Super-resolution Imaging of Chemical Synapses in the Brain (original) (raw)
. Author manuscript; available in PMC: 2011 Jun 9.
Abstract
Determination of the molecular architecture of synapses requires nanoscopic image resolution and specific molecular recognition, a task that has so far defied many conventional imaging approaches. Here we present a super-resolution fluorescence imaging method to visualize the molecular architecture of synapses in the brain. Using multicolor, three-dimensional stochastic optical reconstruction microscopy, the distributions of synaptic proteins can be measured with nanometer precision. Furthermore, the wide-field, volumetric imaging method enables high-throughput, quantitative analysis of a large number of synapses from different brain regions. To demonstrate the capabilities of this approach, we have determined the organization of ten protein components of the presynaptic active zone and the postsynaptic density. Variations in synapse morphology, neurotransmitter receptor composition, and receptor distribution were observed both among synapses and across different brain regions. Combination with optogenetics further allowed molecular events associated with synaptic plasticity to be resolved at the single-synapse level.
INTRODUCTION
Chemical synapses are specialized cell-cell junctions of several hundred nanometers in size, in which an elaborate protein machinery orchestrates neurotransmitter-mediated signal transmission between neurons and their target cells. Ultrastructural analysis has provided fundamental insights into the organization and function of synapses. In most central synapses, the pre- and postsynaptic terminals are closely apposed, electron dense structures separated by the synaptic cleft. The presynaptic active zone (PAZ) is characterized by an accumulation of synaptic vesicles and a cytomatrix that facilitates neurotransmitter release (Jin and Garner, 2008; Schoch and Gundelfinger, 2006). The postsynaptic terminal consists of a dense protein scaffolding meshwork referred to as the postsynaptic density (PSD), which anchors neurotransmitter receptors as well as various signaling molecules that relay the neurotransmitter signals (Okabe, 2007; Sheng and Hoogenraad, 2007). In addition to the major protein components of the pre- and postsynaptic specializations, recent proteomic and genomic studies have suggested that hundreds of other protein species are localized in synapses (Collins et al., 2006; Fernandez et al., 2009).
Correlating synaptic structure with function requires a detailed characterization of the organization and concerted action of synaptic proteins. In turn, the dissection of the protein architecture of synapses demands imaging with nanometer-scale resolution as well as highly specific and efficient molecular identification, a task difficult to accomplish by conventional imaging techniques. Ultrastructural analyses of synapses have so far relied primarily on electron microscopy (EM) (Arthur et al., 2007; Chen et al., 2008b; Peters et al., 1991), with the detection of specific molecules accomplished by immuno-gold labeling. However, the challenges to achieve high-density immuno-gold labeling, to image several molecular species simultaneously, and to obtain three-dimensional (3D) immuno-EM reconstructions make it difficult to precisely assess the relative positions of different synaptic proteins. While EM tomography may overcome these limitations and ultimately yield a high-resolution 3D molecular architecture of the synapse (Chen et al., 2008b), only a few synaptic proteins have been identified with this technology so far. Fluorescence microscopy, on the other hand, readily allows multiple protein species to be efficiently labeled and simultaneously imaged in 3D. However, it has been difficult to study sub-synaptic structures with fluorescence microscopy because of the small size of synapses, which is near the diffraction-limited resolution of light microscopy.
Therefore, determining the protein architecture of the synapse calls for a multicolor, 3D imaging method with molecular-scale resolution such that the precise spatial relationship between different synaptic proteins can be mapped out. Moreover, it is desirable to perform this type of imaging in brain tissues, which allow for the study of synaptic organization as a part of intact neural circuits and the assessment of structural and molecular variations in different synapse types and brain regions. The recent development of super-resolution fluorescence microscopy, which enables imaging with sub-diffraction-limit resolution (Heintzmann and Gustafsson, 2009; Hell, 2007; Huang et al., 2009), offers promising solutions to these challenges. These super-resolution approaches include ensemble imaging methods, such as stimulated emission depletion (STED and related RESOLFT) microscopy (Hell, 2007; Klar and Hell, 1999) and saturated structured-illumination microscopy (Gustafsson, 2005), as well as single-molecule imaging methods, such as stochastic optical reconstruction microscopy (STORM) or (fluorescence) photoactivation localization microscopy ((F)PALM) (Betzig et al., 2006; Hess et al., 2006; Rust et al., 2006). Among these approaches, STED microscopy has been applied to imaging the morphology of dendritic spines (Ding et al., 2009; Nagerl et al., 2008) and to observing the organization of a few synaptic components individually (Kittel et al., 2006; Meyer et al., 2009; Willig et al., 2006). High-density single-particle tracking facilitated by photoactivation has revealed distinct foci of enhanced actin polymerization in the dendritic spines of cultured neurons (Frost et al., 2010).
