GATA-dependent recruitment of MEF2 proteins to target promoters (original) (raw)
Abstract
The myocyte enhancer factor-2 (MEF2) proteins are MADS-box transcription factors that are essential for differentiation of all muscle lineages but their mechanisms of action remain largely undefined. In mammals, the earliest site of MEF2 expression is the heart where the MEF2C isoform is detectable as early as embryonic day 7.5. Inactivation of the MEF2C gene causes cardiac developmental arrest and severe downregulation of a number of cardiac markers including atrial natriuretic factor (ANF). However, most of these promoters contain no or low affinity MEF2 binding sites and they are not significantly activated by any MEF2 proteins in heterologous cells suggesting a dependence on a cardiac-enriched cofactor for MEF2 action. We provide evidence that MEF2 proteins are recruited to target promoters by the cell-specific GATA transcription factors, and that MEF2 potentiates the transcriptional activity of this family of tissue-restricted zinc finger proteins. Functional MEF2/GATA-4 synergy involves physical interaction between the MEF2 DNA-binding domain and the carboxy zinc finger of GATA-4 and requires the activation domains of both proteins. However, neither MEF2 binding sites nor MEF2 DNA binding capacity are required for transcriptional synergy. The results unravel a novel pathway for transcriptional regulation by MEF2 and provide a molecular paradigm for elucidating the mechanisms of action of MEF2 in muscle and non-muscle cells.
Keywords: GATA factors/heart development/MEF2 proteins/muscle transcription
Introduction
Members of the myocyte enhancer factor-2 (MEF2) family of MADS (MCM1, Agamous, Deficiens, Serum response factor)-box transcription factors are evolutionarily conserved proteins that are expressed at high levels in all muscle cells. MEF2 proteins are also found in non-muscle cells including brain and lymphoid tissue (reviewed in Black and Olson, 1998). In mammals, the MEF2 family is composed of four members, MEF2A, MEF2B, MEF2C and MEF2D, which form homo- and heterodimers that bind the consensus DNA sequence (T/C)TA(A/T)4TA(G/A) present in many muscle and non-muscle promoters. MEF2 proteins contain a conserved N-terminal 56 amino acid MADS domain and an adjacent 29 amino acid MEF2 domain, which together mediate DNA binding and dimerization.
Genetic studies have provided evidence for an essential role of MEF2 proteins in muscle-specific gene expression and differentiation of all three muscle lineages. In Drosophila, mutation of the D-mef2 gene results in embryos lacking differentiated skeletal, cardiac and visceral muscle cells (Bour et al., 1995; Lilly et al., 1995; Ranganayakulu et al., 1995). In mice, inactivation of the MEF2C gene, which is the first MEF2 isoform expressed during embryonic development, leads to cardiac morphogenetic defects, vascular abnormalities and lethality by embryonic day 9.5 (Lin et al., 1997, 1998; Bi et al., 1999). The mechanisms by which MEF2 proteins regulate myogenesis of both striated and smooth muscle cells and the identity of their downstream targets in these various tissues are only starting to be elucidated. At present, the mechanisms of action of MEF2 have been analyzed mostly in skeletal muscle where MEF2 appears to act as cofactors for the myogenic basic helix–loop–helix (bHLH) proteins, MyoD, Myf5, myogenin and MRF4 (Kaushal et al., 1994; Molkentin et al., 1995; Black et al., 1998). Thus, MEF2 proteins strongly potentiate the transcriptional activity of the bHLH myogenic factors and cooperate with them for inducing and maintaining the skeletal muscle phenotype. This cooperativity is mediated by direct interaction between the DNA-binding domains of MEF2 and myogenic proteins, and necessitates a DNA-binding site for only one of the two factors. Therefore, in skeletal myocytes, MEF2 may modulate transcription by two distinct pathways: one involving DNA binding to MEF2 sites and another one involving recruitment of MEF2 to E-boxes in target promoters via the myogenic bHLH factors. Whether similar mechanisms underlie the action of MEF2 in cardiac and visceral muscle cells where the MyoD family of transcription factors is not expressed remains unknown.
MEF2-binding sites have been reported in several cardiac promoters and their mutation was shown to decrease promoter activity in cardiomyocytes; they include the MEF2 sites in the ventricular myosin light chain (MLC2V), cardiac troponin T, cardiac troponin I, α-myosin heavy chain (αMHC) and Desmin (Iannello et al., 1991; Zhu et al., 1991; Yu et al., 1992; Molkentin and Markham, 1993; Kuisk et al., 1996; Di Lisi et al., 1998). Analysis of cardiac gene expression in mice with targeted mutation of the MEF2C gene confirmed that some of these genes, like αMHC, required MEF2C for optimum transcription (Lin et al., 1997). In addition to αMHC, two other cardiac-specific genes not previously associated with MEF2 proteins, atrial natriuretic factor (ANF) and α-cardiac actin (α-CA), were completely absent in the hearts of MEF2C-deficient embryos. How MEF2C regulates transcription of these target genes remains unclear; the two αMHC MEF2 sites are low-affinity MEF2-binding sites (Yu et al., 1992; Molkentin and Markham, 1993), and MEF2 proteins are unable to activate αMHC-driven reporters in cotransfection assays although they can potentiate transactivation of the αMHC promoter by the thyroid hormone receptor (Lee et al., 1997). Moreover, ectopic expression of MEF2 proteins in explanted Xenopus ectoderm failed to activate endogenous αMHC or α-CA genes (Chambers et al., 1994; Fu and Izumo, 1995). However, forced expression of MEF2 proteins in whole Xenopus embryos results in precocious expression of endogenous αMHC and enlarged hearts (Fu and Izumo, 1995). Together, these studies suggest that MEF2 regulates transcription of αMHC and possibly other cardiac genes in conjunction with a cell-specific cofactor present in embryonic mesoderm (or endoderm) but not in ectoderm.
The ANF promoter, a known downstream target for the cardiac-specific transcription factors GATA-4 and Nkx2-5, does not contain MEF2 consensus binding sites (Durocher et al., 1996, 1997; Charron et al., 1999); this suggests an indirect action of MEF2 on ANF transcription possibly through modulation of GATA-4, Nkx2-5 or other ANF regulators. However, neither GATA-4 nor Nkx2-5 levels are altered in MEF2C–/– embryos (Lin et al., 1997).
In this study, we provide evidence that MEF2 proteins are recruited by the cardiac-specific transcription factor GATA-4 to synergistically activate ANF and several other MEF2C target promoters including αMHC and α-CA. The MEF2–GATA-4 synergy is mediated by physical interaction between the respective DNA-binding domains and requires the transactivation domains of both factors. GATA-binding sites are necessary and sufficient for cooperativity with MEF2. Other GATA factors, including GATA-6, which is expressed in cardiac and smooth muscle cells, and GATA-2 and -3, which are present in hemopoietic and neuronal cells, are also able to cooperate with MEF2 proteins. Together, the data suggest that in addition to cooperating with the myogenic bHLH proteins in skeletal muscle differentiation, MEF2 proteins act as cofactors for the tissue-restricted zinc finger GATA proteins in cardiac myogenesis and raise the possibility that GATA factors may be essential components of MEF2 action in several other cell types.
