FHL2, a novel tissue-specific coactivator of the androgen receptor (original) (raw)
Abstract
The control of target gene expression by nuclear receptors requires the recruitment of multiple cofactors. However, the exact mechanisms by which nuclear receptor–cofactor interactions result in tissue-specific gene regulation are unclear. Here we characterize a novel tissue-specific coactivator for the androgen receptor (AR), which is identical to a previously reported protein FHL2/DRAL with unknown function. In the adult, FHL2 is expressed in the myocardium of the heart and in the epithelial cells of the prostate, where it colocalizes with the AR in the nucleus. FHL2 contains a strong, autonomous transactivation function and binds specifically to the AR in vitro and in vivo. In an agonist- and AF-2-dependent manner FHL2 selectively increases the transcriptional activity of the AR, but not that of any other nuclear receptor. In addition, the transcription of the prostate-specific AR target gene probasin is coactivated by FHL2. Taken together, our data demonstrate that FHL2 is the first LIM-only coactivator of the AR with a unique tissue-specific expression pattern.
Keywords: androgen/coactivator/LIM-only/nuclear receptor/transcription
Introduction
Steroid hormone receptors such as the androgen receptor (AR) constitute a subfamily of the nuclear receptor superfamily of ligand-activated transcription factors that play important roles in development, differentiation and homeostasis (Wilson et al., 1991; Cato and Peterzierl, 1998). Members of the steroid receptor family bind as dimers to palindromic or direct repeat response elements (Evans, 1988; Beato et al., 1995). They share a common modular structure and are composed of several domains that mediate DNA binding, dimerization, ligand binding and transcriptional activity (Mangelsdorf et al., 1995). The ligand binding domain (LBD) performs a number of functions including ligand binding, transcriptional activation or repression and contributes to receptor dimerization. Upon ligand binding several major structural changes are induced within the LBD of nuclear receptors (Moras and Gronemeyer, 1998). One obvious difference between the unliganded (apo)- and liganded (holo)-LBD structures is a positional reorientation of helix 12 (H12) (Moras and Gronemeyer, 1998). H12 is indispensable for the transcriptional activation function (AF) of the LBD and harbours the so-called AF–2 AD core motif (Moras and Gronemeyer, 1998). Like other steroid receptors the AR contains an additional N–terminal ligand-independent activation function, the AF–1 (Evans, 1988; Beato et al., 1995). Recent data show direct interaction between the N– and the C–terminus of the AR, supporting the requirement of both the AF–1 and the AF–2 for transcriptional activation (Jenster et al., 1995; Doesburg et al., 1997; Ikonen et al., 1997). It is suggested that the ligand-induced conformational changes of the LBD in concert with physical interactions with the N–terminus result in the formation of novel surfaces, which allow direct protein–protein interactions of the liganded AR with transcriptional cofactors.
Distinct classes of ligand-dependent transcriptional cofactors have been described. They include CBP/p300, the p160 family, pCAF/GCN5 and TRAP/DRIP (Xu et al., 1999). According to the current model these cofactors are organized in multi-protein complexes and facilitate the access of nuclear receptors and the RNA polymerase II core machinery to their target DNA by chromatin remodelling and histone modification. Most of these coactivator complexes not only interact with members of the nuclear receptor family, but also with several other sequence-specific DNA-binding transcription factors (Xu et al., 1999). Under various experimental conditions nuclear receptors are capable of recruiting any of the different cofactor complexes. However, in vivo, only distinct complexes may be required for selective and tissue-specific gene activation (Xu et al., 1999). This raises the question of how target gene selection, and tissue- and development-specific gene activation are achieved in the organism. One attractive possibility to regulate transcription in a specific manner is the restricted expression of cofactors, such as the cold-inducible PPARγ coactivator PGC–1 (Puigserver et al., 1998). These tissue-specific cofactors may allow the recruitment of distinct transcriptional complexes tethered to DNA via the nuclear receptor, finally resulting in selective expression of target genes.
During embryogenesis the AR is essential for the development and differentiation of sexual organs (Jenster, 1999). The development and maintenance of the prostate require androgens and the AR (Trapman and Brinkmann, 1996; Jenster, 1999). The AR also controls the production of proteins that are secreted in the prostatic fluid such as prostate-specific antigen or probasin (Matuo et al., 1985; Sinha et al., 1987; Jenster, 1999). In the prostate, the AR is expressed in the epithelial cells, the secretory cells of the prostate, which respond to androgens (Aumüller et al., 1998). Interestingly, the prostate-specific probasin gene is a selective target gene of the AR, whereas other steroid hormone receptors such as the glucocorticoid receptor fail to activate probasin (Claessens et al., 1996). According to the current model, the AR also plays an important role in the development of prostate cancer, since growth and survival of primary prostatic cancer cells are largely dependent on androgens (Gregory et al., 1998). Consequently, androgen ablation therapies are used clinically for treating patients with prostate cancer.
Expression of the AR is also detected in many other tissues, mainly in male sexual organs, but for example also in liver and cardiac muscle (Schleicher et al., 1989; Kimura et al., 1993). The physiological function of the AR in these tissues still needs to be defined in detail. However, the observation that androgens can mediate cardiac hypertrophy suggests a role of the AR in heart function (Marsh et al., 1998).
The receptors for glucocorticoids (GR), progesterone (PR) and mineralocorticoids (MR) recognize the same DNA element as the AR (Cato and Peterzierl, 1998). In situations where ligands for all steroid receptors are present, differential receptor regulation is necessary to control target gene expression selectively. This could be achieved by interaction with specific coregulators. As demonstrated for most of the other nuclear receptors the AR interacts with ubiquitously expressed coactivators such as members of the p160 family or CBP/p300 (Aarnisalo et al., 1998; Berrevoets et al., 1998). Additional coactivators of the AR have been described. However, for none of these AR cofactors has a selective interaction with the AR been demonstrated, and most of these cofactors also interact with related steroid receptors (Moilanen et al., 1998; Alen et al., 1999; Fujimoto et al., 1999; Heinlein et al., 1999; Hsiao and Chang, 1999; Hsiao et al., 1999; Kang et al., 1999). Therefore, differential regulation of target genes by the nuclear receptors remains unclear.
Although it is suggested that AR activation requires the interaction of the N– and C–terminus, all screens for AR cofactors have been performed with isolated receptor domains so far. In order to identify AR cofactors that bind to the surface generated by the interaction of the holo-LBD together with the N–terminus, we employed a modified yeast two-hybrid system. As the bait protein we used a human AR in which the DNA-binding domain (DBD) was replaced by the Gal4 DBD. We cloned a LIM-only protein identical to a previously reported protein FHL2/DRAL with unknown function (Genini et al., 1997; Chan et al., 1998). Our data show that FHL2 is a selective agonist-dependent coactivator of the AR, but not of other nuclear receptors. FHL2 interacts specifically with the AR in vitro and in vivo. FHL2 harbours a strong, autonomous transactivation function and selectively increases the transcriptional activity of the AR in an agonist- and AF-2-dependent manner. FHL2 is therefore the first LIM-only coactivator for a particular member of the nuclear receptor family. Expression analysis of FHL2 mRNA shows strong expression in the ventricles of the heart. Substructure analysis demonstrates FHL2 protein expression in the myocardium of the heart, as well as in the epithelial cell layer of the prostate. Moreover, protein expression of the AR and FHL2 overlaps in the nuclei of these secretory and androgen-sensitive cells of the prostate. Expression of FHL2 results in an agonist- and AR-dependent transcriptional coactivation of the prostate-specific AR target gene probasin, suggesting an important role for the AR–FHL2 complex in prostate function.