Here, we establish STORM as an effective tool to analyze the molecular architecture of sub-cellular structures in tissue samples. By determining the position of each molecule with nanometer precision, STORM allows a high-resolution structural reconstruction of molecular assemblies. We devised a systematic three-color, 3D STORM imaging approach to map out the protein organization in synapses. This wide-field, volumetric imaging approach allows a large number of synapses in different brain regions to be imaged in a relatively rapid manner, facilitating a systematic comparative analysis. Using this technology, we determined the spatial relationship of ten pre- and postsynaptic proteins, revealing a highly oriented organization of presynaptic scaffolding proteins and a differential compartmental distribution of PSD components. Significant variations in the neurotransmitter receptor composition and distribution were observed both from synapse to synapse and across brain regions. When combined with optogenetics, this approach further allowed activity-dependent changes in protein organization to be examined at the single-synapse level.
RESULTS
Multicolor, 3D super-resolution imaging of brain tissue
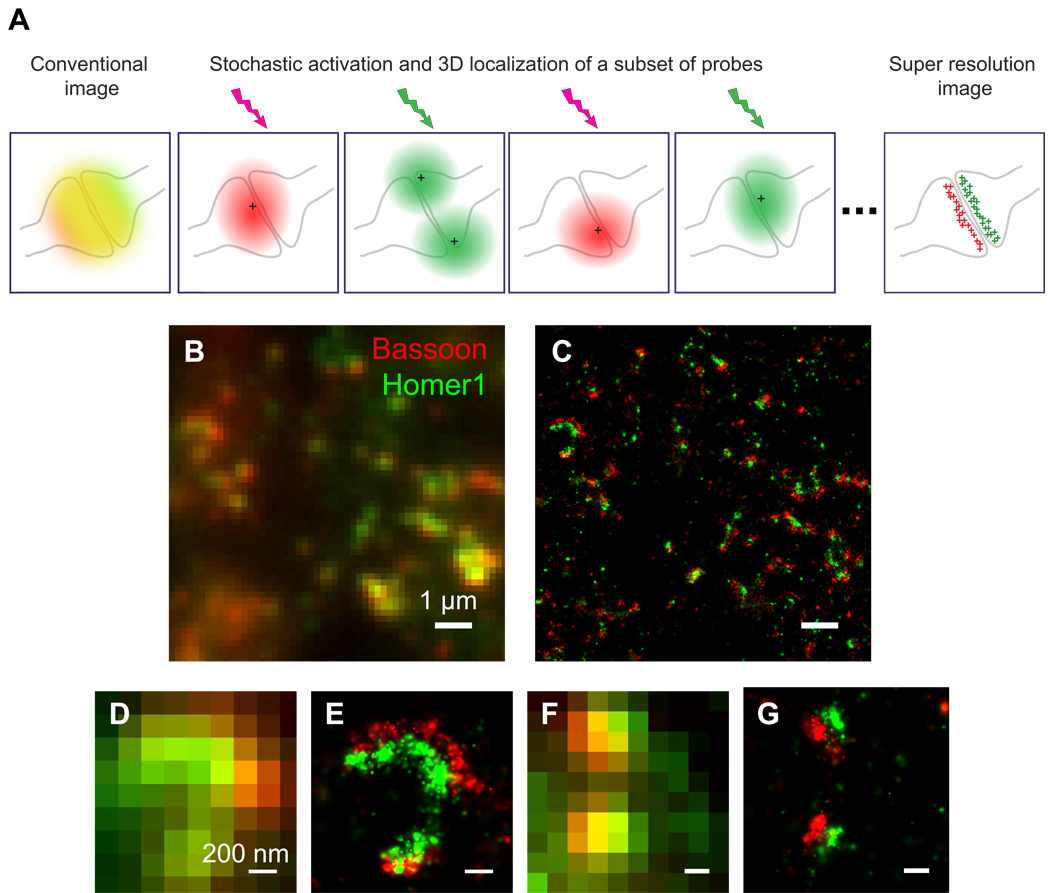
STORM acquires its high image resolution based on single-molecule imaging of photoswitchable fluorescent probes. A typical STORM imaging experiment is often comprised of many imaging cycles (Figure 1A). Within each cycle, only a sparse subset of fluorophores are activated, imaged and deactivated. The low density of the activated fluorophores allows the images of these fluorophores to be readily separated from each other. As a result, the position of each individual fluorophore can be determined to a precision substantially beyond the diffraction-limited resolution by finding the centroid of its image. Iterating this procedure to obtain the localizations of many fluorophores then allows the reconstruction of a sub-diffraction-limit image from these localization points (Betzig et al., 2006; Hess et al., 2006; Rust et al., 2006). For 3D STORM, the x, y and z coordinates of each activated fluorophore can be determined using astigmatism imaging (Huang et al., 2008b). In this scheme, a cylindrical lens is inserted in the detection path of the microscope to render the image of a single fluorophore elliptical (Figure 1A). The lateral (x, y) and axial (z) coordinates of the fluorophore can then be determined from the centroid position and ellipiticity of the image, respectively (Huang et al., 2008a; Huang et al., 2008b).
Figure 1. STORM imaging of pre- and postsynaptic scaffolding proteins.