Results
MEF2 proteins activate the ANF promoter via two distinct mechanisms
The absence of ANF transcripts in the heart of mice homozygous for a null mutation of the MEF2C gene (Lin et al., 1997) indicates that ANF is a downstream target for MEF2 proteins. We tested whether the effect of MEF2C was due to direct action on the ANF promoter be it via DNA binding or through recruitment by protein–protein interactions. The first 700 bp of the rat ANF promoter are sufficient to recapitulate cardiac specificity and spatio-temporal regulation of the endogenous gene in cultured cardiomyocytes (Argentin et al., 1994) and in transgenic mice (Durocher et al., 1998).
Sequence analysis of the entire 700 bp rat ANF promoter revealed no consensus MEF2 sites; the closest sequence homologies to MEF2-binding sites mapped to an A/T-rich element sharing similarities with a MEF2 consensus (Figure 1A). To verify whether this A/T-rich element could be recognized by cardiac-derived or recombinant MEF2 proteins, it was used in gel shift assay to compete MEF2 binding on the well characterized muscle creatine kinase (MCK) MEF2 site. As seen in Figure 1B, several A/T-rich ANF elements including the TATA-box, the SRE-like and the CArG-box could not compete the MEF2 binding on the MCK probe even when used at a 500-fold molar excess. On the other hand, the distal A/T-rich element was able to displace the MEF2 binding although at a much lower efficiency than the MCK site. Identical results were obtained using MEF2C and MEF2D or cardiomyocyte nuclear extracts (data not shown). Consistent with its ability to recognize MEF2 proteins, the ANF A/T-rich probe was able to bind all three recombinant MEF2 proteins tested (MEF2A, MEF2C and MEF2D) albeit with low affinity (Figure 1B and data not shown). Finally, when cloned upstream of a minimal promoter, the ANF A/T-rich element could be transactivated 4-fold by cotransfection with MEF2 expression vectors in heterologous cells (Figure 2C). These results suggest that the ANF promoter contains a very low-affinity MEF2-binding site that could mediate MEF2 action.
Fig. 1. The ANF promoter harbors a low-affinity MEF2-binding site. (A) Schematic representation of the ANF promoter. Regulatory elements are boxed and their location relative to the transcription start site is indicated. All these elements are evolutionarily conserved on the ANF promoter from many species. SRE-like is a low-affinity serum response element; the GATAd and GATAp are the distal and proximal GATA-binding sites, respectively. The consensus MEF2-binding site is also shown. The A/T-rich mut sequence indicates the mutations introduced to abolish the A/T-rich element. rANF and hANF are the rat and human ANF promoter, respectively. (B) The A/T-rich element is a low-affinity MEF2-binding site. EMSAs were performed on the MEF2 element of the MCK promoter (MEF2-MCK, left panel) or the A/T-rich element of ANF (A/T-rich, right panel) using in vitro translated MEF2A. In the left panel, the MEF2A binding was competed with different unlabeled ANF probes described in Materials and methods. Only the A/T-rich element of the ANF promoter was able to compete the MEF2A binding. Similar results were obtained with in vitro translated MEF2C and MEF2D.
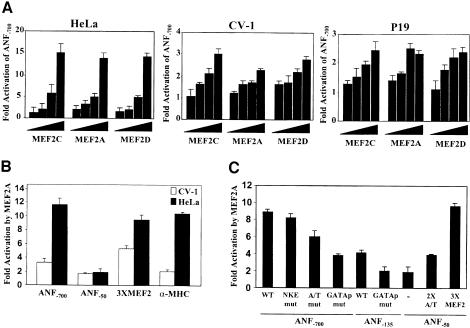
Fig. 2. The low-affinity A/T-rich and the proximal GATA elements contribute to MEF2-dependent ANF promoter activation. (A) Dose-dependent ANF–700 promoter activation by MEF2A, MEF2C and MEF2D in HeLa, CV-1 and P19 cell lines. Transient transfections were performed using 50 ng, 100 ng, 500 ng and 1 µg of MEF2 expression vector. Note the fold-activation difference between HeLa and CV-1 or P19 cells. (B) Preferential activation of the ANF–700 and αMHC promoters, but not an artificial MEF2 reporter, in HeLa cells. Transfections were performed using 1 µg of MEF2A expression vector. Similar results were obtained using MEF2C and MEF2D. (C) The low-affinity A/T-rich and the proximal GATA elements contribute to MEF2-dependent ANF promoter activation. Transfections were performed in HeLa cells using 1 µg of MEF2A expression vector. Similar results were obtained using MEF2C and MEF2D. 3XMEF2 and 2XA/T are the MEF2-MCK and the ANF A/T-rich elements trimerized and dimerized, respectively, in front of the ANF–50 minimal promoter.
Indeed, MEF2C and other MEF2 proteins activate the ANF promoter in a dose-dependent manner in several non-cardiac cells (Figure 2A). Interestingly, the magnitude of activation was much greater (4- to 6-fold) in HeLa cells than in most cell lines tested (including CV1, P19 and C2C12), with maximal ANF promoter induction of 15-fold. This difference in MEF2 responsiveness was also observed with the cardiac αMHC promoter, which was induced by 10-fold in HeLa cells and was barely responsive in CV1 or P19 cells (Figure 2B and data not shown); in contrast, a synthetic promoter harboring a multimerized MCK MEF2 site upstream of a minimal ANF promoter was more similarly activated by MEF2 in HeLa (9-fold) and CV1 (6-fold) cells (Figure 2B). The transfected MEF2 vectors produced similarly high levels of MEF2 proteins in all cell lines as assessed by gel shift assays (data not shown). Thus, the differences observed in the level of MEF2-dependent ANF and αMHC promoter activation may reflect cooperative interaction between transfected MEF2 proteins and other cellular factors bound to the promoters.
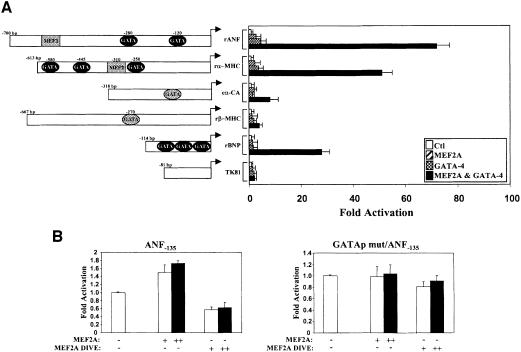
Mutational analysis was used to test which DNA elements on the ANF promoter are required for activation by MEF2 (Figure 2C). Consistent with its characterization as a weak-affinity MEF2 site, the A/T-rich element was necessary for maximal MEF2 activation but its deletion or mutation reduced promoter activation by only 30%. Surprisingly, mutation of the proximal GATA element that can bind endogenous GATA-2 protein present in HeLa cells (Grépin et al., 1994) had a more drastic effect on MEF2 responsiveness, suggesting that GATA factors may cooperate with MEF2. In fact, the proximal ANF promoter (ANF–135), which lacks any MEF2-binding site, was induced 4- to 5-fold by MEF2 proteins, and mutation of the GATA site therein abrogated MEF2 responsiveness (Figure 2C). This element is a high-affinity binding site for GATA factors (Charron et al., 1999) but does not bind MEF2 proteins (Figure 1B). Together, these results suggest that MEF2 could act as a cofactor of promoter-bound GATA proteins to activate the ANF and possibly other cardiac promoters.
