-6 | Habitat | thermal treated sewage sludge | TAS [2] |
| MIGS-15 | Biotic relationship | free living | NAS |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [17] | |
| Isolation | thermal treated sewage sludge | TAS [2] | |
| MIGS-4 | Geographic location | Munich, Germany | TAS [2] |
| MIGS-5 | Sample collection time | between 1973 and 1988, probably 1984 | TAS [1] |
| MIGS-4.1,MIGS-4.2 | Latitude, Longitude | 48.139, 11.58 | NAS |
| MIGS-4.3 | Depth | 0 m | NAS |
| MIGS-4.4 | Altitude | 518 m | NAS |
Chemotaxonomy
Acid hydrolysates of the cell wall of strain S 6022T yielded a ratio of glutamic acid to ornithine to alanine to β-alanine to muramic acid to glucosamine = 1:1.1:1.2:1.6:0.9:1.1. The murein structure type belongs to the murein variation A3β [19] with cross-linking via β-alanine [1]. The cell wall is unusually rich in protein content [1]. The principal isoprenoid quinone is an unsaturated menaquinone of type MK-8/0. MK-6/0, MK-7/0, MK-10/0 appear as minor constituents (4.8%, 7.7%, 12.8%) MK-6/0 [1]. Nothing is known about the spectrum of cellular fatty acids in the organism.
Genome sequencing information
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genomes OnLine Database [8] and the complete genome sequence in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-13 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: two Sanger libraries - 8 kb pMCL200 and fosmid pcc1Fos - and one 454 Pyrosequence standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX, Illumina GA |
| MIGS-31.2 | Sequencing coverage | 7.4× Sanger; 28.5× Pyrosequence |
| MIGS-30 | Assemblers | Newbler version 1.1.02.15, Arachne |
| MIGS-32 | Gene calling method | Prodigal, GenePRIMP |
| INSDC ID | CP001823 (chromosome), CP001824 (plasmid) | |
| GenBank date of release | November 23, 2009 | |
| GOLD ID | Gc01151 | |
| NCBI project ID | 21087 | |
| Database: IMG-GEBA | 2502082099 | |
| MIGS-13 | Source material identifier | DSM 20745 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
S. thermophilus S 6022T, DSM 20745, was grown in DSMZ medium 467 [20] at 55°C. DNA was isolated from 1-1.5 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions with modification st/FT for cell lysis according to Wu et al. [21].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger, 454 and Illumina sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/. 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 4,435 overlapping fragments of 1,000 bp and entered into the assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the Arachne assembler. Possible mis-assemblies were corrected and gaps between contigs were closed by custom primer walks from sub-clones or PCR products. A total of 109 Sanger finishing reads were produced. Illumina reads were used to improve the final consensus quality using an in-house developed tool (the Polisher – publication in preparation). The final assembly consists of 35,091 Sanger and 516,954 Roche/454 reads. The error rate of the completed genome sequence is less than 1 in 100,000. Together all sequence types provided 35.9× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [22] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline (http://geneprimp.jgi-psf.org/) [23]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (http://img.jgi.doe.gov/er) platform [24].
Genome properties
The two replicons containing genome is 3,993,764 bp long with a 68.1% GC content (Table 3 and Figure 3). Of the 3,582 genes predicted, 3525 were protein coding genes, and 57 RNAs; 40 pseudogenes were also identified. The majority of the protein-coding genes (72.3%) were assigned a putative function while those remaining were annotated as hypothetical proteins. The properties and the statistics of the genome are summarized in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,993,764 | 100.00% |
| DNA coding region (bp) | 3,461,586 | 86.67% |
| DNA G+C content (bp) | 2,720,128 | 68.11% |
| Number of replicons | 2 | |
| Extrachromosomal elements | 0 | |
| Total genes | 3,582 | 100.00% |
| RNA genes | 57 | 1.59% |
| rRNA operons | 2 | |
| Protein-coding genes | 3,525 | 98.41% |
| Pseudo genes | 40 | 1.12% |
| Genes with function prediction | 2,591 | 72.33% |
| Genes in paralog clusters | 677 | 18.90% |
| Genes assigned to COGs | 2,619 | 73.12% |
| Genes assigned Pfam domains | 2,679 | 74.79% |
| Genes with signal peptides | 709 | 19.79% |
| Genes with transmembrane helices | 908 | 25.35% |
| CRISPR repeats | 1 |
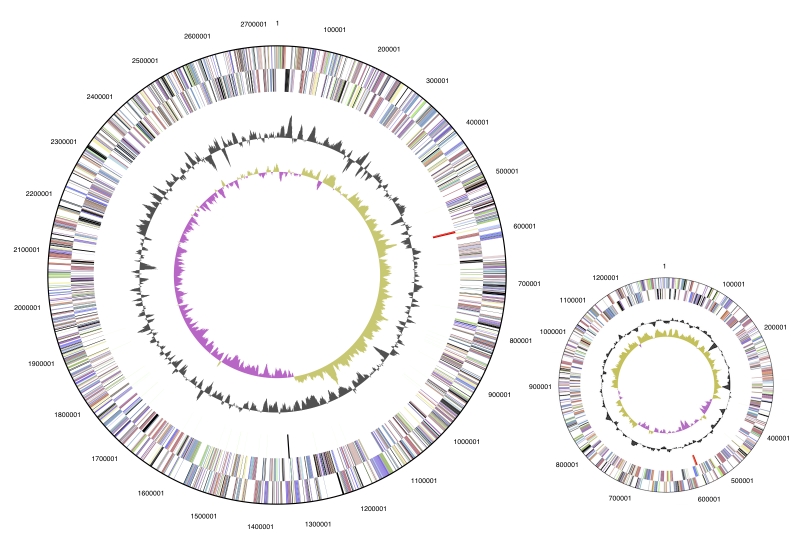
Figure 3.