(A) Schematic of 3D STORM. For molecules that give overlapping images (represented by the colored region in the left panel), STORM resolves these molecules by stochastically activating them at different times during image acquisition. At any time, only a sparse, optically resolvable subset of molecules are activated, allowing their images (represented by the red and green ellipsoids in the middle panels) to be separated from each other and their 3D positions (represented by the crosses in the middle panels) to be precisely determined from the centroid positions and ellipiticities of these images. Iteration of this process allows the positions of many molecules to be determined and a super-resolution image to be reconstructed from these positions (represented by the red and green crosses right panel).
(B–G) Presynaptic protein Bassoon and postsynaptic protein Homer1 in the mouse MOB glomeruli were identified by immunohistochemistry using Cy3-A647 and A405-A647 conjugated antibodies, respectively. The conventional fluorescence image (B) shows punctate patterns that are partially overlapping, whereas the STORM image (C) of the same area clearly resolves distinct synaptic structures. Further zoom-in of the conventional images (D, F) does not reveal detailed structure of the synapses whereas the corresponding STORM images (E, G) distinguish the presynaptic Bassoon and postsynaptic Homer1 clusters.
To date, single-molecule-localization based super-resolution microscopy has only been demonstrated on cultured cells, which have relatively simple sample preparation and imaging requirements. To study sub-cellular structures in the brain, it is however important to perform imaging directly on brain tissue samples. Here, we set out to implement STORM imaging of tissue sections.
Compared to imaging cultured cells, tissue imaging presents unique challenges, especially for single-molecule detection. One major challenge is the fluorescence background of tissue samples. In particular, because brain sections have a very high density of synapses, a significant amount of background fluorescence is generated from molecules outside the focal plane, which directly impairs the localization precision of individual molecules. We solved this problem with a two-step imaging approach. First, samples were illuminated with the imaging/deactivation laser at a low incidence angle in order to deactivate fluorophores over a large depth above and below the focal plane. Next the incident angles of the activation and imaging lasers were increased such that only fluorophores within a few microns of the focal plane were reactivated and imaged (See Experimental Procedure, “STORM image acquisition” section for more details). This procedure greatly reduced the fluorescence background from out-of-focus fluorophores.