Synergistic activation of the ANF promoter by MEF2 and GATA factors
Two members of the GATA family of zinc finger transcription factors, GATA-4 and GATA-6, are expressed in cardiac muscle cells and bind to and activate the ANF promoter (Charron et al., 1999). To test whether either factor could recruit MEF2 proteins to target promoters, the effect of co-expressing them with MEF2 in heterologous cells was assayed on ANF promoter activity. Cotransfection of GATA-4 with MEF2A, MEF2C or MEF2D leads to a synergistic 40- to 50-fold activation of the ANF promoter (Figure 3A). MEF2 proteins were also able to cooperate to varying degrees with other GATA factors including the hemopoietic GATA-1, -2 and -3, and the other cardiac GATA factor, GATA-6, but not GATA-5 (Figure 3B). The inability of MEF2C and MEF2A to synergize with GATA-5 and the more modest synergy achieved with GATA-2 and -3 are not due to different levels of GATA proteins produced as all expression vectors have been shown to produce similar protein levels (Viger et al., 1998; Nemer et al., 1999).
Fig. 3. The MEF2 and GATA transcription factors cooperatively activate the ANF–700 promoter. (A) MEF2A, MEF2C and MEF2D functionally interact with GATA-4. Cotransfections were performed in HeLa cells using the ANF-luc–700 construct and 1 µg of MEF2A, MEF2C or MEF2D expression vector in the absence (–) or presence (+) of 1.5 µg of GATA-4 expression vector. (B) MEF2 proteins functionally interact with a subset of GATA proteins. Cotransfections were performed as in (A) using 1.5 µg of various GATA expression vectors in the absence (–) or presence (+) of 1 µg of MEF2C expression vector. Note that cooperative interaction between MEF2A and the different GATA factors was identical to the one shown here for MEF2C and GATA-1 to -6. Similar results were also obtained in the CV1 cell line.
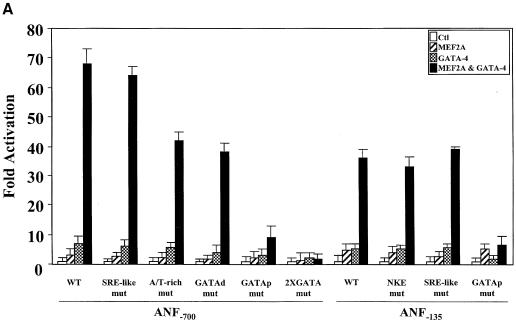
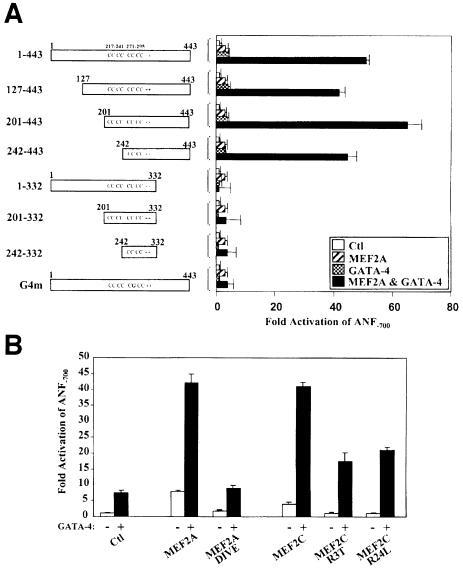
In order to map the promoter element(s) required for the synergy between MEF2 and GATA-4, various ANF promoter mutations were tested. Mutation of the low-affinity MEF2-binding element (A/T-rich) or the distal GATA element reduced maximal MEF2–GATA-4 synergy by 35–40% (Figure 4A). Mutation of the proximal GATA element in the context of the –700 or the –135 bp promoter completely abolished synergy, indicating that this element is essential for MEF2–GATA-4 cooperation. Interestingly, the –135 bp ANF promoter was sufficient to produce 50% of the maximal synergy obtained with the longer promoter (ANF–700) and displayed the same response to the various MEF2–GATA combinations as the ANF–700 promoter (Figure 3), suggesting that binding of GATA factors to the proximal GATA element may be sufficient to recruit MEF2 proteins to the promoter.
Fig. 4. The proximal GATA element is necessary and sufficient for MEF2–GATA synergy. Cotransfections were performed in HeLa cells using various promoter constructs and 1 µg of MEF2A and/or 1.5 µg of GATA-4 expression vectors. The ANF promoter constructs used are described in Materials and methods. 3XMEF2 and 2XA/T are the MEF2-MCK and the ANF A/T-rich elements trimerized and dimerized, respectively, in front of the ANF–50 minimal promoter. 2XGATA is a dimer of the BNP GATA elements in front of the minimal BNP promoter.
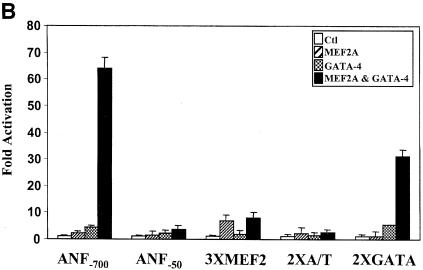
Indeed, an artificial reporter driven by multimerized GATA-binding sites could be synergistically activated by GATA and MEF2 proteins; however, neither the high-affinity MCK MEF2-binding site nor the lower affinity ANF MEF2 element was sufficient to support MEF2–GATA cooperativity (Figure 4B).
MEF2 factors physically interact in vitro and in vivo with GATA-4
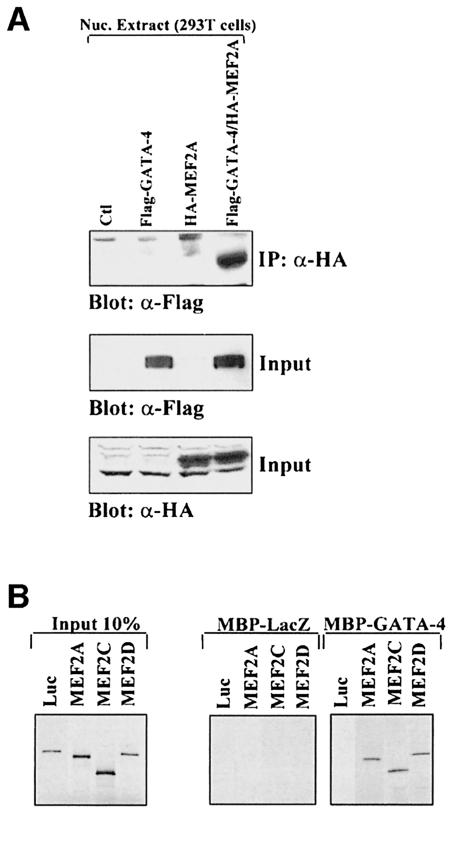
The observation that MEF2A–GATA-4 synergy required only the GATA-binding site implied that GATA-4 recruits MEF2 proteins to the ANF promoter through protein–protein interaction. Indeed, MEF2A and GATA-4 could be co-immunoprecipitated in vivo (Figure 5A), suggesting physical interaction between the two proteins.