Results
Yeast two-hybrid screen
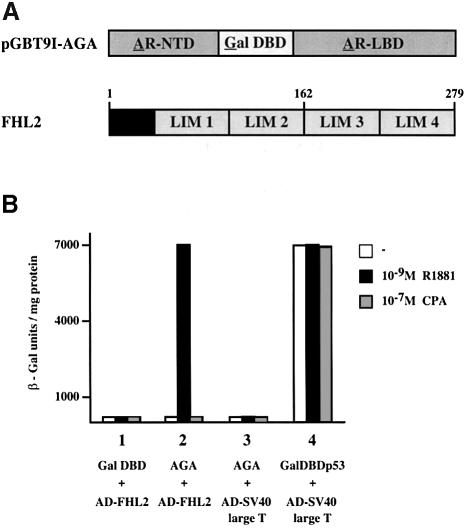
We employed a modified yeast two-hybrid system to identify proteins that interact with the human AR (Wilson et al., 1991). As the bait a hybrid protein (AGA) was used in which the Gal4 DBD replaces the AR DBD (Figure 1A). This approach was chosen in order to isolate AR cofactors, which bind to the surface generated by the interaction of the holo-LBD together with the N–terminus. The transcriptional activity of the bait protein AGA in yeast, especially in the yeast strain HF7c that we used for the two-hybrid assays, is very weak. The screening of a human prostate library was performed in the presence of the synthetic AR agonist R1881. We identified a 1.4 kb cDNA containing an 840 bp open reading frame. The cDNA encodes a protein of 279 amino acids with a predicted molecular weight of 32 kDa (Figure 1A). Sequence similarity searches revealed that it is identical to the previously reported protein FHL2/DRAL with unknown function (Genini et al., 1997; Chan et al., 1998). FHL2 is a LIM–only protein that contains four-and-a-half LIM domains. LIM domains are defined by double zinc finger motifs and mediate protein–protein interactions (Dawid et al., 1998; Jurata and Gill, 1998).
Fig. 1. FHL2 interacts with the AR in yeast in an agonist-dependent manner. (A) Schematic representation of the AGA bait protein and the FHL2 protein. (B) β-galactosidase activity in yeast expressing AGA and Gal4AD–FHL2 in the presence or absence of the agonist R1881 or the antagonist CPA (bar 2). Bars 1 and 3 show the negative and bar 4 shows the positive controls, respectively.
To quantify the in vivo interaction between FHL2 and the AR in yeast, liquid β–galactosidase assays were performed (Figure 1B). FHL2 fused to the activation domain of the yeast transcription factor Gal4 associates with the AR bait protein AGA in an agonist-dependent manner, thereby increasing β-galactosidase reporter gene activity as strongly as the positive control. In the presence of the antagonist cyproterone acetate (CPA), the AR bait protein fails to interact with FHL2 (Figure 1B).
Tissue-specific FHL2 mRNA expression
To analyse the expression pattern of FHL2 mRNA, Northern blot analyses of human and mouse tissues were performed. Multiple tissue Northern blot analyses of human fetal RNA show specific expression of FHL2 in heart (Figure 2A). In addition, Figure 2A shows that FHL2 expression in mouse adult tissues is also restricted to heart and confirms FHL2 mRNA expression in adult human heart (Genini et al., 1997; Chan et al., 1998). To investigate FHL2 expression in human muscle tissues, an expression analysis of RNA from different adult human striated and smooth muscle tissues was performed (Figure 2A). FHL2 is exclusively expressed in heart muscle, since no FHL2 signal could be detected in skeletal muscle or in smooth muscle from different tissues including the stromal smooth muscle cells of the prostate (Figure 2A). To investigate further the expression pattern of FHL2 in human heart, mRNA from explanted human hearts was subjected to Northern blot analysis. Figure 2B shows that FHL2 is mainly expressed in the right and left ventricles. A similar ventricle restricted expression pattern is observed in non-failing human hearts (data not shown).
Fig. 2. Analysis of FHL2 expression. (A) FHL2 is specifically expressed in the heart. Northern blots of human tissues (fetal, adult and adult muscle) and adult mouse tissues were probed with FHL2. β–actin was used as an internal control. (B) FHL2 is mainly expressed in the heart ventricles. Northern blot analysis of mRNA of left and right ventricles (LV, RV) and left and right atria (LA, RA) of an explanted human heart. GAPDH was used as a control. (C) FHL2 and AR protein expression in human heart. Extract (200 μg) from a left ventricle was analysed in a Western blot. Controls are shown in lane 2 (10 μg of 293 cell extract transfected with human AR) and lane 3 (100 ng of purified His-tagged FHL2). The upper panel was decorated with an α–AR antibody, the lower panel with an α–FHL2-specific antibody. (D) Immunohistochemical staining of FHL2 in cardiac muscle cells (a and b) or FHL2 and AR in prostate epithelium (c–f). Controls are shown in b, d and f. In the heart, FHL2 antibodies stained specifically myocardial fibres (my), but not vascular smooth muscle cells (sm) (a). Two hematoxilin–eosin stained arteriolae (at) shown in (a) entirely lack FHL2 immunoreactivity. Both cytoplasmic and nuclear FHL2 immunoreactivity was detected in the secretory epithelium of the prostate (c). AR immunoreactivity was confined to the nucleus of the secretory epithelium (e). Abbreviations: at, arteriola; br, brain; co, colon; ep, epithelial cells; ht, heart; kd, kidney; li, liver; lm, lumen; lu, lung; my, myofibres; pan, pancreas; pl, placenta; pr, prostate; skm, skeletal muscle; si, small intestine; sm, smooth muscle; sp, spleen; st, stomach; str, stroma; te, testis; ut, uterus.
To analyse specifically protein expression of FHL2 in human tissues we generated a monoclonal antibody against FHL2. The α-FHL2 antibody is highly specific for FHL2 and does not interact with any other LIM domain-containing protein tested (data not shown). In human heart the α-Flirt antibody detects a protein of 32 kDa, as exemplified in an extract of the left ventricle (Figure 2C, lower panel). Notably, in the same heart extract AR protein is also observed with an α-AR-specific antibody (Figure 2C, upper panel), demonstrating coexpression of both proteins in heart. Additional immunohistochemical analyses show that FHL2 protein is specifically detected in myofibres of heart myocardium (Figure 2D, a and b). No FHL2 protein is detected in aorta, endo- or epicardium (data not shown). Furthermore, in the smooth muscle cells surrounding the arteriolae FHL2 protein is not expressed (Figure 2D, a). In the prostate FHL2 protein is detected in the epithelial cells (Figure 2D, c). In agreement with our Northern blot data (Figure 2B) FHL2 protein is not expressed in the stromal smooth muscle cells of the prostate. Importantly, in the secretory and androgen-sensitive epithelial cells nuclear FHL2 clearly colocalizes with nuclear AR immunoreactivity (Figure 2D, c and e).
