Outcome Prediction of Children with Neuroblastoma using a Multigene Expression Signature, a Retrospective SIOPEN/COG/GPOH Study (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 25.
Abstract
BACKGROUND
More accurate prognostic assessment of patients with neuroblastoma is required to improve the choice of risk-related therapy. The aim of this study is to develop and validate a gene expression signature for improved outcome prediction.
METHODS
Fifty-nine genes were carefully selected based on an innovative data-mining strategy and profiled in the largest neuroblastoma patient series (n=579) to date using RT-qPCR starting from only 20 ng of RNA. A multigene expression signature was built using 30 training samples, tested on 313 test samples and subsequently validated in a blind study on an independent set of 236 additional tumours.
FINDINGS
The signature accurately classifies patients with respect to overall and progression-free survival (p<0·0001). The signature has a performance, sensitivity, and specificity of 85·4% (95%CI: 77·7–93·2), 84·4% (95%CI: 66·5–94·1), and 86·5% (95%CI: 81·1–90·6), respectively to predict patient outcome. Multivariate analysis indicates that the signature is a significant independent predictor after controlling for currently used riskfactors. Patients with high molecular risk have a higher risk to die from disease and for relapse/progression than patients with low molecular risk (odds ratio of 19·32 (95%CI: 6·50–57·43) and 3·96 (95%CI: 1·97–7·97) for OS and PFS, respectively). Patients with increased risk for adverse outcome can also be identified within the current treatment groups demonstrating the potential of this signature for improved clinical management. These results were confirmed in the validation study in which the signature was also independently statistically significant in a model adjusted for MYCN status, age, INSS stage, ploidy, INPC grade of differentiation, and MKI. The high patient/gene ratio (579/59) underlies the observed statistical power and robustness.
INTERPRETATION
A 59-gene expression signature predicts outcome of neuroblastoma patients with high accuracy. The signature is an independent risk predictor, identifying patients with increased risk in the current clinical risk groups. The applied method and signature is suitable for routine lab testing and ready for evaluation in prospective studies.
FUNDING
The Belgian Foundation Against Cancer, found of public interest (project SCIE2006-25), the Children Cancer Fund Ghent, the Belgian Society of Paediatric Haematology and Oncology, the Belgian Kid’s Fund and the Fondation Nuovo-Soldati (JV), the Fund for Scientific Research Flanders (KDP, JH), the Fund for Scientific Research Flanders (grant number: G•0198•08), the Institute for the Promotion of Innovation by Science and Technology in Flanders, Strategisch basisonderzoek (IWT-SBO 60848), the Fondation Fournier Majoie pour l’Innovation, the Instituto Carlos III,RD 06/0020/0102 Spain, the Italian Neuroblastoma Foundation, the European Community under the FP6 (project: STREP: EET-pipeline, number: 037260), and the Belgian program of Interuniversity Poles of Attraction, initiated by the Belgian State, Prime Minister's Office, Science Policy Programming.
Introduction
Few tumours have engendered as much fascination and frustration for clinicians and scientists as neuroblastoma (NB). This tumour is one of the most frequent solid malignancies in children and, in contrast to many other paediatric malignancies, remains fatal in almost half of the patients despite advances in multimodal anti-cancer therapies. Current therapeutic stratification of NB patients is based on risk estimation according to combinations of age, tumour stage, MYCN status, DNA ploidy status, and histopathology.1 Clinical experience with this system suggests that the stratification of patients for treatment is useful, but patients with the same clinicopathological parameters, receiving the same treatment, can have markedly different clinical courses. Consequently, patients with an intrinsic poor prognosis classified as low- and intermediate-risk based on the current stratification system will receive inappropriately mild treatment and this could lead to a loss of valuable time prior to installing the required, more intensive treatment. On the other hand, patients with an intrinsic good prognosis but recognised as high-risk with current stratification will undergo a toxic therapy putting them unnecessarily at risk for potential long term side effects. In addition, survival rates remain disappointingly low in the current high-risk treatment group. Therefore, the challenge remains to identify additional tumour-specific prognostic markers for improved risk estimation at the time of diagnosis. Only then can patients receive the most appropriate therapy, be monitored more intensively if needed, and become eligible for new experimental therapies.
In analogy with the successful identification of gene expression signatures in other tumour entities2–5, we sought to develop, validate, and implement a robust multigene expression signature for more accurate assessment of prognosis in children with NB. Here, for the first time to our knowledge, in contrast to previously published gene expression studies in NB, we aimed for a high patient/gene ratio, testing a carefully selected small number of genes (59) on a large panel of tumour samples (579), which underlies the observed statistical power and robustness. We further validated the signature in an independent set of tumours whereby laboratory analyses were performed blinded to clinical and outcome data.
Methods
Robust selection of prognostic genes
Using a unique data-mining strategy we re-analysed seven published microarray gene expression studies6–12 containing nearly 700 NB patients in order to identify genes that correlate with patient outcome (Supplemental Material 1). Briefly, all probes and clinical patient information were updated before re-analysis and a uniform risk definition was applied to select training patients across the different studies. Whereas in each of the published studies a different data-mining method was used, an important step in this procedure was the use of a uniform method, namely prediction analysis of microarrays13 (PAM), generating seven new prognostic gene sets.