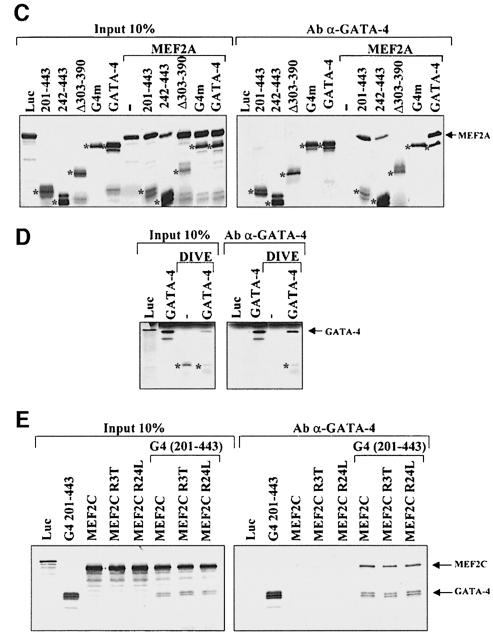
Fig. 5. MEF2 proteins physically interact with GATA-4. (A) MEF2A interacts in vivo with GATA-4. Nuclear extracts from 293T cells transfected with empty vectors (Ctl), Flag-GATA-4 and/or HA-MEF2A were immunoprecipitated using an anti-HA antibody, separated on 10% SDS–PAGE, transferred to PVDF membranes, and subjected to immunoblotting using an anti-Flag antibody (top panel). The lower two panels are Western blots carried out on the same nuclear extracts using either HA (to reveal tagged MEF2A proteins) or Flag (to reveal tagged GATA-4 proteins) antibodies. (B) MEF2A proteins interact in vitro with GATA-4. Pull-down assays were performed using immobilized, bacterially produced MBP fusions (MBP-GATA-4 and MBP-LacZ as control) and in vitro translated 35S-labeled MEF2A, MEF2C, MEF2D or luciferase (luc) protein. The protein complexes were resolved on 10% SDS–PAGE. (C) The physical interaction between GATA-4 and MEF2 requires the C-terminal zinc finger DNA-binding domain of GATA-4. Full-length GATA-4 and various GATA-4 mutants (depicted in Figure 6A) were in vitro cotranslated with MEF2A and co-immunoprecipitated using an antibody directed against the extreme C-terminus of GATA-4. The protein complexes were resolved on 15% SDS–PAGE. The asterisks highlight GATA protein bands. (D) The DNA-binding domain of MEF2 is sufficient for interaction with GATA-4. MEF2A DIVE (aa 1–86) retains the MADS and MEF2 domains. Co-immunoprecipitations were performed as described in (C). The asterisks highlight the MEF2A DIVE band. The protein complexes were resolved on 20% SDS–PAGE. (E) MEF2 DNA-binding-defective mutants interact with GATA-4. MEF2C R3T and MEF2C R24L do not bind DNA but are still able to dimerize. A deleted GATA-4 construct [G4 (201–443)] was used to differentiate between GATA-4 and MEF2C, which have similar electrophoretic mobility. Co-immunoprecipitations were performed as described in (C). The protein complexes were resolved on 10% SDS–PAGE.
To determine whether this interaction was direct, we performed in vitro pull-down assays using immobilized MBP-GATA-4 and in vitro translated 35S-labeled MEF2 proteins. MBP-GATA-4 was able to retain specifically MEF2A, MEF2C and MEF2D but not the control luciferase (Figure 5B), confirming that GATA-4 and MEF2 directly interact.
In order to map the interaction domain between MEF2 and GATA proteins, different mutants of GATA-4 were in vitro cotranslated with MEF2A and co-immunoprecipitated using an antibody directed against the extreme C-terminus of GATA-4 (present in all mutants tested). MEF2A was able to interact with the full-length GATA-4 and the N-terminal activation domain-deleted mutant (201–443) (Figure 5C). Deletion of the N-terminal zinc finger of GATA-4 (242–443) reduced but did not abrogate interaction with MEF2A. However, MEF2A was unable to interact with the G4m [which harbors a point mutation in the C-terminal zinc finger, abolishing DNA binding (Charron et al., 1999)] or the Δ303–390 mutant, indicating that the C-terminal zinc finger structure and the basic region are essential for physical interaction with MEF2. The same approach was also used to map the GATA-4 interaction domain on MEF2 and revealed that the DNA-binding domain consisting of the MADS and MEF2 domains (MEF2A DIVE, aa 1–86) is sufficient for interaction with GATA-4 (Figure 5D). Interestingly, within the MADS domain, interaction with GATA factors and binding to DNA could be segregated as two DNA-binding-defective mutants (MEF2C R3T and MEF2C R24L) retained the ability to bind GATA-4 (Figure 5E). These results suggest that GATA-4 and MEF2 physically interact through their DNA-binding domains.
MEF2–GATA synergy does not require MEF2 DNA binding
To determine whether the activation domains of either or both GATA and MEF2 proteins are required, various GATA-4 mutants were tested for their capacity to activate the ANF promoter synergistically with MEF2A. The GATA-4 mutants that delete the N-terminal region (127–443 and 201–443) and the first zinc finger (242–443) were able to synergize with MEF2A (Figure 6A). However, the GATA-4 mutants that delete the C-terminal transactivation domain (1–332, 201–332 and 242–332) were all unable to support MEF2 synergy. Consistent with a requirement for GATA-4 DNA binding (Figure 4A) and the GATA-4 DNA-binding domain for physical interaction with MEF2 (Figure 5C), a point mutation in the second zinc finger that destroys DNA binding (G4m) abrogated MEF2 synergy (Figure 6A).
Fig. 6. Mapping of the GATA-4 and MEF2 domains required for synergy. (A) The C-terminal activation domain of GATA-4 is required for MEF2 synergy. Cotransfections were performed in HeLa cells on the ANF–700 promoter construct using 1 µg of MEF2 and 1.5 µg of GATA-4 expression vectors. (B) The C-terminal activation domain of MEF2, but not its DNA-binding capacity, is required for synergy with GATA-4, as shown by the ability of MEF2C R3T and MEF2C R24L to synergize with GATA-4. Note that the DNA-binding domain (MEF2A DIVE) is not sufficient to support functional synergy although it interacts physically with GATA-4 as shown in the previous figure.
Functional synergy also required the activation domain of MEF2, as deletion of the C-terminal activation domain (MEF2A DIVE) completely abolished the synergy with GATA-4 (Figure 6B). However, consistent with the requirement for GATA- but not MEF2-binding sites, MEF2C mutants that are DNA-binding defective retained the ability to synergize with GATA-4 (Figure 6B). These results indicate that GATA-4 is able to recruit DNA-binding-defective MEF2 proteins to transcriptionally active complexes.
MEF2–GATA synergy: a mechanism for MEF2 action in the heart
We next tested whether transcription of other cardiac genes is cooperatively activated by MEF2 and GATA-4. As seen in Figure 7A, in addition to ANF, the αMHC, α-CA and B-type natriuretic peptide (BNP) promoters are also synergistically activated by MEF2 and GATA-4. Both αMHC and α-CA are downregulated in MEF2C null mice and neither contain a high-affinity MEF2-binding site although both are GATA targets (Sepulveda et al., 1998; Charron et al., 1999). However, as shown by the βMHC promoter, not all GATA target promoters are synergistically activated by MEF2, suggesting that functional GATA–MEF2 synergy is promoter context dependent and may be targeted to a specific subset of cardiac genes.