FHL2 binds selectively to the AR
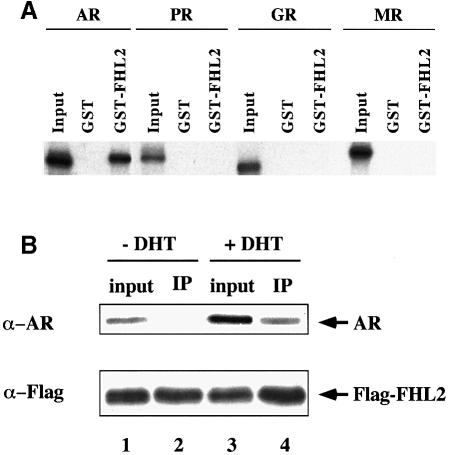
To analyse whether FHL2 fulfils the characteristics of a transcriptional coactivator we first investigated the AR–FHL2 protein–protein interaction. GST pulldown experiments were performed using GST–FHL2 and various in vitro translated [35S]methionine-labelled nuclear receptors (Figure 3A). Figure 3A shows that GST–FHL2 binds specifically to the full-length AR but fails to interact with the control GST protein. Although GST–FHL2 binds full-length AR both in the absence (data not shown) and in the presence of ligand, we included agonist in all further pulldown assays to ensure comparability. GST–FHL2 does not associate with the most homologous steroid hormone receptors GR, PR or MR in the presence of their cognate ligands. FHL2 also fails to associate with receptors of the retinoic acid receptor/thyroid hormone receptor subfamily (data not shown). These results indicate that the interaction between the AR and FHL2 is highly specific for this particular member of the nuclear receptor superfamily.
Fig. 3. FHL2 interacts with the AR in vitro and in vivo. (A) Selective interaction of FHL2 with the AR. GST pulldown assays were performed with in vitro translated, labelled AR, PR, GR or MR in the presence of their cognate ligands and GST–FHL2 fusion protein. GST protein was used as a control. (B) AR coimmunoprecipitates with FHL2 in the presence of the natural agonist DHT. Nuclear extracts of 293 cells transfected with AR and Flag-FHL2 were immunoprecipitated with α–Flag antibody. Ten percent of the extract used for immunoprecipitation was loaded as input in lanes 1 and 3. The immunoprecipitate (IP) is loaded in lanes 2 and 4. Western blots were either decorated with an α–Flag- or an α–AR-specific antibody.
Association between the AR and FHL2 is also revealed by coimmunoprecipitation (Figure 3B). Nuclear extracts from 293 cells transfected with the AR and Flag-epitope-tagged FHL2 were immunoprecipitated using α-Flag antibody. Western blot analysis shows that the AR–Flag–Flirt complex is efficiently immunoprecipitated in the presence of the natural AR agonist dihydrotestosterone (DHT). No AR was found in immunoprecipitated complexes using untagged FHL2 (data not shown) or in the absence of agonist (Figure 3B), demonstrating specificity and agonist dependence of the AR–FHL2 interaction.
Mapping of the AR and FHL2 interaction domains
To delineate the domains in the AR that mediate the protein–protein interaction with FHL2 in vitro, GST pulldown experiments were performed (Figure 4). GST–FHL2 binds full-length AR, but interacts neither with the AR LBD (AR624–919) nor with the AR1–626, which lacks the LBD (Alen et al., 1999). This confirms that the interaction with FHL2 requires the structurally intact AR. When H12 of the AR LBD, which harbours the activation function 2 core motif, was deleted (ARΔH12), no interaction with FHL2 was observed (Figure 4A). In other nuclear receptors helix 3 (H3) is part of the coactivator binding surface (Nolte et al., 1998). To identify specific amino acids within the H3 region that could potentially affect interaction with FHL2, we assayed a series of point mutations in the AR H3 region (Alen et al., 1999). GST–FHL2 binds to the AR mutants K720A, W718A, A719K and to the double mutant V715A/V716A (data not shown). However, FHL2 did not bind the mutant ARL712R (Figure 4A). These results suggest that the H12 and the H3 regions of the AR are an important part of the FHL2 interaction surface.
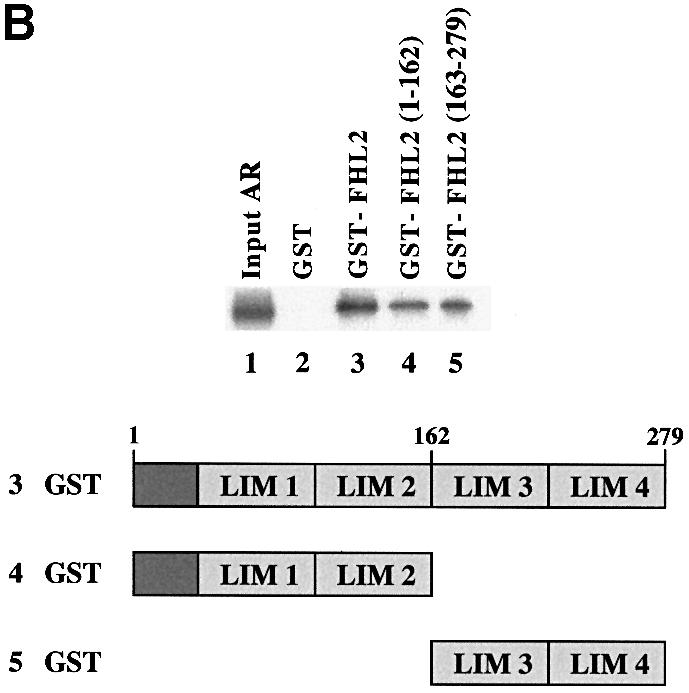
Fig. 4. AR associates with FHL2 in an AF-2-dependent manner. (A) GST pulldown assays were performed using GST–FHL2 fusion protein and in vitro translated, labelled AR or AR mutants in the presence of the agonist R1881. (B) Interaction between the AR and either GST, GST–FHL2, GST–FHL2(1–162) or GST–FHL2(163–279) fusion proteins in the presence of the agonist R1881. Numbers above the scheme indicate the amino acid position.
To determine which region of the FHL2 protein binds to the AR, a series of mutant GST–FHL2 proteins was tested for their ability to interact with full-length AR (Figure 4B). Deletion of either the FHL2 N–terminus or the C–terminal LIM domains 3–4 reduced but did not abolish the ability of the FHL2 protein to bind to the AR (Figure 4B). These results suggest that both the N– and C–terminal LIM domains contribute to the interaction with the AR.