Graphical circular map of the genome. Chromosome (left), plasmid (right), drown not in scale. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 162 | 4.6 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 162 | 4.6 | Transcription |
| L | 121 | 3.4 | Replication, recombination and repair |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| D | 26 | 0.7 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 61 | 1.7 | Defense mechanisms |
| T | 109 | 3.1 | Signal transduction mechanisms |
| M | 173 | 4.9 | Cell wall/membrane biogenesis |
| N | 36 | 1.0 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 47 | 1.3 | Intracellular trafficking and secretion |
| O | 108 | 3.1 | Posttranslational modification, protein turnover, chaperones |
| C | 226 | 6.4 | Energy production and conversion |
| G | 157 | 4.5 | Carbohydrate transport and metabolism |
| E | 404 | 11.5 | Amino acid transport and metabolism |
| F | 60 | 1.7 | Nucleotide transport and metabolism |
| H | 145 | 4.1 | Coenzyme transport and metabolism |
| I | 111 | 3.1 | Lipid transport and metabolism |
| P | 163 | 4.6 | Inorganic ion transport and metabolism |
| Q | 79 | 2.2 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 373 | 10.6 | General function prediction only |
| S | 207 | 5.9 | Function unknown |
| - | 963 | 27.3 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Gabriele Gehrich-Schröter for growing S. thermophilus cultures and Susanne Schneider for DNA extraction and quality analysis (both at DSMZ). This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Demharter W, Hensel R, Smida J, Stackebrandt E. Sphaerobacter thermophilus gen. nov., sp. nov. A deeply rooting member of the actinomycetes subdivision isolated from thermophilically treated sewage sludge. Syst Appl Microbiol 1989; 11:261-266 [Google Scholar]
- 2.Hensel R, Demharter W, Hilpert R. The microflora involved in aerobic-thermophilic sludge stabilization. Syst Appl Microbiol 1989; 11:312-319 [Google Scholar]
- 3.Jackson TJ, Ramaley RF, Meinschein WG. Thermomicrobium, a new genus of extremely thermophilic bacteria. Int J Syst Bacteriol 1973; 23:28-36 [Google Scholar]
- 4.Wu D, Raymond J, Wu M, Chatterji S, Ren Q, Graham JE, Bryant DA, Robb F, Colman A, Tallon L, et al. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS ONE 2009; 4:e4207 10.1371/journal.pone.0004207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 6.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 7.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 8.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes OnLine Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottow JCG. Detection of hippurate hydrolase among Bacillus species by thin layer chromatography and other methods. J Appl Bacteriol 1974; 37:15-30 [DOI] [PubMed] [Google Scholar]
- 10.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes: the “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugenholtz P, Stackebrandt E. Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description). Int J Syst Evol Microbiol 2004; 54:2049-2051 10.1099/ijs.0.03028-0 [DOI] [PubMed] [Google Scholar]
- 13.Garrity GM, Holt JG. Class I. Thermomicrobia class. nov. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 447. [Google Scholar]
- 14.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Validation List no. 85. Int J Syst Evol Microbiol 2002; 52: 685-690; 10.1099/ijs.0.02358-0 [DOI] [PubMed] [Google Scholar]
- 15.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479-491 [Google Scholar]
- 16.Garrity GM, Bell JA, Lilburn T. Taxonomic outline of the Procaryotes In: Garrity GM, Bell JA, Lilburn TG (eds), Taxonomic Outline of the Procaryotes, Bergey's Manual of Systematic Bacteriology, Second Edition. Release 4.0, Fourth Edition, Springer-Verlag, New York, 2003, p. 1-39. [Google Scholar]
- 17.Biological Agents. Technical rules for biological agents www.baua.de TRBA 466.
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 1972; 36:407-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/ media_list.php
- 21.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anonymous. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. Oak Ridge National Laboratory and University of Tennessee 2009 http://compbio.ornl.gov/prodigal/
- 23.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. (Submitted). [DOI] [PubMed] [Google Scholar]
- 24.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]