Fig. 7. The MEF2–GATA-4 synergy: a mechanism for MEF2 action in the heart. (A) The MEF2–GATA-4 synergy is not limited to the ANF promoter. HeLa cells were cotransfected with 1 µg of MEF2A and 1.5 µg of GATA-4 expression vectors together with various cardiac promoters. Except for the cardiac α-actin promoter that was from chicken, all other promoters used are from rat and are described in Materials and methods. TK81 is the thymidine kinase –81 bp promoter. Elements shaded in black and gray are high- and low-affinity sites, respectively, as determined by DNA-binding assays. (B) A dominant-negative MEF2 protein decreases ANF promoter activity in cardiomyocytes. Primary culture of cardiomyocytes was transfected with the wild-type ANF–135 (left panel) or GATA-mutated ANF–135 promoter (GATAp mut/ANF–135, right panel) and no (–), 50 ng (+) or 1000 ng (++) of MEF2A or a dominant-negative form of MEF2A (MEF2A DIVE). The results shown represent the mean ± SD of two independent experiments each carried out in duplicate.
Finally, to ascertain whether, in cardiac cells, MEF2 proteins are GATA cofactors, the effect of a dominant-negative MEF2 protein on the activity of the proximal ANF promoter was determined. This promoter contains a GATA- but no MEF2-binding site. Cotransfection of a MEF2 mutant that retains the ability to associate physically with GATA-4 but lacks the activation domain reduces by 50% the activity of the ANF promoter in primary cardiomyocyte cultures; in contrast, cotransfection with wild-type MEF2A induces promoter activity by 80% (Figure 7B). Both activation by wild-type MEF2 and inhibition by its dominant-negative form are blunted by point mutation of the GATA-binding site (Figure 7B). These data are consistent with a role for MEF2 proteins as co-activators of GATA factors in cardiac muscle cells and point to a novel GATA-dependent pathway for transcriptional activation by MEF2.
Discussion
The MEF2 transcription factors are key regulators of cardiac myogenesis and morphogenesis, but the molecular basis for their actions is poorly understood. The data presented here provide evidence that, in cardiac myocytes, MEF2 proteins are recruited by the cardiac-specific GATA transcription factors to target promoters and functionally synergize with this family of tissue-restricted zinc finger proteins. This observation is reminiscent of the cooperative interaction between MEF2 proteins and the myogenic bHLH factors in skeletal muscles, and suggests that MEF2 proteins are able to interact with and potentiate the action of other classes of cell-specific transcription factors. Given the co-expression of MEF2 and GATA factors in several cell types including smooth muscle, neuronal and T cells, the GATA-dependent MEF2 pathway described in this work may provide a molecular paradigm for understanding the mechanisms of action of MEF2 in many target cells.
GATA proteins are evolutionarily conserved cell-restricted transcription factors that play crucial roles in differentiation. In vertebrates, six GATA factors have been identified and they are all expressed in a lineage-restricted and developmentally controlled manner. GATA-1, -2 and -3 are predominantly expressed in hemopoietic cells while GATA-4, -5 and -6 are largely restricted to the heart and gut. Genetic and biochemical studies have revealed crucial roles for specific family members in hemopoietic, cardiac, neuronal and endodermal cells (Pevny et al., 1991; Tsai et al., 1994; Grépin et al., 1995; Pandolfi et al., 1995; Molkentin et al., 1997; Morrisey et al., 1998). In addition to their essential roles in development, GATA factors are also required for the proper function of adult organs. GATA-binding sites are present on many hemopoietic and cardiac promoters, which are potently activated by GATA factors (reviewed in Charron and Nemer, 1999). Moreover, GATA proteins act cooperatively with other classes of transcription factors including several zinc finger proteins, such as SP1 and FOG-1 (friend of GATA-1) and homeodomain-containing factors like the cardiac-specific Nkx2-5 (Durocher et al., 1997; Tsang et al., 1997). The differential interactions of GATA proteins with other transcription factors are likely to be important for functional specificity of GATA proteins during embryonic development and in differentiated and adult cells.
In this study, we report for the first time that, in addition to interacting with homeodomain- and zinc finger-containing proteins, GATA factors are also able to interact with members of the MADS-box family of transcription factors. This interaction involves physical contact between the C-terminal zinc finger DNA-binding domain and the adjacent basic region of GATA-4 and the MADS domain of MEF2. This, in turn, leads to synergistic activation of the ANF and other MEF2 target promoters independently of the DNA-binding activity of MEF2. Synergy is observed with two of the three cardiac GATA factors: GATA-4 and -6 but not with GATA-5; MEF2 synergy was also detected with GATA-1, -2 and -3 although at varying levels. Thus, all GATA factors are not equally competent to synergize with MEF2. The observation that GATA-4 and -6 but not -5 synergize with MEF2 is interesting given that GATA-4 and -5 but not -6 synergize with the cardiac homeodomain protein Nkx2-5 (Durocher et al., 1997); this suggests that while MEF2 and Nkx2-5 interact with the same domain of GATA-4, they apparently do not recognize the same molecular determinants.
Although GATA factors physically contact MEF2 proteins through the DNA-binding domain, DNA binding and GATA interaction are dissociable and neither physical nor functional interaction with GATA-4 on natural promoters requires MEF2 DNA binding. The ability of MEF2 to cooperate with MyoD in skeletal myogenesis and to synergize with MyoD in activating an artificial promoter driven by multimerized MyoD-binding sites was also shown to be independent of MEF2 DNA-binding capacity (Molkentin et al., 1995). DNA binding has been shown to be dispensable for some of the actions of two other sequence-specific DNA-binding proteins, the glucocorticoid receptor (Reichardt et al., 1998) and the cell-specific homeodomain protein Pit1 (Dasen et al., 1999). However, in both cases, DNA-binding-independent activities involved transcriptional repression. In the present study, we show that transcriptional activation of several natural promoters by MEF2 proteins is independent of MEF2-binding sites and MEF2 DNA-binding ability. Thus, both suppressive and activating functions of transcription factors may involve DNA-binding-independent pathways.
In addition to the MADS domain, the activation domain of MEF2 is required for functional synergy with GATA-4, suggesting that MEF2 proteins potentiate transcriptional activity of GATA factors through recruitment and/or stabilization of co-activators in the GATA transcription complex. Such co-activators may include the CBP/p300 family, as both MEF2 and GATA proteins have been shown to interact independently with these co-activators (Sartorelli et al., 1997; Blobel et al., 1998; Kakita et al., 1999). Alternatively, MEF2–GATA interaction may displace or overcome a corepressor of either or both factors. For example, MEF2 interaction with GATA-4 or -6 in the heart may displace FOG-2, a GATA-associated cofactor that represses GATA-4 activity (Lu et al., 1999; Svensson et al., 1999). Conversely, recruitment of MEF2 by GATA factors may displace the MEF2-associated corepressors MITR or the HDAC4 deacetylase (Miska et al., 1999; Sparrow et al., 1999).
In addition to cardiomyocytes, MEF2 proteins are co-expressed with members of the GATA family in several other cell types. Most notable is the presence of MEF2 proteins with GATA-6 in smooth muscle cells (Narita et al., 1996; Suzuki et al., 1996), and with GATA-3 in T lymphocytes (Zheng and Flavell, 1997), somites (George et al., 1994) and brain (Pandolfi et al., 1995). Given the demonstrated role of GATA factors in cell differentiation, the GATA–MEF2 synergy provides a general paradigm for understanding the role of MEF2 proteins as determining factors in diverse cell lineages.