We extended this study by an extensive literature screening for single candidate prognostic genes (~800 abstracts). In total, we composed a list of 59 prognostic genes that were independently identified in at least two of the seven prognostic gene sets or literature gene list (Supplemental Table 1).
Study population
The initial cohort was comprised of 343 NB patients from the International Society of Pediatric Oncology, European Neuroblastoma Group (SIOPEN). Patients were only included provided primary untreated NB tumour RNA (at least 60% tumour cells and confirmed histological diagnosis of NB) was available and of sufficient quality. Almost all patients (n=324, 95%) were uniformly treated according to the SIOPEN protocols: HR-NBL1 (High-Risk Neuroblastoma Study, n=66) (https://www.siopen-r-net.org), INES14,15 (Infant Neuroblastoma European Study, n=177; NB99·study (resectable tumours), NB99·1 (unresectable tumours), NB99·2 (Stage 4S & stage 4 (no bone, lung, pleura, or CNS)), NB99·3 (Stage 4 with bone, pleura, lung, or CNS involvement), or NB99·4 (Stage 2, 3, 4 & 4S MYCN amplified tumours)), EUNB16 (European Unresectable Neuroblastoma Study, n=22), or LNESG117 (Localized Neuroblastoma European Study Group, n=59). Thirty-three patients from the Gesellschaft für Paediatrische Onkologie und Haematologie (GPOH) with localised tumour treated with surgery alone were also used in the study and included in the LNESG1 group if older than 12 months at diagnosis and in the INES group (NB99.study) if younger than 12 months. The remainder (n=19) was treated according to similar protocols. The median follow-up was 63 months (range 1–180 months) and >24 months for the majority of the patients (91%). At the time of analysis, 290 out of 343 patients were alive (Supplemental Data, Vermeulen.rdml).
The validation cohort was comprised of 236 patients from the Children’s Oncology Group (COG-United States) (67 low-risk, 56 intermediate-risk, and 113 high-risk patients) with at least 24 months of follow-up for patients without event. The treatments received are not known for most COG patients as 32% (76/236) of patients enrolled only on a nontherapeutic/biology COG study to have risk biomarkers determined and tumour banked. The rest of the COG patients were enrolled on 13 different therapeutic studies, receiving an array of treatments per COG protocols according to risk over that time period. All laboratory analyses were performed blinded to clinical and outcome data. All patients were consented and enrolled on at least one COG study and institutions had Institutional Review Board approval for the COG studies (Supplemental Data, Vermeulen.rdml).
This study was approved by the Ghent University Hospital Ethical Committee (EC2008/159).
Sample preparation
Total RNA extraction from primary NB tumour samples was performed in the individual collaborating laboratories by three different methods and starting from 20 ng of total RNA, a sample pre-amplification method was applied (WT-Ovation, NuGEN) (Supplemental Material 2). Based on the assessment of RNA purity and integrity as detailed in Supplemental Material 3, we retained approximately 80% of the samples with an acceptable quality18 (RNA Quality Index ≥5 as determined by the Experion (software version 3.0, Bio-Rad) and absence of enzymatic inhibitors19).
High-throughput real-time quantitative PCR based gene expression
A real-time quantitative polymerase chain reaction (RT-qPCR) assay was designed for each of the 59 prognostic genes and five reference genes by PrimerDesign and validated through an extensive in silico analysis pipeline.20 PCR plates were prepared using a 96-well head pipetting robot (Sciclone ALH3000, Calliper) and RT-qPCR was performed on a high-throughput 384-well plate instrument (LC480, Roche). To detect and correct for inter-run variation and allow future data comparison with other labs, we used absolute standards (Biolegio), run in parallel with patient samples. Further details on gene expression analysis and on data pre-processing are available in Supplemental Material 4.
Statistical analysis
The multigene expression signature was built using 30 training samples, tested on the remaining SIOPEN samples and validated in a blind manner using COG samples (Supplemental Material 5).
For the SIOPEN cohort, the R-language for statistical computing (version 2·6·2) was used to train and test the prognostic signature, to evaluate its performance by receiver operating characteristic (ROC) curve and area under the curve (AUC) analyses and for Kaplan-Meier survival analyses using the Bioconductor MCRestimate, the ROC and survival packages, respectively. Multivariate logistic regression analyses were performed using SPSS (version 16). Currently used risk factors such as age at diagnosis (≥12 months vs. <12 months), INSS (International Neuroblastoma Staging System) stage (stage 4 vs. not stage 4), and MYCN status (amplified vs. not amplified) were tested and variables with p<0·05 were retained in the model. Since an interaction between the signature and risk factors was not expected to occur, interaction terms were not included in the models. For ROC and multivariate analyses, only patients with an event and patients with sufficient follow-up time (≥36 months) were included if no event occurred since 95% of events in neuroblastoma are expected to occur within the 36 months after diagnosis.