In addition, spherical aberration and light scattering of tissue samples cause significant photon loss from each fluorophore and thus a further reduction in the localization precision. To counter this problem, we identified Alexa Fluor 647 (A647), a far-red photoswitchable dye with exceptional brightness, as the optimal primary fluorescent label. For three-color imaging, we immunolabeled the target proteins with three spectrally distinct activator-reporter dye pairs, A405-A647, Cy2-A647, or Cy3-A647, which allows the bright A647 dyes to be used for all three channels (Bates et al., 2007). This color scheme further eliminated chromatic aberration as the same reporter fluorophores were imaged in different color channels. During image acquisition, the reporter dye A647 was activated with one of three activation lasers (405 nm, 457 nm and 532 nm) and subsequently imaged and deactivated with a red imaging laser (657 nm). Different probes were distinguished by the wavelength of the activation laser that they each responded to, which was determined by the activator dyes (Figure S1A). Together, the reduction of fluorescence background and the brightness of A647 (~2500 detected photons per switching cycle in the tissue sample) allowed a localization precision of 14 nm in the xy plane and 35 nm in the z direction, experimentally determined from repetitive localizations of individual isolated probes in the tissue samples.
The requirement of multicolor imaging presents additional challenges for tissue imaging. Although the brightness of A647 enables high resolution in multicolor imaging, the red imaging laser itself can activate the dark-state A647 in an activator-independent manner, albeit inefficiently. In a densely labeled brain section, these “color-blind”, nonspecific activations can result in significant cross-talk between the different color channels. We devised an image analysis algorithm to automatically subtract this color cross-talk. This algorithm takes advantage of the fact that the probability of nonspecific activation caused by the imaging laser in any local region of the sample should remain constant across imaging frames, regardless of whether the frame is preceded by an activation laser pulse. We thus designed an imaging sequence such that each activation laser pulse was followed by three imaging frames (Figure S1B). The molecules that appeared fluorescent in the imaging frames immediately following the activation laser pulses could have been activated either specifically by the activation laser or nonspecifically by the imaging laser. Localizations of these molecules were colored by their preceding activation pulses and plotted in a raw STORM image. The molecules that became fluorescent in the subsequent frames distant from the activation pulses could only be a result of the nonspecific activation by the imaging laser. These subsequent frames then allowed us to derive the nonspecific activation contribution in any local region of the sample and to estimate the corresponding number of nonspecific localizations in the raw image, considering the constant probability of nonspecific activation across frames. This contribution was then statistically subtracted from the raw image to obtain color-specific contributions (See Experimental Procedure, “Multicolor 3D imaging and crosstalk subtraction” section for more details). This local cross-talk subtraction approach enabled us to reduce errors associated with labeling variability across different regions of a sample or across different samples.
Fast ultrastructural imaging of synapses
Until now, ultrastructural imaging in the brain has relied primarily on transmission electron microscopy (TEM). Obtaining sufficient contrast under TEM necessitates ultrathin sectioning and stringent sample preparation requirements. 3D reconstruction further demands serial sectioning and section alignment. The low throughput of these sample preparation and imaging processes hampers detailed comparative analysis across a large number of samples and brain areas. 3D STORM, on the other hand, allows relatively fast volumetric imaging without the requirements of ultrathin sectioning or sample scanning (imaging a volume of ~30 µm × 30 µm × 1 µm by STORM takes only a few minutes or less time). Here, we demonstrate that STORM imaging can be adapted to both ultra-high resolution and relatively high-throughput data collection.