Finally, it is tempting to speculate on the role of the MEF2–GATA synergy as a nuclear target of several signaling cascades including calcineurin and p38 MAP kinase. Both pathways, which are highly relevant to many human disorders such as ischemia, heart failure and inflammatory diseases, have been shown to activate MEF2 in cardiac (Kolodziejczy et al., 1999), skeletal muscle (Zetser et al., 1999), neuronal (Mao et al., 1999) and T cells (Han et al., 1997; Blaeser et al., 2000). GATA factors have also been suggested as downstream targets of calcineurin in cardiac and skeletal muscle hypertrophy in connection with the calcineurin-activated NFAT factor (Molkentin et al., 1998; Musaro et al., 1999; Semsarian et al., 1999). It would be interesting to test whether calcineurin or p38-dependent post-translational modifications of MEF2 or GATA proteins modulate the MEF2–GATA physical interaction and/or the resulting functional synergy.
Materials and methods
Cell cultures and transfections
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Transfections were carried out using calcium phosphate 24 h after plating. At 36 h post-transfection, cells were harvested and luciferase activity was assayed with a Berthold LB953 luminometer. The amount of reporter was kept at 1.5 µg per 35 mm dish and the total amount of DNA was kept constant (usually 7 µg). The amount of expression vector used is indicated in the figure legends. Primary cardiomyocyte cultures were prepared from 4-day-old Sprague–Dawley rats as previously described (Charron et al., 1999). The results shown are the mean ± SD of at least two independent experiments carried out in duplicate.
Plasmids
ANF-luciferase promoter constructs were cloned in the PXP-2 vector as described previously (Argentin et al., 1994; Durocher et al., 1996). The BNP-luc constructs were described in Grépin et al. (1994), the βMHC-luc and cardiac α-actin-luc reporters were described in Abdellatif et al. (1994). The αMHC-luc vector was kindly provided by P.M.Buttrick (Buttrick et al., 1993). The construction of the various pCG-GATA-4 vectors was based on the original rat GATA-4 cDNA as previously described (Grépin et al., 1994). The various deletions or point mutations of the ANF promoter and the pRSET-GATA-4 derivatives were generated as described previously (Durocher et al., 1997; Charron et al., 1999). ANF constructs with mutations in the GATA elements or in the NKE were previously described (Durocher et al., 1996; Charron et al., 1999). The A/T-rich mutation is shown in Figure 1; the SRE-like mutation replaces the TTT of the ANF-SRE by GGG thus destroying SRF binding. Heterologous promoters were generated by multimerizing the relevant oligonucleotides flanked by _Bam_HI and _Bgl_II sites upstream of the minimal (–50 bp) ANF-luciferase reporter. pcDNA-MEF2A DIVE was constructed by insertion of the _Xba_I–_Bam_HI fragment of the corresponding pCGN-MEF2A DIVE construct into the _Xba_I–_Bam_HI sites of the pcDNA-3 vector. MEF2 plasmids were kindly provided by E.N.Olson (Molkentin et al., 1996a) and K.Walsh (Andres et al., 1995). The MBP-GATA-4 plasmid was prepared by subcloning a _Xba_I–_Bam_HI rat GATA-4 cDNA fragment containing the entire open reading frame and 1.2 kb of 3′ untranslated sequences (Grépin et al., 1994) into the MBP-expressing pMalc-2 vector (New England Biolabs, Beverly, MA) cut with _Nhe_I–_Bam_HI.
Recombinant protein production
Recombinant MBP-GATA-4 was obtained according to our previously described protocol (Durocher et al., 1997). Essentially, individual colonies were picked and grown in 500 ml of LB up to an OD of 0.6 at 600 nm. Induction of the recombinant protein was carried out by adding isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.5 mM for 2 h at 37°C. The cultures were centrifuged and the bacteria were resuspended and lysed by sonication. Purification on amylose columns (New England Biolabs, Beverly, MA) was performed according to the manufacturer’s instructions.
In vitro transcribed/translated 35S-labeled MEF2 and GATA proteins were produced in rabbit reticulocyte lysates using the TNT-coupled in vitro transcription/translation system (Promega Corp., Madison, WI) from pcDNA-MEF2 derivatives using either T7 or Sp6 RNA polymerase.
In vitro protein–protein interactions
In vitro binding studies were performed using purified MBP-GATA-4 immobilized on an amylose–Sepharose resin (New England Biolabs) and in vitro transcribed/translated MEF2 proteins. Typically, 2–6 µl of 35S-labeled MEF2 proteins were incubated in the presence of 300 ng of immobilized GATA-4 fusion protein in 500 µl of binding buffer [150 mM NaCl, 50 mM Tris–Cl pH 7.5, 0.3% Nonidet P-40, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and 0.25% bovine serum albumin (BSA)] for 2 h at 4°C with agitation and then centrifuged for 2 min at 15 000 r.p.m. at room temperature. The resin was washed three times by vortexing in 500 µl of binding buffer at room temperature and three more times by vortexing in 500 µl of binding buffer without BSA. The protein complexes were released from the resin after boiling in Laemmli buffer and resolved by SDS–PAGE. Labeled proteins were visualized and quantified by autoradiography on phospho storage plates (PhosphorImager, Molecular Dynamics).
To determine the domains of GATA-4 and MEF2 required for physical interaction, full-length GATA-4 or mutated GATA-4 plasmids were used for in vitro cotranscription/cotranslation with wild-type or mutant MEF2A and MEF2C. The cotranslated proteins were incubated in 500 µl of binding buffer with 1 µl of GATA-4 antibody (Santa-Cruz Biotechnology) for 2 h at 4°C with agitation and for an additional 2 h with 20 µl of protein A/G Plus–agarose added (Santa Cruz Biotechnology). Bound immunocomplexes were washed and visualized as mentioned above.
Electrophoretic mobility shift assays (EMSAs)
Three microliters of the in vitro translated MEF2A, MEF2C and MEF2D proteins were used for the binding reactions performed essentially as previously described for GATA binding (Charron et al., 1999) except that 100 ng of dI-dC were included in the binding reaction. Reactions were loaded on a 4% polyacrylamide gel and run at 200 V at room temperature in 0.25× Tris-borate-EDTA. The MEF2-MCK probe is as described in Molkentin et al. (1996b). The rat ANF probes used were as follows: TATA-box, –46TCAGGGAGCTGGGGGCTATAAAAACGGGAGACGCC–11; SRE-like, –124GATCCACTGATAACTTTAAAAGGGCATCTTCA–99; CArG, –417GATCCTCCCGCCCTTATTTGGAGCCCCTGA–390; A/T-rich, –597GAT CCATACTCTAAAAAAATATAATAGCTCTTTCA–567.
Immunoprecipitations and immunoblots
Co-immunoprecipitations of Flag-GATA-4 and HA-MEF2A were carried out using nuclear extracts of 293T cells overexpressing the relevant proteins. Nuclear extracts were prepared as follows: five million 293T cells transfected with 15 µg of expression vectors were harvested 48 h post-transfection in ice-cold phosphate-buffered saline (PBS) containing 1 mM sodium orthovanadate and 1 mM EDTA. The cells were resuspended in hypotonic buffer (20 mM HEPES pH 7.9, 20 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM EDTA, 1 mM EGTA, 0.25 mM sodium molybdate, 10 µg/ml leupeptin, 10 µg/ml aprotinin, 10 µg/ml pepstatin, 2 mM DTT, 0.5 mM PMSF and 100 nM okadaic acid) and swollen on ice for 15 min. Twenty-five microliters of 10% NP-40 were added and the microtubes were vortexed vigorously. The nuclei were then pelleted by centrifugation at 7000 r.p.m. at 4°C. The nuclear pellet was resuspended in 50–100 µl of high salt buffer (hypotonic buffer containing 20% glycerol and 0.4% NaCl) and shaken vigorously at 4°C for 1 h. The nuclear extracts were cleared by centrifugation at 15 000 r.p.m. for 15 min at 4°C and the protein concentration was determined by the Bradford assay. Co-immunoprecipitation reactions were carried out on 50 µg of nuclear extracts using 1 µl of 12CA5 antibody in 500 µl of binding buffer without BSA, and bound immunocomplexes were washed and subjected to SDS–PAGE, as described previously (Durocher et al., 1997). Proteins were transferred on Hybond-PVDF membrane and subjected to immunoblotting. Anti-Flag M5 (Sigma) and 12CA5 (anti-HA) monoclonal antibodies were used at a dilution of 1/8000, revealed with an anti-mouse-HRP (Sigma) at a dilution of 1/50 000 and visualized using ECL Plus (Amersham Pharmacia Biotechnology).