For the validation of the signature on the COG cohort, a case-control study was set up (Supplemental Material 6). This was done in order to ensure a sufficient number of events in each risk group, i.e., to increase the power from what would have resulted from a random sample. A case was defined as failure (relapse, progression, or death from disease for progression-free survival (PFS), and death for overall survival (OS)) prior to two years and control as non-failure prior to two years in patients with at least two years of follow-up. Controls and cases with complete data were selected 2 to 1 to increase the sample size and power. Multivariate logistic regression analyses were performed to determine if the signature was a significant independent predictor after controlling for known risk factors. Statistical analyses were conducted in SAS (version 9).
Role of the funding source
The funding sources of the study had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. All authors had access to the raw data as they are publically available and can be downloaded from http://medgen.ugent.be/jvermeulen. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
Establishment and testing of a 59-gene expression signature
Based on an innovative strategy consisting of re-analysis of seven published microarray gene expression studies6–12 combined with an extensive literature screening, a set of 59 genes with prognostic power in at least two independent studies was selected (Supplemental Material 1 and Table 1).
A prognostic multigene signature was subsequently built based on the expression of the 59 genes using 15 deceased high-risk and 15 low-risk patients with a long progression-free survival time. Patients with a low- or high-risk based on the expression of the 59 genes will be defined as low or high molecular risk respectively throughout the rest of the text.
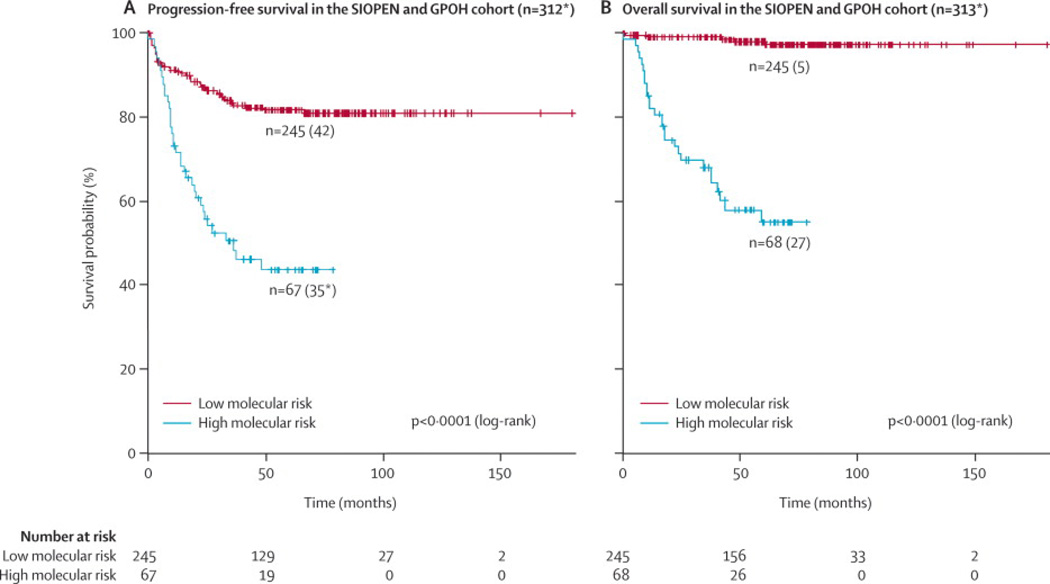
This multigene expression signature significantly distinguished the remaining 313 (missing relapse date for one high molecular risk case) patients with respect to PFS and OS (p<0·0001) (Figure 1). PFS at five years from the date of diagnosis was 81·2% (95%CI: 76·8–87·0) for the group of patients at low molecular risk compared to 43·6% (95%CI: 32·4–58·6) for the group of patients at high molecular risk. The five-year OS was 98·0% (95%CI: 96·1–100) and 55·0% (95%CI: 43·1–70·1) respectively.
Figure 1.