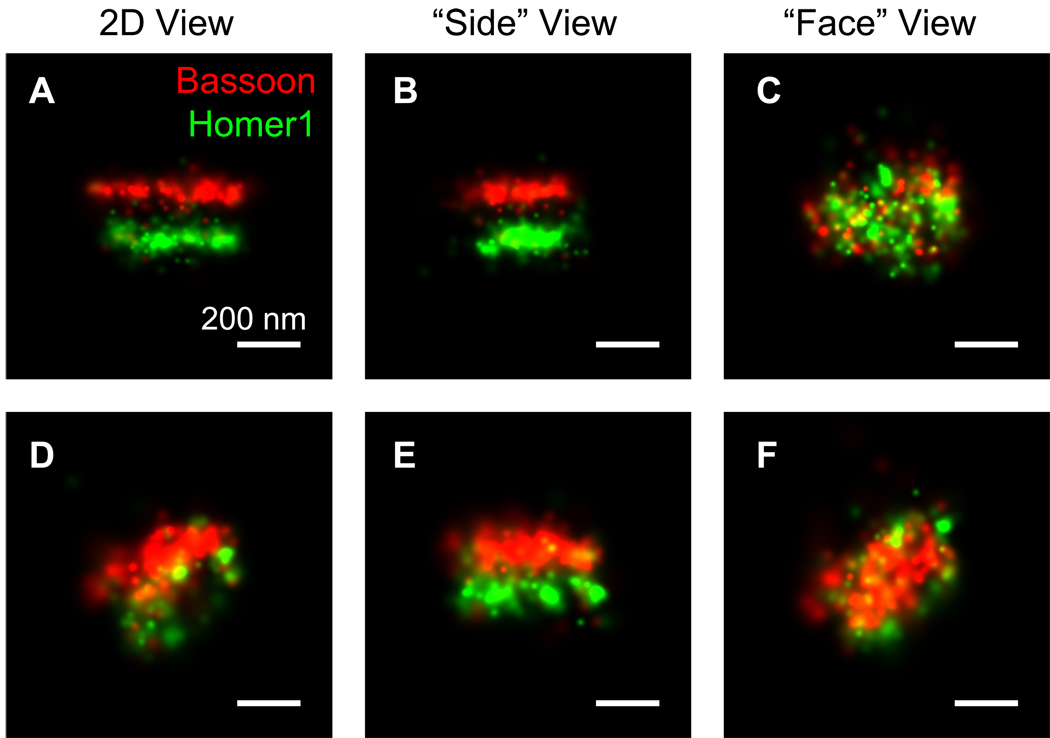
As an initial test of the ability of STORM to resolve closely positioned synaptic components, we imaged a pair of pre- and postsynaptic scaffolding proteins, Bassoon and Homer1, respectively. Imaging was performed in the glomerular layer of the main olfactory bulb (MOB), where olfactory sensory neurons form synapses with mitral cell dendrites at high density (Shepherd, 2004). The two proteins were immunostained with antibodies against the N-terminal region of Bassoon (Bassoon[N]) and the C-terminal region of Homer1 isoform b and c (Homer1[C]), respectively. In conventional wide-field fluorescence images (Figure 1B, D, F), Bassoon and Homer1 appeared as partially overlapping puncta without any discernable structural feature. In contrast, STORM images (Figure 1C, E, G) clearly resolved the two protein distributions as juxtaposed pairs of dense molecular structures separated by the synaptic cleft. 3D STORM further allowed the shape of the PAZ and PSD to be determined. Figure 2 demonstrates two such examples: Whether the synapse was oriented with its trans-synaptic axis in (Figure 2A–C) or out of (Figure 2D–F) the imaging focal plane, 3D images enabled us to resolve Bassoon and Homer1 structures flanking the synaptic cleft in the “side” views (Figures 2B, 2E), and their elliptical disk shape in the “face” views (Figures 2C, 2F). A full 3D perspective of a synapse can be visualized in Supplemental Movie S1. In the following analysis, closely apposed Bassoon and Homer1 structures (center positions separated by less than 250 nm) were identified as excitatory synapses.
Figure 2. 3D STORM images of synapses.
Despite different orientations of synapses (A–C and D–F), 3D imaging allows the visualization of presynaptic Bassoon and postsynaptic Homer1 as well-separated elliptical disks in different viewing angles. The “side” views represent projection images with the trans-synaptic axis rotated into the viewing plane, and the “face” views represent projection images with trans-synaptic axis rotated perpendicular to the viewing plane.