Acknowledgments
Acknowledgements
We are grateful to Anne Aries and Pierre Paradis for generating some ANF mutants, and to Eric Olson, Jeffrey Molkentin and Ken Walsh for the gift of MEF2 vectors. We thank Lise Laroche for secretarial assistance, and members of the Nemer laboratory for discussions and critical reading of this manuscript. This work was supported by grants from the Canadian Medical Research Council (MRC). S.M. holds a studentship from the MRC; F.C. was funded by a research traineeship from the Heart and Stroke Foundation of Canada and is presently a recipient of a National Cancer Institute of Canada studentship. M.N. is a senior MRC scientist.
References
- Abdellatif M., MacLellan,W.R. and Schneider,M.D. (1994) p21 Ras as a governor of global gene expression. J. Biol. Chem., 269, 15423–15426. [PubMed] [Google Scholar]
- Andres V., Fisher,S., Wearsch,P. and Walsh,K. (1995) Regulation of GAX homeobox gene transcription by a combination of positive factors including myocyte-specific enhancer factor 2. Mol. Cell. Biol., 15, 4272–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentin S., Ardati,A., Tremblay,S., Lihrmann,I., Robitaille,L., Drouin,J. and Nemer,M. (1994) Developmental stage-specific regulation of atrial natriuretic factor gene transcription in cardiac cells. Mol. Cell. Biol., 14, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Drake,C.J. and Schwarz,J.J. (1999) The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev. Biol., 211, 255–267. [DOI] [PubMed] [Google Scholar]
- Black B.L. and Olson,E.N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol., 14, 167–196. [DOI] [PubMed] [Google Scholar]
- Black B.L., Molkentin,J.D. and Olson,E.N. (1998) Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell. Biol., 18, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaeser F., Ho,N., Prywes,R. and Chatila,T.A. (2000) Ca2+-dependent gene expression mediated by MEF2 transcription factors. J. Biol. Chem., 275, 197–209. [DOI] [PubMed] [Google Scholar]
- Blobel G.A., Nakajima,T., Eckner,R., Montminy,M. and Orkin,S.H. (1998) CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl Acad. Sci. USA, 95, 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour B.A., O’Brien,M.A., Lockwood,W.L., Goldstein,E.S., Bodmer,R., Taghert,P.H., Abmayr,S.M. and Nguyen,H.T. (1995) Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev., 9, 730–741. [DOI] [PubMed] [Google Scholar]
- Buttrick P.M., Kaplan,M.L., Kitsis,R.N. and Leinwand,L.A. (1993) Distinct behavior of cardiac myosin heavy chain gene constructs in vivo. Discordance with in vitro results. Circ. Res., 72, 1211–1217. [DOI] [PubMed] [Google Scholar]
- Chambers A.E., Logan,M., Kotecha,S., Towers,N., Sparrow,D. and Mohun,T.J. (1994) The RSRF/MEF2 protein SL1 regulates cardiac muscle-specific transcription of a myosin light-chain gene in Xenopus embryos. Genes Dev., 8, 1324–1334. [DOI] [PubMed] [Google Scholar]
- Charron F. and Nemer,M. (1999) GATA transcription factors and cardiac development. Semin. Cell Dev. Biol., 10, 85–91. [DOI] [PubMed] [Google Scholar]
- Charron F., Paradis,P., Bronchain,O., Nemer,G. and Nemer,M. (1999) Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell. Biol., 19, 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen J.S., O’Connell,S.M., Flynn,S.E., Treier,M., Gleiberman,A.S., Szeto,D.P., Hooshmand,F., Aggarwal,A.K. and Rosenfeld,M.G. (1999) Reciprocal interactions of Pit1 and GATA2 mediate signaling gradient-induced determination of pituitary cell types. Cell, 97, 587–598. [DOI] [PubMed] [Google Scholar]
- Di Lisi R., Millino,C., Calabria,E., Altruda,F., Schiaffino,S. and Ausoni,S. (1998) Combinatorial _cis_-acting elements control tissue-specific activation of the cardiac troponin I gene in vitro and _in vivo._J. Biol. Chem., 273, 25371–25380. [DOI] [PubMed] [Google Scholar]
- Durocher D., Chen,C.Y., Ardati,A., Schwartz,R.J. and Nemer,M. (1996) The ANF promoter is a downstream target for Nkx-2.5 in the myocardium. Mol. Cell. Biol., 16, 4648–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Charron,F., Warren,R., Schwartz,R.J. and Nemer,M. (1997) The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J., 16, 5687–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Grépin,C. and Nemer,M. (1998) Regulation of gene expression in the endocrine heart. In Conn,P.M. (ed.), Recent Progress in Hormone Research. The Endocrine Society Press, Bethesda, MD, pp. 7–23. [PubMed] [Google Scholar]
- Fu Y.C. and Izumo,S. (1995) Cardiac myogenesis—overexpression of xcsx2 or xmef2a in whole Xenopus embryos induces the precocious expression of xmhc-α gene. Rouxs Arch. Dev. Biol., 205, 198–202. [DOI] [PubMed] [Google Scholar]
- George K.M., Leonard,M.W., Roth,M.E., Lieuw,K.H., Kioussis,D., Grosveld,F. and Engel,J.D. (1994) Embryonic expression and cloning of the murine GATA-3 gene. Development, 120, 2673–2686. [DOI] [PubMed] [Google Scholar]
- Grépin C., Dagnino,L., Robitaille,L., Haberstroh,L., Antakly,T. and Nemer,M. (1994) A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol. Cell. Biol., 14, 3115–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grépin C., Robitaille,L., Antakly,T. and Nemer,M. (1995) Inhibition of transcription factor GATA-4 expression blocks in vitro cardiac muscle differentiation. Mol. Cell. Biol., 15, 4095–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Jiang,Y., Li,Z., Kravchenko,V.V. and Ulevitch,R.J. (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature, 386, 296–299. [DOI] [PubMed] [Google Scholar]
- Iannello R.C., Mar,J.H. and Ordahl,C.P. (1991) Characterization of a promoter element required for transcription in myocardial cells. J. Biol. Chem., 266, 3309–3316. [PubMed] [Google Scholar]
- Kakita T., Hasegawa,K., Morimoto,T., Kaburagi,S., Wada,H. and Sasayama,S. (1999) p300 protein as a coactivator of GATA-5 in the transcription of cardiac-restricted atrial natriuretic factor gene. J. Biol. Chem., 274, 34096–34102. [DOI] [PubMed] [Google Scholar]
- Kaushal S., Schneider,J.W., Nadal-Ginard,B. and Mahdavi,V. (1994) Activation of the myogenic lineage by mef2a, a factor that induces and cooperates with myod. Science, 266, 1236–1240. [DOI] [PubMed] [Google Scholar]
- Kolodziejczy S.M., Wang,L., Balazsi,K., DeRepentigny,Y., Kothary,R. and Megeney,L.A. (1999) MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr. Biol., 9, 1203–1206. [DOI] [PubMed] [Google Scholar]
- Kuisk I.R., Li,H., Tran,D. and Capetanaki,Y. (1996) A single MEF2 site governs desmin transcription in both heart and skeletal muscle during mouse embryogenesis. Dev. Biol., 174, 1–13. [DOI] [PubMed] [Google Scholar]