Kaplan-Meier and log-rank analysis for progression-free (a) and overall (b) survival of the entire SIOPEN/GPOH cohort
* missing relapse date for one HR case
LR: low molecular risk
HR: high molecular risk
PFS: progression-free survival
OS: overall survival
(Legend: number of patients in predicted subgroups, between brackets: number of patients with event)
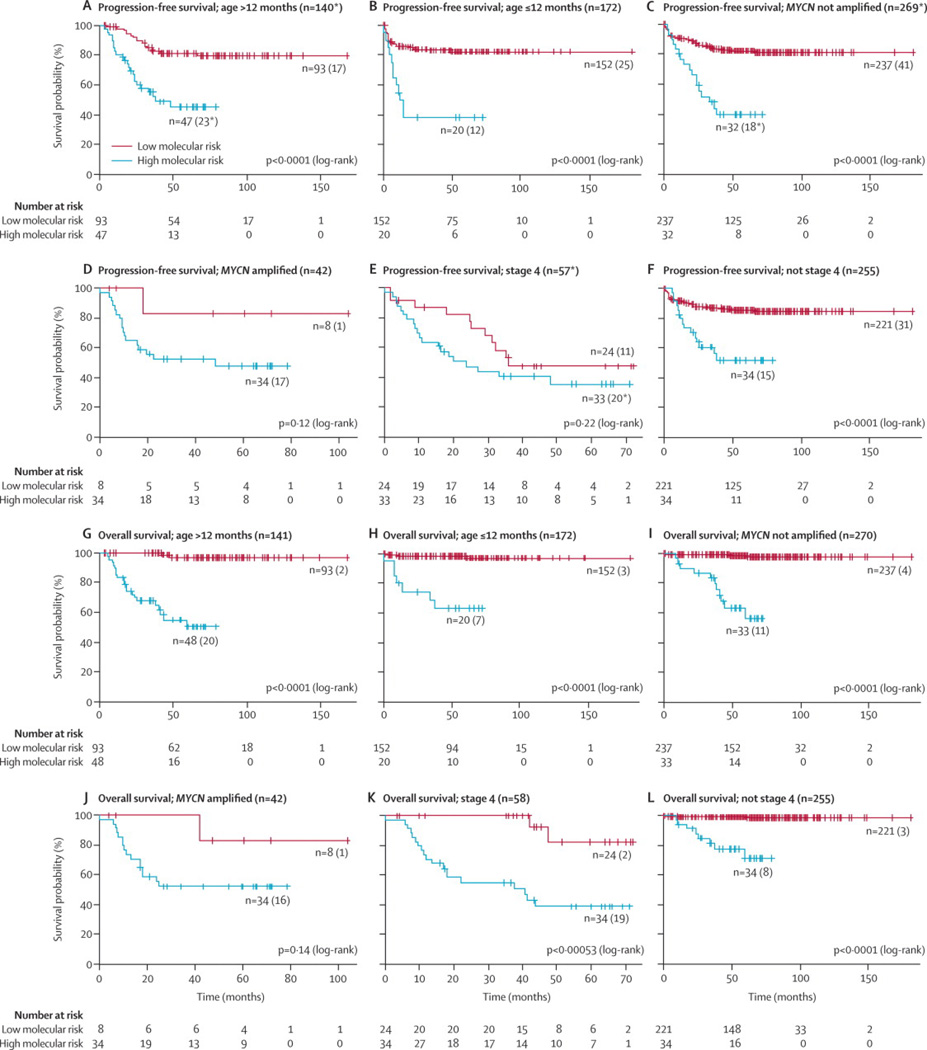
Patients with increased risk for both a shorter PFS and OS could also be identified after stratification by currently used European risk factors such as age, MYCN status, and INSS stage (Figure 2).
Figure 2.
Kaplan-Meier and log-rank analysis for progression-free (a) and overall (b) survival of the stratified SIOPEN cohort according to currently used risk factors
* missing relapse date for one HR case
LR: low molecular risk
HR: high molecular risk
PFS: progression-free survival
OS: overall survival
(Legend: number of patients in predicted subgroups, between brackets: number of patients with event)
Missing MYCN status for 1 HR case
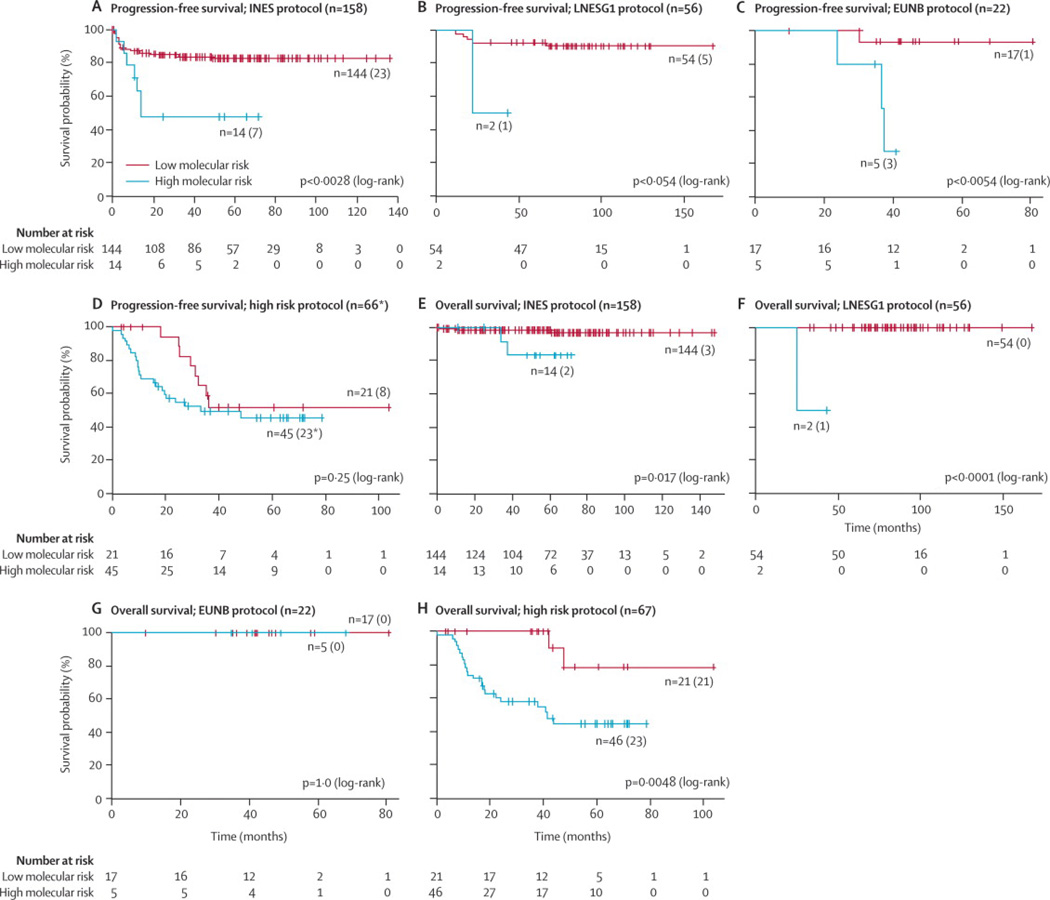
Subsequently, we tested the signature within each SIOPEN treatment protocol. In the group of patients treated according to the INES (NB99·study, NB99·1, NB99·2, and NB99·3), LNEGS1 protocols, and HR-NBL1 protocol (with inclusion of patients sharing the same high-risk features as described in Supplemental Material 5 and treated according to similar protocols) patients with increased risk for death could be identified (p=0·017, p<0·0001, and p=0·0048 respectively). While the signature was useful in identifying those patients at risk of a progression or relapse amongst patients treated according to the INES, LNESG1, and EUNB protocols (p=0·0028, p=0·054, and p=0·0054 respectively), there was no difference in PFS between patients at high and low molecular risk treated according to the HR-NBL1 protocol (Figure 3).