FHL2 contains an autonomous transcriptional activation domain
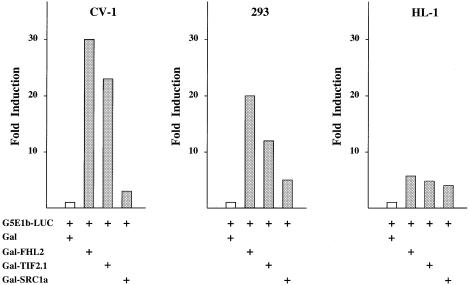
Next, we investigated the transcriptional properties of FHL2 in transient transfection experiments. Since we could not observe DNA binding of FHL2 (data not shown), plasmids expressing the Gal4 DBD fused to full-length FHL2 (Gal–FHL2) were generated and tested in comparison with the known coactivators Gal–TIF2.1 (Voegel et al., 1996) and Gal–SRC1 (Onate et al., 1998). Figure 5 shows that FHL2 tethered to DNA robustly induces the transcriptional activity of the G5E1b-LUC reporter gene in various cell lines. Importantly, the activity of Gal–FHL2 and Gal–TIF2.1 is comparable, whereas Gal-SRC1 seems to be a weaker activator in the cell lines tested (Figure 5). Taken together, these data show that FHL2 contains an autonomous transcriptional activation function.
Fig. 5. FHL2 contains an autonomous transactivation function. The Gal–FHL2 fusion protein transactivates the G5E1b-LUC reporter gene in different cell lines (CV-1, 293 and HL-1). The coactivators Gal–TIF2.1 and Gal–SRC1a were used as controls.
FHL2 acts as a coactivator of AR-dependent transcription
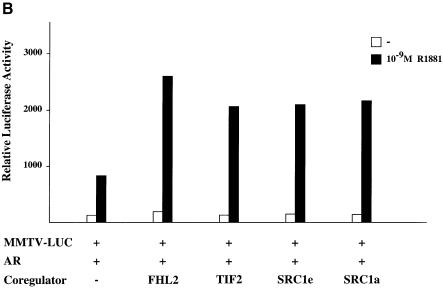
To examine the potential regulation of the AR by FHL2 on a naturally occurring gene the MMTV promoter was chosen because the steroid hormone receptors AR, PR, GR and MR are known to regulate its expression in a ligand-dependent fashion (Schüle et al., 1990). Accordingly, we cotransfected each of these steroid hormone receptors together with FHL2 and the MMTV-LUC reporter gene into 293 cells (Figure 6A). Coexpression of the AR and FHL2 results in a ligand-dependent coactivation, which is not promoted in the presence of an AR antagonist. Importantly, the ligand-dependent transcriptional activity of GR, PR and MR remained unchanged in the presence of FHL2 (Figure 6A). The same result was obtained when the coactivation assay was performed with sub-saturating agonist concentrations for the various receptors (data not shown). In addition, the transcriptional activity of all other nuclear receptors tested, for example ERs, RXR, RAR, TR, VDR, GCNF and RORs, is not coactivated by FHL2 (data not shown). As expected, coactivation of AR is AF-2-dependent as demonstrated by the mutant ARΔH12 (Figure 6A). To compare the ability of FHL2 to coactivate the AR with the ability of previously described coactivators we tested TIF2, SRC1a or SRC1e together with the AR and the MMTV-LUC reporter gene in 293 cells. Figure 6B shows that similar to the other coactivators FHL2 potently coactivates the AR in an agonist-dependent manner.
Fig. 6. FHL2 is a specific coactivator of the AR. (A) FHL2 coactivates the AR in an agonist- and AF-2-dependent manner. Expression plasmids coding for AR, ARΔH12, PR, GR or MR were cotransfected with the MMTV-LUC reporter with or without FHL2 in 293 cells. (B) FHL2, TIF2, SRC1e or SRC1a coactivate the AR to a similar extent in 293 cells. (C) FHL2 functions as a coactivator of the AR-specific probasin-LUC reporter (PB-LUC). AR and FHL2 were cotransfected as indicated in the presence of two different AR agonists (R1881, DHT) or the antagonist CPA in CV-1 cells.
FHL2 coactivates the transcription of the prostate-specific AR target gene probasin
The promoter of the cellular probasin gene, which is expressed in the epithelial cells of the prostate, is selectively regulated by the AR (Claessens et al., 1996). Therefore, we tested FHL2 coactivator function on a probasin reporter (PB-LUC) (Figure 6C). FHL2 enhances the activity of the AR in the presence of either synthetic (R1881) or natural (DHT) agonists in CV-1 cells. Antagonists such as CPA, however, completely block transactivation. Taken together, these data clearly demonstrate that the coactivator function of FHL2 for the AR is AF-2- and agonist-dependent in different cell types and promoter environments (Figure 6B and C). The fact that FHL2 coactivates a well known AR target gene supports a role of the AR–FHL2 complex in prostate function.
Discussion
Recent studies suggest that tissue-restricted gene expression can be controlled by tissue-specific coactivators of transcription factors (Xu et al., 1999). Using a modified yeast two-hybrid screen we identified a novel tissue-specific coactivator of the AR. Sequence comparison showed that it is identical to the previously described FHL2/DRAL protein of unknown function (Genini et al., 1997; Chan et al., 1998). FHL2 is a LIM-only protein that contains four LIM domains and an N–terminal half LIM domain. LIM domains are characterized by a cysteine-composed double zinc finger motif: C–X2-C-X16–23-H-X2-(C/H)-X2-C–X2-C-X16–23-C–X2-(C/H/D) (Jurata and Gill, 1998). LIM domains can mediate protein–protein interaction with LIM domain-containing proteins or various other classes of proteins (Dawid et al., 1998).
FHL2 is a selective coactivator of the AR
We show that FHL2 is a bona fide coactivator of the AR. FHL2 interacts specifically with the AR in vitro and in vivo and harbours a strong, autonomous transactivation function. FHL2 selectively increases the transcriptional activity of the AR in an agonist- and AF-2-dependent manner. In yeast, interaction of the AR bait AGA and FHL2 is strictly dependent on the presence of agonist and fails in the presence of antagonist (Figure 1B). In mammalian cells AR–FHL2 interaction is also strictly agonist-dependent in all assays (Figures 3B and 6A, B). Despite the fact that the AR and FHL2 interact in a strictly agonist-dependent manner when tested in vivo, in vitro binding of recombinant proteins was agonist-independent. This may be due to the presence of an as yet unknown factor in cells that, unless ligand is present, prevents the agonist-dependent interaction of FHL2 and the AR in vivo. Our findings are similar to recently published observations that the interaction of PPARγ and the coactivator PGC–1 (Puigserver et al., 1998) or the interaction of thyroid receptor β and histone deacetylase 2 (Sasaki et al., 1999) is agonist-dependent in vivo, yet ligand-independent in vitro. In synopsis, our data show that FHL2 can bind AR directly. However, the formal possibility might still exist that the FHL2 effects on AR are indirect or require an additional factor.