- Lee Y., Nadal-Ginard,B., Mahdavi,V. and Izumo,S. (1997) Myocyte-specific enhancer factor 2 and thyroid hormone receptor associate and synergistically activate the α-cardiac myosin heavy-chain gene. Mol. Cell. Biol., 17, 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B., Zhao,B., Ranganayakulu,G., Paterson,B.M., Schulz,R.A. and Olson,E.N. (1995) Requirement of MADS domain transcription factor D-MEF2 for muscle formation in _Drosophila._Science, 267, 688–693. [DOI] [PubMed] [Google Scholar]
- Lin Q., Schwarz,J., Bucana,C. and Olson,E.N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor mef2c. Science, 276, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Lu,J., Yanagisawa,H., Webb,R., Lyons,G.E., Richardson,J.A. and Olson,E.N. (1998) Requirement of the MADS-box transcription factor MEF2C for vascular development. Development, 125, 4565–4574. [DOI] [PubMed] [Google Scholar]
- Lu J.R., McKinsey,T.A., Xu,H.T., Wang,D.Z., Richardson,J.A. and Olson,E.N. (1999) FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol., 19, 4495–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Bonni,A., Xia,F., Nadal-Vicens,M. and Greenberg,M.E. (1999) Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science, 286, 785–790. [DOI] [PubMed] [Google Scholar]
- Miska E.A., Karlsson,C., Langley,E., Nielsen,S.J., Pines,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D. and Markham,B.E. (1993) Myocyte-specific enhancer-binding factor (MEF-2) regulates α-cardiac myosin heavy chain gene expression in vitro and _in vivo._J. Biol. Chem., 268, 19512–19520. [PubMed] [Google Scholar]
- Molkentin J.D., Black,B.L., Martin,J.F. and Olson,E.N. (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell, 83, 1125–1136. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Firulli,A.B., Black,B.L., Martin,J.F., Hustad,C.M., Copeland,N., Jenkins,N., Lyons,G. and Olson,E.N. (1996a) MEF2B is a potent transactivator expressed in early myogenic lineages. Mol. Cell. Biol., 16, 3814–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Black,B.L., Martin,J.F. and Olson,E.N. (1996b) Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol., 16, 2627–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Lin,Q., Duncan,S.A. and Olson,E.N. (1997) Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev., 11, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Lu,J.R., Antos,C.L., Markham,B., Richardson,J., Robbins,J., Grant,S.R. and Olson,E.N. (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell, 93, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey E.E., Tang,Z., Sigrist,K., Lu,M.M., Jiang,F., Ip,H.S. and Parmacek,M.S. (1998) GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev., 12, 3579–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaro A., McCullagh,K.J., Naya,F.J., Olson,E.N. and Rosenthal,N. (1999) IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature, 400, 581–585. [DOI] [PubMed] [Google Scholar]
- Narita N., Heikinheimo,M., Bielinska,M., White,R.A. and Wilson,D.B. (1996) The gene for transcription factor GATA-6 resides on mouse chromosome 18 and is expressed in myocardium and vascular smooth muscle. Genomics, 36, 345–348. [DOI] [PubMed] [Google Scholar]
- Nemer G., Qureshi,S.A., Malo,D. and Nemer,M. (1999) Functional analysis and chromosomal mapping of GATA5, a gene encoding a zinc finger DNA-binding protein. Mamm. Genome, 10, 993–999. [DOI] [PubMed] [Google Scholar]
- Pandolfi P.P., Roth,M.E., Karis,A., Leonard,M.W., Dzierzak,E., Grosveld,F.G., Engel,J.D. and Lindenbaum,M.H. (1995) Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nature Genet., 11, 40–44. [DOI] [PubMed] [Google Scholar]
- Pevny L., Simon,M.C., Robertson,E., Klein,W.H., Tsai,S.F., D’Agati,V., Orkin,S.H. and Costantini,F. (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature, 349, 257–260. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G., Zhao,B., Dokidis,A., Molkentin,J.D., Olson,E.N. and Schulz,R.A. (1995) A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in _Drosophila._Dev. Biol., 171, 169–181. [DOI] [PubMed] [Google Scholar]
- Reichardt H.M. et al. (1998) DNA binding of the glucocorticoid receptor is not essential for survival. Cell, 93, 531–541. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Huang,J., Hamamori,Y. and Kedes,L. (1997) Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol., 17, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semsarian C., Wu,M.J., Ju,Y.K., Marciniec,T., Yeoh,T., Allen,D.G., Harvey,R.P. and Graham,R.M. (1999) Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature, 400, 576–581. [DOI] [PubMed] [Google Scholar]
- Sepulveda J.L., Belaguli,N., Nigam,V., Chen,C.Y., Nemer,M. and Schwartz,R.J. (1998) GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol. Cell. Biol., 18, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow D.B., Miska,E.A., Langley,E., Reynaud-Deonauth,S., Kotecha,S., Towers,N., Spohr,G., Kouzarides,T. and Mohun,T.J. (1999) MEF-2 function is modified by a novel co-repressor, MITR. EMBO J., 18, 5085–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E., Evans,T., Lowry,J., Truong,L., Bell,D.W., Testa,J.R. and Walsh,K. (1996) The human GATA-6 gene: structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics, 38, 283–290. [DOI] [PubMed] [Google Scholar]
- Svensson E.C., Tufts,R.L., Polk,C.E. and Leiden,J.M. (1999) Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl Acad. Sci. USA, 96, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.Y., Keller,G., Kuo,F.C., Weiss,M., Chen,J., Rosenblatt,M., Alt,F.W. and Orkin,S.H. (1994) An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature, 371, 221–226. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crosslely,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- Viger R.S., Mertineit,C., Trasler,J.M. and Nemer,M. (1998) Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Müllerian inhibiting substance promoter. Development, 125, 2665–2675. [DOI] [PubMed] [Google Scholar]
- Yu Y.T., Breitbart,R.E., Smoot,L.B., Lee,Y., Mahdavi,V. and Nadal-Ginard,B. (1992) Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev., 6, 1783–1798. [DOI] [PubMed] [Google Scholar]
- Zetser A., Gredinger,E. and Bengal,E. (1999) p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem., 274, 5193–5200. [DOI] [PubMed] [Google Scholar]
- Zheng W. and Flavell,R.A. (1997) The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell, 89, 587–596. [DOI] [PubMed] [Google Scholar]
- Zhu H., Garcia,A.V., Ross,R.S., Evans,S.M. and Chien,K.R. (1991) A conserved 28-base-pair element (HF-1) in the rat cardiac myosin light-chain-2 gene confers cardiac-specific and α-adrenergic-inducible expression in cultured neonatal rat myocardial cells. Mol. Cell. Biol., 11, 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]