Figure 3.
Kaplan-Meier and log-rank analysis for progression-free (a) and overall (b) survival of the stratified SIOPEN cohort according to treatment group
* missing relapse date for one HR case
LR: low molecular risk
HR: high molecular risk
PFS: progression-free survival
OS: overall survival
(Legend: number of patients in predicted subgroups, between brackets: number of patients with event)
Multivariate logistic regression analysis of the SIOPEN patients was performed within a subset of the overall SIOPEN cohort as described in the patients and methods section. Table 1a shows that the signature and INSS stage were the only significant independent predictors (odds ratio of 19·32 (95%CI: 6·50–57·43) and 3·96 (95%CI: 1·97–7·97) for OS and PFS, in case of an adverse outcome signature). Further, within the INES and HR protocols, multivariate logistic regression analysis demonstrated that the signature was the only significant independent predictor for OS (odds ratio of 7·00 (95%CI: 1·04–46·95) and 9·20 (95%CI: 1·80–47·06), respectively).
Table 1.
Multivariate logistic regression analysis
| a. SIOPEN/GPOH cohort | |||
|---|---|---|---|
| backward-selected logistic regression model for PFS - entire cohort | |||
| variable (n = 257)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | <0·00012 | 3·96 | (1·97, 7·97) |
| INSS stage (stage 4 vs. not stage 4) | <0·00049 | 3·76 | (1·79, 7·92) |
| backward-selected logistic regression model for OS - entire cohort | |||
| variable (n = 254)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | <0·0001 | 19·32 | (6·50, 57·43) |
| INSS stage (stage 4 vs. not stage 4) | 0·0016 | 4·84 | (1·82, 12·91) |
| backward-selected logistic regression model for PFS - HR protocol | |||
| variable (n = 52)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | NS | NA | NA |
| INSS stage (stage 4 vs. not stage 4) | 0·019 | 5·06 | (1·30, 19·72) |
| backward-selected logistic regression model for OS - HR protocol | |||
| variable (n = 52)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | 0·0080 | 9·20 | (1·80, 47·06) |
| backward-selected logistic regression model for PFS - INES protocol | |||
| variable (n = 122)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | 0·0080 | 5·30 | (1·54, 18·23) |
| backward-selected logistic regression model for OS - INES protocol | |||
| variable (n = 120)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | 0·045 | 7·00 | (1·04, 46·95) |
| b. COG cohort | |||
|---|---|---|---|
| backward-selected logistic regression model for PFS | |||
| variable (n = 209)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | <0·0001 | 3·68 | (2·01, 6·71) |
| logistic regression models for OS with separate forced adjustment for variables | |||
|---|---|---|---|
| variable (n = 111)* | p-value | odds ratio | 95% CI on odds ratio |
| expression signature (high-risk vs. low-risk) | <0·0001 | 10·53 | (4·00, 27·78) |
| age (≥ 18 months vs. < 18 months) | NS | NA | NA |
| expression signature (high-risk vs. low-risk) | <0·0001 | 7·14 | (2·70, 18·87) |
| INSS stage (stage 4 vs. not stage 4) | 0·015 | 3·52 | (1·28, 9·71) |
| expression signature (high-risk vs. low-risk) | 0·0048 | 4·81 | (1·62, 14·29) |
| MYCN status (amplified vs. not amplified) | 0·0084 | 4·93 | (1·51, 16·13) |
| expression signature (high-risk vs. low-risk) | <0·0001 | 7·19 | (2·71, 18·87) |
| ploidy (diploid vs. hyperdiploid) | 0·014 | 3·37 | (1·28, 8·85) |
The probability that a patient will be correctly classified by the signature based on a ROC-curve analysis (AUC) was 85·4% (95%CI: 77·7–93·2) and 66·9% (95%CI: 59·2–74·6) for OS and PFS, outperforming current risk factors (age (62·3% (95%CI: 52·2–72·4) and 53·5% (95%CI: 45·8–61·2)), INSS stage (77·0% (95%CI: 66·8–87·1) and 65·4% (95%CI: 57·6–73·2)), and MYCN status (72·7% (95%CI: 61·7–83·8) and 57·2% (95%CI: 49·3–65·2))). For prediction of OS, the signature had a sensitivity of 84·4% (27/32) (95%CI: 66·5–94·1) (= the percentage of patients at high molecular risk that had an adverse outcome) and a specificity of 86·5% (192/222) (95%CI: 81·1–90·6) (= the percentage of patients at low molecular risk that had a good outcome).