The structure–function analysis shows that both the N– and C–terminus of the AR are required for the interaction with FHL2. Deletion of H12 of the AR, which harbours the AF-2 AD core motif, eliminates FHL2 binding and consequently abolishes coactivation. Furthermore, a single point mutation in the H3 of the AR (ARL712R) also abrogates FHL2 binding. This AR mutant binds hormone, although with reduced affinity (Alen et al., 1999). These observations show that the AR–FHL2 interaction is not merely dependent on the ability of AR to bind hormone, but that in the holo-LBD conformation H3 and H12 constitute an important part of the interaction surface. Although no crystal structure of the AR is available to date, one might speculate that L712 in H3 is either directly involved in the AR–FHL2 interaction or, alternatively, is essential for generating the correct three-dimensional interaction surface. Obviously, the holo-LBD conformation is necessary but not sufficient for FHL2 binding and function. In addition, the AR N–terminus is required to generate a functional interaction surface, since its deletion abolishes FHL2 binding. Ubiquitously expressed coactivators such as TIF2 contact the AR LBD with the LXXLL motif (Berrevoets et al., 1998). Since FHL2 does not contain an LXXLL motif, it will be interesting to analyse the AR–FHL2 interface on a structural basis.
FHL2 contains a strong and autonomous transactivation function. Importantly, the activity of Gal–FHL2 is similar to Gal–TIF2.1, whereas Gal–SRC1 seems to be a weaker activator in the cell lines tested. Since TIF2 and SRC1 are ubiquitously expressed and enhance the transcriptional activity of several nuclear receptors (Onate et al., 1995; Voegel et al., 1996), it will be of interest to elucidate whether FHL2 and other coactivators act in concert or in different coactivator complexes on the AR. When tested on different natural promoters such as MMTV or the AR-specific probasin, FHL2 acts as a coactivator of the AR in several cell lines. FHL2 selectively enhances AR-dependent transactivation in an agonist-dependent manner, but neither stimulates the transcriptional activity of the related steroid hormone receptors PR, MR, GR nor that of any other nuclear receptor tested. The exceptional selectivity of the AR is a unique property of the LIM-only protein FHL2, since the other described AR coactivators (ARA54, ARA55, ARA70 and ARA160) show no selectivity for the AR (Fujimoto et al., 1999; Heinlein et al., 1999; Hsiao and Chang, 1999; Kang et al., 1999). Therefore, to our knowledge, FHL2 is the first AR-specific coactivator.
The emerging importance of tissue-specifically expressed LIM-only proteins is further stressed by ACT, a four-and-a-half LIM domain-containing coactivator for CREM/CREB expressed in testis (Fimia et al., 1999). The LIM-only protein MLP may act as an integrator of MyoD-, E47- and MEF2-mediated transcription, thereby controlling myogenesis (Kong et al., 1997). In contrast to FHL2, the two LIM domain-containing protein MLP does not contain a transcriptional activation domain (Kong et al., 1997). Therefore, the functional difference between LIM-only proteins may depend either on the number or structure of their LIM domains. LIM-only proteins such as MLP may either serve as a scaffold to facilitate the assembly of transcriptional complexes or act themselves as coactivators participating actively in transactivation, as demonstrated for FHL2.
FHL2 is expressed in heart myocardium and prostate epithelial cells
In human fetal tissue FHL2 mRNA expression is only observed in heart, resembling the adult mRNA expression, where FHL2 is predominantly expressed in human and mouse heart (Figure 2A). Although expression of FHL2 was described in skeletal muscle, we could not detect FHL2 mRNA in skeletal or smooth muscle (Figure 2A), even when analysing RNA from various muscle tissues. Moreover, our Northern blot data demonstrate that FHL2 is detected in the heart ventricles (Figure 2B). In addition, the immunohistochemical analyses show that FHL2 protein expression is restricted to the myocardium (Figure 2A). In agreement with our Northern blot analyses, FHL2 protein could be detected neither in smooth muscle surrounding the arteriolae nor in aorta (Figure 2A; data not shown). In contrast to the expression of FHL2 in the myocardium, other related LIM-only family members such as SLIM1 (Brown et al., 1999) and SmLIM/CRP2 are expressed in aorta or smooth muscles, respectively (Jain et al., 1998). MLP/CRP3, another member of the CRP subfamily of LIM-only proteins, is expressed in striated muscle and heart (Arber et al., 1994).
Since we isolated FHL2 from a prostate library, we used an α–FHL2-specific monoclonal antibody to analyse FHL2 expression in normal human prostate. The immunohistochemical analysis shows that FHL2 is expressed in the epithelial cells of the prostate, where it overlaps with nuclear expression of the AR. In stromal smooth muscle cells of the prostate no FHL2 protein is detected. This is in agreement with our Northern blot data, where no FHL2 mRNA is found in prostate smooth muscle cells (Figure 2B). In summary, our expression data demonstrate tissue-specific expression of FHL2 in heart myocardium and prostate epithelial cells.
FHL2 function in heart and prostate
Immunohistochemical studies show that FHL2 protein is expressed in myocardial cells (Figure 2D). In protein extracts of human ventricle the presence of the AR and FHL2 in the same extract is demonstrated (Figure 2C). In addition, biochemical and molecular analyses showed expression of the AR in fetal and adult myocardium (Schleicher et al., 1989; Kimura et al., 1993). However, little is known about the function of the AR in heart. Indirect evidence for AR function is suggested by the observation that androgens can mediate cardiac hypertrophy (Marsh et al., 1998). Preliminary data suggest that FHL2 may play a role in the pathogenesis of human heart disease, since FHL2 expression is lowered in patients suffering from dilated cardiomyopathy (data not shown). Further studies will analyse the function of FHL2 in physiological and pathological situations of the heart.
The prostate is a male sexual organ where high concentrations of androgens such as testosterone or dihydrotestosterone are present. The AR is mainly expressed in the epithelial cells of the prostate and controls the expression of several target genes, such as the probasin gene. Moreover, the AR selectively upregulates the probasin gene, whereas GR and PR fail to do so (Rennie et al., 1993; Claessens et al., 1996). Our data demonstrate that FHL2 can coactivate the AR-mediated activity of a probasin reporter in an AR- and agonist-dependent manner (Figure 6C). Importantly, FHL2 and AR protein expression in prostate colocalizes in the nuclei of epithelial cells (Figure 2D, c and e), underlining the potential role of FHL2 in prostate gene expression.
According to the current model (Jenster, 1999) the AR is not only essential for normal prostate development, but is also highly involved in prostate neoplastic transformation. The prevalence of prostate carcinomas increases with age and is rated among the two leading causes of death of cancer among men (Landis et al., 1999). Initially, tumour growth is largely dependent on androgens, but in later stages most tumours grow androgen-independently (Gregory et al., 1998). In a subpopulation of prostate carcinomas mutations and deletions of the AR are observed, which might account for androgen-independent tumour growth (Trapman and Brinkmann, 1996; Culig et al., 1998; McPhaul, 1999). However, no explanation has been found for the androgen-insensitivity in the vast majority of prostate tumours, which still express wild-type AR. One hypothesis to explain this clinically disastrous phenomenon is an altered composition, structure or regulation of AR–coactivator complexes. Therefore, it will be interesting to evaluate the role of FHL2 on the AR and tumour characteristics in these prostate carcinomas.