Blind validation of the multigene expression signature
In order to validate the multigene expression signature in a completely independent patient cohort, 236 COG tumours were tested in a blind manner. The same signature as used for the SIOPEN cohort identified COG patients who were at greater risk for progression, relapse, or death. Multivariate logistic regression analysis showed that the signature was independently statistically significant in a model adjusted for MYCN status, age, INSS stage, ploidy, INPC grade of differentiation, and MKI. The signature was the only independent significant predictor for PFS, with complete data for 139 controls and 70 cases. Patients at high molecular risk had a greater risk for relapse or progression (odds ratio of 3·68 (95%CI: 2·01–6·71)). In terms of OS, there were not enough deaths to power the fit of a logistic regression model with forced inclusion of all factors. Therefore, separate models testing the signature with adjustment for one risk factor at a time were fit, with complete data for 74 controls and 37 cases. In each model comparing the signature to a given risk factor, the odds ratio of the expression signature always had a higher significance (smaller P-value) than any other variable (Table 1b).
Discussion
Identification of more specific and sensitive markers for outcome prediction and response to therapy is required in order to further improve the choice of risk-related therapy for children with NB. Using a carefully selected set of 59 prognostic genes based on an innovative data-mining strategy, we performed a gene expression study on the largest NB patient series to date, covering 579 patients in total. Our robust prognostic multigene expression signature was tested on a large set of SIOPEN tumours from uniformly treated patients and validated on an independent set of COG tumours. The signature is a strong independent risk predictor, able to identify patients with increased risk in the current risk groups.
Our study is unique in that a carefully selected set of only 59 genes was tested on a large panel of 579 tumour samples, thus increasing statistical power and robustness through this high patient/gene ratio. Several previous studies have attempted to identify prognostic signatures in NB based on genome-wide mRNA expression profiles. However, an important limitation of most published gene expression studies is the lack of statistical power due to extremely low patient/gene ratio. As such, there are inherent but often overlooked statistical issues, such as data over-fitting, unstable gene lists, and lack of study power.21 Consequently, for any small set of tumours, a gene classifier can be easily established, with little or no utility if not validated on an independent patient cohort.
After having established and successfully tested our robust prognostic signature on the total patient cohort, we next assessed the value of the signature in relation to the currently used risk factors, using multivariate logistic regression analysis and survival analysis after stratification of the patients based on the currently used risk factors. The signature significantly discriminates patients in most of the clinical risk subgroups. Possible reasons for absence of discrimination in some subgroups might be the relatively low number of patients in these subgroups or not-sufficiently long follow-up times. Most importantly, the multivariate analysis attributes independent significant value to the signature. Based on this signature, patients with higher risk for death by disease can be identified (odds ratio of 19·32 (95%CI: 6·50–57·43)), indicating that our gene signature clearly outperforms the other risk factors. This demonstrates the potential of this gene expression signature for improved clinical management of NB patients.
Of further interest is that survival analyses within the groups of patients treated according to the current European treatment protocols clearly demonstrate that the signature enables the discrimination of patients with different disease outcome. This is an important finding, especially within the current high-risk category of patients treated according to the HR-NBL1 protocol, as currently no information on genomic aberrations or other factors is available in order to identify a group of patients with worse outcome in this subgroup of NB patients. Strikingly, all but one patient who achieved second complete response in this subgroup were classified as having low molecular risk and had encountered late relapses (median: 31·6, mean: 32·2 months) compared to the group of patients who did not achieve second complete response of which all but two were classified as having high molecular risk and had encountered early relapses (median: 10·0, mean: 13·6 months). Further, death from disease also occurred earlier in the group of patients with high molecular risk (average: 12·3, median: 9·6 months) compared to patients with low molecular risk (average: 27·1, median: 27·1 months). Consequently, patients with high molecular risk within this subgroup could be the future candidates for new and hopefully more effective targeted therapies and some of those with a low molecular risk might possibly be allowed for no transplant. On the other hand, patients who have a high molecular risk and who are currently treated with surgery alone or mild chemotherapy might benefit from more appropriate therapies, i.e. according to the current HR-NBL1 protocol.
An essential step in the validation procedure of our signature is its performance assessment on an independent set of COG tumours whereby all analyses were performed blinded to clinical and outcome data. Similar performance of the expression signature was observed, indicating that the signature can yield reproducible results in independent patient cohorts. Moreover, irrespective of possible confounding factors related to patient ethnicity, treatment with other drugs, and RNA extraction with different standard operating procedures, the success of this validation study also confirms the robustness of the signature. In comparison to existing NB classifiers, our signature has comparable performance within the total cohort of patients, but here, for the first time to our knowledge, the added value of the signature in comparison to currently used risk stratification systems has been confirmed on a totally independent set of tumours in a blind study.
In order to reduce the gene set to a smaller robust gene subset we used several methods including Spearman’s rank correlation clustering and selection of one or two genes in each gene cluster, top ranking univariate cox and logistic regression analyses and the rank product method. Although similar classification performance could be obtained, the 59-gene list always slightly outperformed the reduced lists.
Inspection of the genes being part of the signature reveals seven genes that have previously been linked to NB biology (MYCN, NTRK1, ODC122) or have been proposed as positional candidate genes including CAMTA123 and CHD524 on 1p, BIRC525,26 on 17q, and CADM1 (IGSF4)27,28 on 11q. Gene Ontology analysis of the signature and comparison of the gene list with the super PCNA gene (proliferation signature) (unpublished data; Detours et al.) revealed that only very few genes are involved in cell cycle regulation and proliferation. This remarkable finding is in contrast to signatures in many other cancer entities (e.g. breast cancer29) in which typically more than two thirds of the genes are implicated in proliferation. In line with this, there appear to be very few genes involved in inflammation, also typically seen in other cancers.30 Additional Gene Ontology analysis of the prognostic gene list showed that genes implicated in neuronal differentiation such as PTN, NRCAM, DPYSL3, SCG2, DDC, FYN, NTRK1, MAPT, PMP22, CHD5, and MTSS1, are enriched amongst the genes higher expressed in low-risk tumours. Further scrutinizing of the prognostic gene list and functional analyses might reveal genes that play a role in neuroblastoma pathogenesis and therefore could serve as potential therapeutic targets or point at pathways involved in cancer that could be targeted by new therapies.
Important features of the applied RT-qPCR quantification strategy for marker gene expression analysis are speed, accuracy, cost-effectiveness, applicability in routine laboratories, and requirement of minimal amounts of RNA. As the tumour sample size is often very limited, the applied RNA amplification procedure offers great advantage to accessibility of material for diagnostic and prognostic work-up. Another key success factor of our strategy is the possibility of using universally applicable, quantifiable absolute standards. These synthetic standards not only allow careful monitoring and correction of inter-run variation but also enable the exchange of data between different laboratories, irrespective of the use of a different PCR instrument or reagents. Further validation of this strategy will enable the performance of large multicentre studies conducted at different sites (unpublished data; Vermeulen et al.). An important critical issue of all gene expression studies is RNA quality. The accuracy of gene-expression profiling might indeed be influenced by this metric depending on the quantification method used, the number of genes included in the classifier, expression differences, intra-group variability, and expression levels of the marker genes. Impact of RNA quality on gene-expression has extensively been discussed in the literature and conflicting data exist. In order to not compromise the conclusions of this study, we had stringent RNA quality and purity requirements and excluded ~20% of the samples. Moving forward, further studies should evaluate the impact of RNA quality on classification performance and establish a cut-off designating sufficient quality for reliable class prediction. At the same, standard operation procedures should be introduced to maximize the extraction and storage of high quality ribonucleic acids.
In conclusion, we established and validated a robust prognostic multigene expression signature in the largest NB population till now. The signature can act as an independent risk predictor enabling the identification of patients with increased risk in the current treatment groups. Important advantages of this signature compared to previously published gene expression classifiers are the need of smaller amounts of starting material, the lower number of genes, higher cost-efficiency and speed of the quantification method, and the possibility of cross lab data comparison. This study should form the basis for future investigations such as large well-defined prospective studies with international collaboration. A further challenge is the performance of an integrated analysis for determining the prognostic performance of combining this expression signature with other genomic features of the tumour including microRNA and gene copy number profiling and epigenetic markers along with the currently used clinicobiological factors for risk stratification.