In summary, FHL2 represents the first tissue-specific LIM-only coactivator of the AR. In general, coactivators such as FHL2 may provide a molecular basis for the spatial regulation of nuclear receptor function. Our data also show that FHL2 establishes a new group of tissue-specific LIM-only coactivators that are selective for one particular nuclear receptor. Finally, FHL2 might also serve as an integrator for several signal pathways, thereby fine-tuning the response to androgens in the heart and the prostate.
Materials and methods
Plasmids
The yeast expression plasmid pGBT9I-AGA was generated by inserting the AGA fragment [AR(1–503)-Gal4(1–147)-AR(624–919)] at the _Sal_I–_Xho_I sites of pGBT9 (Clontech) lacking the Gal4 DBD. To obtain pCMX–FHL2, pCMXGal–FHL2, GST–FHL2 and His–FHL2 the corresponding plasmids were constructed by inserting a PCR amplification product of the FHL2 cDNA at the _Bam_HI site of pCMX, pCMXGal4DBD (Umesono et al., 1991), pGEX-5X-2 (Pharmacia) or pRSETC (Invitrogen). For generating the mutants GST–FHL2(1–162) and GST–FHL2(163–279) the corresponding fragments were PCR amplified and inserted at the _Sma_I–_Eco_RI sites of pGEX-4T-1 (Pharmacia). pCMX-Flag-FHL2 was constructed by inserting the Flag epitope (MDYKDDDKDPEFPGSEFPGIP) in-frame with the FHL2 cDNA. The AR mutant pSG5-hARΔH12(1–892) was generated by PCR cloning of hAR(503–892) into pSG5hAR digested with _Kpn_I–_Bgl_II. pCMXhAR624–919 was constructed by inserting the AR624–919 fragment at the _Bam_HI–_Nhe_I sites of pCMX-KOZAK-ATG (Greschik et al., 1999). pSG5hAR, pSG5hAR1-626, pSG5-hARL712R (Alen et al., 1999), pSG5TIF2, pSG5GalTIF2.1 (Voegel et al., 1996), pABGal147-SRC1 (Onate et al., 1998), GR (RSVhGR) (Schüle et al., 1990), PR (pSG5hPR) (Misrahi et al., 1987), MR (RSVhMR) (Arriza et al., 1987), probasin-LUC (PB–LUC) (Claessens et al., 1996), MMTV-LUC (Schüle et al., 1990) and Gal5×-E1bTATA-LUC (G5E1b-LUC) (Hagen et al., 1995) were described previously. To construct pSG6SRC1a and pSG6SCR1e the corresponding rat cDNA was cloned into pSG6 (L.Klein-Hitpass). Numbers refer to amino acid positions. Details of cloning are available upon request.
Yeast
An adult human prostate cDNA library (Clontech) was screened with the pGBT9I-AGA bait plasmid in HF7c yeast cells as recommended (Clontech Matchmarker Two-Hybrid System protocol) in the presence of 10–9 M R1881 and 10 mM 3-aminotriazole. β-galactosidase assays were performed in the absence or presence of 10–9 M R1881 or 10–7 M CPA. β-galactosidase units show the mean of three independent assays.
RNA analysis
Northern blots (Clontech) were hybridized with an FHL2 probe according to the manufacturer's instructions (Clontech). Total RNA from myocardium was isolated and analysed (Kirfel et al., 1997). FHL2 coding region was labelled with StripEZ (Ambion) and hybridized as recommended. RNA loading was controlled using β-actin or GAPDH as probe. Myocardium was obtained from patients undergoing heart transplantation due to endstage heart failure. The ethical standards of the University of Freiburg were followed in all procedures.
Cell lines and transfections
293 and CV-1 cells were cultured in Dulbecco's modified Eagle's medium. HL-1 were grown in Ex-Cell 320 medium (Claycomb et al., 1998). Medium was supplemented with 10% double stripped fetal calf serum (Schüle et al., 1990). Transient transfection assays were carried out in 24- or 12-well plates (1 × 105, 9 × 104 or 5 × 104 cells per well for HL-1, 293 or CV-1, respectively). 293 cells were transfected as described (Greschik et al., 1999). CV-1 and HL-1 cells were transfected using DOTAP (Boehringer Mannheim) and Effectene (Qiagen), respectively. The total amount of transfected DNA was kept constant (4 μg for 293, 1.5 μg for CV-1, 250 ng for HL-1) by adding pSG5, pCMX and pUC18. Reporter plasmids (500 ng), 25 ng of receptor expression plasmids, 10 ng of Gal-fusion expression plasmids or 200 ng each of CMX-FHL2, pSG5TIF2, pSG6SRC1a and pSG6SRC1e were transfected per well in 293 or CV–1 cells. G5E1b-LUC reporter (125 ng) and 10 ng of Gal-fusion expression plasmids were used in HL–1 transfections. Luciferase activity was assayed as described (Dörflinger et al., 1999). All experiments were repeated at least five times.
GST pulldown assays
Expression of the GST fusion proteins (Pharmacia) and the in vitro transcription–translation reactions (Promega) were performed according to the manufacturer's instructions. GST pulldown assays were performed as described previously (Dörflinger et al., 1999) using buffer containing 50 mM KCl in the absence or presence of 10–9 M R1881. Ten percent of the in vitro translated proteins was loaded as input.
Coimmunoprecipitation assays and Western blot analyses
293 cells were transfected with 5 μg of pSG5hAR and either 5 μg of pCMX-FHL2 or 5 μg of pCMX-Flag-FHL2. Nuclear extracts (Zinck et al., 1993) were prepared after 1 h incubation with or without 10–7 M DHT. Following preclearing with a 40 μl 1:1 slurry of GammaBind-Sepharose (Pharmacia), supernatants were incubated for 2.5 h with M5 α–Flag antibody (Sigma). Beads were washed five times with WB (10 mM Tris–HCl pH 8.0, 250 mM NaCl, 0.5% NP–40, 0.1 μg/μl bovine serum albumin, 0.5 mM Pefabloc) in the presence or absence of DHT and analysed on a 10% SDS gel. Expression and purification of His–FHL2 (Invitrogen) was performed according to the manufacturer's instructions. Heart extract analyses were performed as described previously (Fimia et al., 1999). Western blots were decorated with α–AR 441 (Santa Cruz), M2 (Sigma) or α–FHL2 (see below) antibodies. Secondary antibody and chemiluminescence procedures were performed according to the manufacturer (Amersham).
Immunohistochemistry
A monoclonal antibody α–FHL2 was generated against recombinant full-length FHL2 in mouse. This antibody reacts specifically with FHL2 in Western blots and immunohistochemistry. Immunohistochemical stainings were performed using indirect immunoperoxidase and a protocol for antigen retrieval as described previously (Bocker et al., 1995). Paraffin-embedded 4 μm tissue slides were pretreated in a microwave oven twice for 5 min. α–FHL2 mouse monoclonal antibody was diluted 1:200 and α–AR mouse monoclonal antibody (M3562; Dako, Copenhagen, Denmark) 1:100. Biotin-linked anti-mouse IgG (1:500; Dako) was used as the secondary antibody and immunoreactions were visualized with the ABC complex diluted 1:50 in phosphate-buffered saline (Vectastain; Boehringer-Ingelheim, Heidelberg, Germany). Negative controls were performed using equivalent amounts of non-specific mouse immunoglobulins instead of primary antibodies. Immunohistochemical stainings were counterstained with hematoxilin or hematoxilin–eosin.
Acknowledgments
Acknowledgements
We thank L.Mercep, S.Erhardt and E.Paggen for excellent technical assistance; H.Gronemeyer, B.W.O'Malley, M.Tsai, L.Klein-Hitpass, B.Peeters and J.J.Palvilmo for providing reagents; N.Böhm, H.L.Pahl and H.Greschik for fruitful discussion; D.Willmann and the members of the Schüle laboratory for support and criticism and C.Schüle for providing the artwork and administrative assistance. This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (SFB 388) and Schering AG to R.S.
References
- Aarnisalo P., Palvimo, J.J. and Janne, O.A. (1998) CREB-binding protein in androgen receptor-mediated signaling. Proc. Natl Acad. Sci. USA, 95, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alen P., Claessens, F., Schoenmakers, E., Swinnen, J.V., Verhoeven, G., Rombauts, W. and Peeters, B. (1999) Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1α with multiple steroid receptors and identification of an internally deleted ELE1β isoform. Mol. Endocrinol., 13, 117–128. [DOI] [PubMed] [Google Scholar]
- Arber S., Halder, G. and Caroni, P. (1994) Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell, 79, 221–231. [DOI] [PubMed] [Google Scholar]
- Arriza J.L., Weinberger, C., Cerelli, G., Glaser, T.M., Handelin, B.L., Housman, D.E. and Evans, R.M. (1987) Cloning of human mineralocorticoid receptor complementary DNA: structural and functional kinship with the glucocorticoid receptor. Science, 237, 268–275. [DOI] [PubMed] [Google Scholar]
- Aumüller G., Holterhus, P.M., Konrad, L., von Rahden, B., Hiort, O., Esquenet, M. and Verhoeven, G. (1998) Immunohistochemistry and in situ hybridization of the androgen receptor in the developing human prostate. Anat. Embryol. (Berl.), 197, 199–208. [DOI] [PubMed] [Google Scholar]
- Beato M., Herrlich, P. and Schütz, G. (1995) Steroid hormone receptors: many actors in search of a plot. Cell, 83, 851–857. [DOI] [PubMed] [Google Scholar]
- Berrevoets C.A., Doesburg, P., Steketee, K., Trapman, J. and Brinkmann, A.O. (1998) Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor 2). Mol. Endocrinol., 12, 1172–1183. [DOI] [PubMed] [Google Scholar]
- Bocker T., Bittinger, A., Wieland, W., Buettner, R., Fauser, G., Hofstaedter, F. and Ruschoff, J. (1995) In vitro and ex vivo expression of nucleolar proteins B23 and p120 in benign and malignant epithelial lesions of the prostate. Mod. Pathol., 8, 226–231. [PubMed] [Google Scholar]
- Brown S., Biben, C., Ooms, L.M., Maimone, M., McGrath, M.J., Gurung, R., Harvey, R.P. and Mitchell, C.A. (1999) The cardiac expression of striated muscle LIM protein 1 (SLIM1) is restricted to the outflow tract of the developing heart. J. Mol. Cell. Cardiol., 31, 837–843. [DOI] [PubMed] [Google Scholar]
- Cato A.C. and Peterzierl, H. (1998) The androgen receptor as mediator of gene expression and signal transduction pathways. Trends Endocrinol. Metab., 9, 150–154. [DOI] [PubMed] [Google Scholar]
- Chan K.K., Tsui, S.K., Lee, S.M., Luk, S.C., Liew, C.C., Fung, K.P., Waye, M.M. and Lee, C.Y. (1998) Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene, 210, 345–350. [DOI] [PubMed] [Google Scholar]
- Claessens F., Alen, P., Devos, A., Peeters, B., Verhoeven, G. and Rombauts, W. (1996) The androgen-specific probasin response element 2 interacts differentially with androgen and glucocorticoid receptors. J. Biol. Chem., 271, 19013–19016. [DOI] [PubMed] [Google Scholar]
- Claycomb W.C., Lanson, N.A.,Jr, Stallworth, B.S., Egeland, D.B., Delcarpio, J.B., Bahinski, A. and Izzo, N.J.,Jr (1998) HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl Acad. Sci. USA, 95, 2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culig Z., Hobisch, A., Hittmair, A., Peterziel, H., Cato, A.C., Bartsch, G. and Klocker, H. (1998) Expression, structure and function of androgen receptor in advanced prostatic carcinoma. Prostate, 35, 63–70. [DOI] [PubMed] [Google Scholar]
- Dawid I.B., Breen, J.J. and Toyama, R. (1998) LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet., 14, 156–162. [DOI] [PubMed] [Google Scholar]
- Doesburg P., Kuil, C.W., Berrevoets, C.A., Steketee, K., Faber, P.W., Mulder, E., Brinkmann, A.O. and Trapman, J. (1997) Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry, 36, 1052–1064. [DOI] [PubMed] [Google Scholar]
- Dörflinger U., Pscherer, A., Moser, M., Rummele, P., Schüle, R. and Buettner, R. (1999) Activation of somatostatin receptor II expression by transcription factors MIBP1 and SEF-2 in the murine brain. Mol. Cell. Biol., 19, 3736–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R.M. (1988) The steroid and thyroid hormone receptor superfamily. Science, 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G.M., De Cesare, D. and Sassone-Corsi, P. (1999) CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature, 398, 165–169. [DOI] [PubMed] [Google Scholar]
- Fujimoto N., Yeh, S., Kang, H.Y., Inui, S., Chang, H.C., Mizokami, A. and Chang, C. (1999) Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J. Biol. Chem., 274, 8316–8321. [DOI] [PubMed] [Google Scholar]
- Genini M., Schwalbe, P., Scholl, F.A., Remppis, A., Mattei, M.G. and Schafer, B.W. (1997) Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol., 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Gregory C.W., Hamil, K.G., Kim, D., Hall, S.H., Pretlow, T.G., Mohler, J.L. and French, F.S. (1998) Androgen receptor expression in androgen-independent prostate cancer is associated with increased expression of androgen-regulated genes. Cancer Res., 58, 5718–5724. [PubMed] [Google Scholar]
- Greschik H., Wurtz, J.M., Hublitz, P., Kohler, F., Moras, D. and Schüle, R. (1999) Characterization of the DNA-binding and dimerization properties of the nuclear orphan receptor germ cell nuclear factor. Mol. Cell. Biol., 19, 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G., Dennig, J., Preiss, A., Beato, M. and Suske, G. (1995) Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J. Biol. Chem., 270, 24989–24994. [DOI] [PubMed] [Google Scholar]
- Heinlein C.A., Ting, H.J., Yeh, S. and Chang, C. (1999) Identification of ARA70 as a ligand-enhanced coactivator for the peroxisome proliferator-activated receptor γ. J. Biol. Chem., 274, 16147–16152. [DOI] [PubMed] [Google Scholar]
- Hsiao P.W. and Chang, C. (1999) Isolation and characterization of ARA160 as the first androgen receptor N–terminal-associated coactivator in human prostate cells. J. Biol. Chem., 274, 22373–22379. [DOI] [PubMed] [Google Scholar]