Supplementary Material
01
Acknowledgements
We thank Els De Smet, Nurten Yigit, and Justine Nuytens for their excellent technical assistance. We are grateful to Andy Pearson for the initiation of the SIOPEN collaboration and John Maris for his support in the initiation of the COG validation study. We acknowledge Rob Powel from PrimerDesign for his help with primer design, Biolegio for their collaboration on absolute standard development, and Steve Lefever for his help with the generation of the RDML RT-qPCR data file. We are indebted to all members of the SIOPEN, GPOH and COG for providing tumour samples or the clinical history of patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributors
JV, KDP, GL, FS, and JVDS had substantial contributions to the conception, design, analysis, and interpretation of the data and the drafting of the article for important intellectual content. JV, JH, and LV had substantial contributions to the technical issues. KDP, AN, PMG, and WBL participated in the statistical analysis. NVR, KS, SB, PS, GPT, RN, MP, IJL, OD, VCo, PFA, KB, JB, BM, and MDH had substantial contributions to the management of the tissue banking, MYCN copy number assessment, and provision of the samples. MF, AO, JM, GS, BDB, HR, AC, VCa, JK, UP, RL, and MDH had an important contribution in the management of the patients and databases and in the provision of the clinical information. All authors contributed to the revision of the manuscript for important intellectual content. All authors read and approved the final version of the manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 5.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 6.Oberthuer A, Berthold F, Warnat P, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Diskin S, Rappaport E, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 8.Berwanger B, Hartmann O, Bergmann E, et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2:377–386. doi: 10.1016/s1535-6108(02)00179-4. [DOI] [PubMed] [Google Scholar]
- 9.Ohira M, Oba S, Nakamura Y, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7:337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Schramm A, Schulte JH, Klein-Hitpass L, et al. Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene. 2005;24:7902–7912. doi: 10.1038/sj.onc.1208936. [DOI] [PubMed] [Google Scholar]
- 11.De Preter K, Vandesompele J, Heimann P, et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biol. 2006;7:R84. doi: 10.1186/gb-2006-7-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McArdle L, McDermott M, Purcell R, et al. Oligonucleotide microarray analysis of gene expression in neuroblastoma displaying loss of chromosome 11q. Carcinogenesis. 2004;25:1599–1609. doi: 10.1093/carcin/bgh173. [DOI] [PubMed] [Google Scholar]
- 13.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Bernardi B, Gerrard M, Boni L, et al. Excellent outcome with reduced treatment for infants with disseminated neuroblastoma without MYCN gene amplification. J Clin Oncol. 2009;27:1034–1040. doi: 10.1200/JCO.2008.17.5877. [DOI] [PubMed] [Google Scholar]
- 15.Canete A, Gerrard M, Rubie H, et al. Poor survival for infants with MYCN-amplified metastatic neuroblastoma despite intensified treatment: the International Society of Paediatric Oncology European Neuroblastoma Experience. J Clin Oncol. 2009;27:1014–1019. doi: 10.1200/JCO.2007.14.5839. [DOI] [PubMed] [Google Scholar]
- 16.Kohler J, Rubie H, Castel V, Garaventa A, Haupt Riccardo. The Treatment of Children over the age of One Year with Unresectable Localised Neuroblastoma without MYCN amplification: a SIOPEN study. Paper presented at the Advances in Neuroblastoma Research meeting; May 2008; Chiba. [Google Scholar]
- 17.De Bernardi B, Mosseri V, Rubie H, et al. Treatment of localised resectable neuroblastoma. Results of the LNESG1 study by the SIOP Europe Neuroblastoma Group. Br J Cancer. 2008;99:1027–1033. doi: 10.1038/sj.bjc.6604640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Nolan T, Hands RE, Ogunkolade W, Bustin SA. SPUD: a quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem. 2006;351:308–310. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 20.Lefever S, Vandesompele J, Speleman F, Pattyn F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 2009;37:D942–D945. doi: 10.1093/nar/gkn777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tinker AV, Boussioutas A, Bowtell DD. The challenges of gene expression microarrays for the study of human cancer. Cancer Cell. 2006;9:333–339. doi: 10.1016/j.ccr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Hogarty MD, Norris MD, Davis K, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–9745. doi: 10.1158/0008-5472.CAN-07-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henrich KO, Fischer M, Mertens D, et al. Reduced expression of CAMTA1 correlates with adverse outcome in neuroblastoma patients. Clin Cancer Res. 2006;12:131–138. doi: 10.1158/1078-0432.CCR-05-1431. [DOI] [PubMed] [Google Scholar]
- 24.Fujita T, Igarashi J, Okawa ER, et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MA, Ohashi K, Zhu X, et al. Survivin mRNA levels are associated with biology of disease and patient survival in neuroblastoma: a report from the children's oncology group. J Pediatr Hematol Oncol. 2006;28:412–417. doi: 10.1097/01.mph.0000212937.00287.e5. [DOI] [PubMed] [Google Scholar]
- 26.Albino D, Scaruffi P, Moretti S, et al. Identification of low intratumoral gene expression heterogeneity in neuroblastic tumors by genome-wide expression analysis and game theory. Cancer. 2008;113:1412–1422. doi: 10.1002/cncr.23720. [DOI] [PubMed] [Google Scholar]
- 27.Michels E, Hoebeeck J, De Preter K, et al. CADM1 is a strong neuroblastoma candidate gene that maps within a 3.72 Mb critical region of loss on 11q23. BMC Cancer. 2008;8:173. doi: 10.1186/1471-2407-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowacki S, Skowron M, Oberthuer A, et al. Expression of the tumour suppressor gene CADM1 is associated with favourable outcome and inhibits cell survival in neuroblastoma. Oncogene. 2008;27:3329–3338. doi: 10.1038/sj.onc.1210996. [DOI] [PubMed] [Google Scholar]
- 29.Kim SY, Kim YS. A gene sets approach for identifying prognostic gene signatures for outcome prediction. BMC Genomics. 2008;9:177. doi: 10.1186/1471-2164-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Curr Opin Genet Dev. 2008;18:3–10. doi: 10.1016/j.gde.2008.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01