- Hsiao P.W., Lin, D.L., Nakao, R. and Chang, C. (1999) The linkage of Kennedy's neuron disease to ARA24, the first identified androgen receptor polyglutamine region-associated coactivator. J. Biol. Chem., 274, 20229–20234. [DOI] [PubMed] [Google Scholar]
- Ikonen T., Palvimo, J.J. and Janne, O.A. (1997) Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J. Biol. Chem., 272, 29821–29828. [DOI] [PubMed] [Google Scholar]
- Jain M.K. et al. (1998) Embryonic expression suggests an important role for CRP2/SmLIM in the developing cardiovascular system. Circ. Res., 83, 980–985. [DOI] [PubMed] [Google Scholar]
- Jenster G. (1999) The role of the androgen receptor in the development and progression of prostate cancer. Semin. Oncol., 126, 407–421. [PubMed] [Google Scholar]
- Jenster G., van der Korput, H.A., Trapman, J. and Brinkmann, A.O. (1995) Identification of two transcription activation units in the N–terminal domain of the human androgen receptor. J. Biol. Chem., 270, 7341–7346. [DOI] [PubMed] [Google Scholar]
- Jurata L.W. and Gill, G.N. (1998) Structure and function of LIM domains. Curr. Top. Microbiol. Immunol., 228, 75–113. [DOI] [PubMed] [Google Scholar]
- Kang H.Y., Yeh, S., Fujimoto, N. and Chang, C. (1999) Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J. Biol. Chem., 274, 8570–8576. [DOI] [PubMed] [Google Scholar]
- Kimura N., Mizokami, A., Oonuma, T., Sasano, H. and Nagura, H. (1993) Immunocytochemical localization of androgen receptor with polyclonal antibody in paraffin-embedded human tissues. J. Histochem. Cytochem., 41, 671–678. [DOI] [PubMed] [Google Scholar]
- Kirfel J., Kelter, M., Cancela, L.M., Price, P.A. and Schüle, R. (1997) Identification of a novel negative retinoic acid responsive element in the promoter of the human matrix Gla protein gene. Proc. Natl Acad. Sci. USA, 94, 2227–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Flick, M.J., Kudla, A.J. and Konieczny, S.F. (1997) Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol. Cell. Biol., 17, 4750–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis S.H., Murray, T., Bolden, S. and Wingo, P.A. (1999) Cancer statistics, 1999. CA Cancer J. Clin., 49, 8–31. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.D., Lehmann, M.H., Ritchie, R.H., Gwathmey, J.K., Green, G.E. and Schiebinger, R.J. (1998) Androgen receptors mediate hypertrophy in cardiac myocytes. Circulation, 98, 256–261. [DOI] [PubMed] [Google Scholar]
- Matuo Y., Nishi, N., Muguruma, Y., Yoshitake, Y., Kurata, N. and Wada, F. (1985) Localization of prostatic basic protein (‘probasin’) in the rat prostates by use of monoclonal antibody. Biochem. Biophys. Res. Commun., 130, 293–300. [DOI] [PubMed] [Google Scholar]
- McPhaul M.J. (1999) Molecular defects of the androgen receptor. J. Steroid Biochem. Mol. Biol., 69, 315–322. [DOI] [PubMed] [Google Scholar]
- Misrahi M., Atger, M., d'Auriol, L., Loosfelt, H., Meriel, C., Fridlansky, F., Guiochon-Mantel, A., Galibert, F. and Milgrom, E. (1987) Structure of the human progesterone receptor gene. Biochem. Biophys. Res. Commun., 143, 740–748. [DOI] [PubMed] [Google Scholar]
- Moilanen A.M., Poukka, H., Karvonen, U., Hakli, M., Janne, O.A. and Palvimo, J.J. (1998) Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol., 18, 5128–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D. and Gronemeyer, H. (1998) The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol., 10, 384–391. [DOI] [PubMed] [Google Scholar]
- Nolte R.T. et al. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature, 395, 137–143. [DOI] [PubMed] [Google Scholar]
- Onate S.A., Tsai, S.Y., Tsai, M.J. and O'Malley, B.W. (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science, 270, 1354–1357. [DOI] [PubMed] [Google Scholar]
- Onate S.A., Boonyaratanakornkit, V., Spencer, T.E., Tsai, S.Y., Tsai, M.J., Edwards, D.P. and O'Malley, B.W. (1998) The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem., 273, 12101–12108. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Wu, Z., Park, C.W., Graves, R., Wright, M. and Spiegelman, B.M. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell, 92, 829–839. [DOI] [PubMed] [Google Scholar]
- Rennie P.S. et al. (1993) Characterization of two _cis_-acting DNA elements involved in the androgen regulation of the probasin gene. Mol. Endocrinol., 7, 23–36. [DOI] [PubMed] [Google Scholar]
- Sasaki S. et al. (1999) Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin β gene. EMBO J., 18, 5389–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher G., Stumpf, W.E., Gurley, J.M. and Drews, U. (1989) Differential nuclear binding of [3H]testosterone and its metabolites to androgen and estrogen receptors in brain, pituitary, heart, kidney and accessory sex glands of the mouse: an autoradiographic study. J. Steroid Biochem., 33, 581–587. [DOI] [PubMed] [Google Scholar]
- Schüle R., Rangarajan, P., Kliewer, S., Ransone, L.J., Bolado, J., Yang, N., Verma, I.M. and Evans, R.M. (1990) Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell, 62, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Sinha A.A., Wilson, M.J. and Gleason, D.F. (1987) Immunoelectron microscopic localization of prostatic-specific antigen in human prostate by the protein A–gold complex. Cancer, 60, 1288–1293. [DOI] [PubMed] [Google Scholar]
- Trapman J. and Brinkmann, A.O. (1996) The androgen receptor in prostate cancer. Pathol. Res. Pract., 192, 752–760. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami, K.K., Thompson, C.C. and Evans, R.M. (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid and vitamin D3 receptors. Cell, 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel J.J., Heine, M.J., Zechel, C., Chambon, P. and Gronemeyer, H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J., 15, 3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Wilson E.M., Simental, J.A., French, F.S. and Sar, M. (1991) Molecular analysis of the androgen receptor. Ann. N Y Acad. Sci., 637, 56–63. [DOI] [PubMed] [Google Scholar]
- Xu L., Glass, C.K. and Rosenfeld, M.G. (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- Zinck R., Hipskind, R.A., Pingoud, V. and Nordheim, A. (1993) c-fos transcriptional activation and repression correlate temporally with the phosphorylation status of TCF. EMBO J., 12, 2377–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]