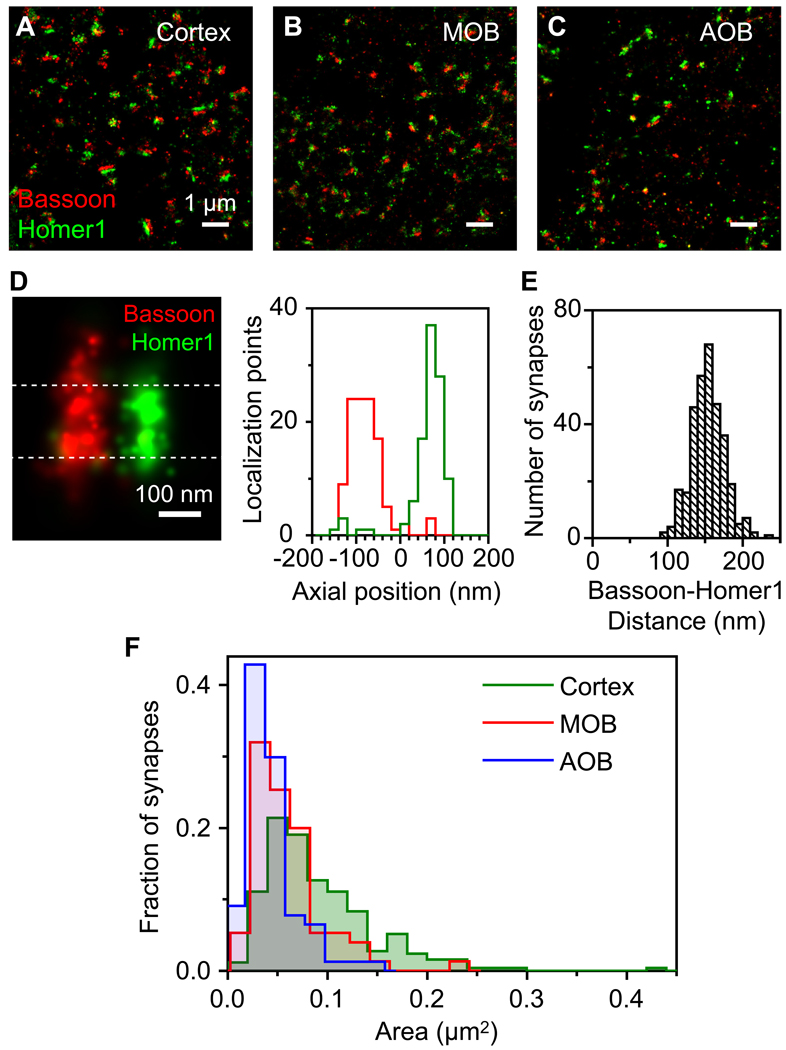
The fast sample preparation and imaging by 3D STORM offers a unique opportunity to perform high throughput ultrastructural analysis of synapses across large brain areas. Here we analyzed synapse morphology in three regions of the mouse brain (Figures S2, 3A–C): the glomerular regions of the main and accessory olfactory bulbs (MOB and AOB), and the ventral orbital cortex (Cortex). The distance between Bassoon and Homer1 (measured as the distance between the peak positions of the two distributions of localization points, Figure 3D) appeared uniform in synapses within the same region as well as across different brain areas. Statistics of 64 synapses from the MOB region within the same sample revealed a relatively narrow distribution with a mean of 153.4 nm, a standard deviation (SD) of 17.3 nm, and a standard error of the mean (SEM) of 2.2 nm. Six independent experiments from different adult mouse brains, in which a total of 327 synapses were imaged, provided an overall Bassoon-Homer1 distance value of 153.8 nm with 1.2 nm SEM (Figure 3E). The narrow distribution centered at ~ 150 nm with very few synapses exhibiting Bassoon-Homer1 distance larger than 200 nm justifies the empirically cutoff value of 250 nm for synapse identification. The Bassoon-Homer1 distances measured from synapses in the AOB and Cortex regions showed similar distributions (155 ± 21 nm and 150 ± 20 nm (± SD), respectively). The cross-section area of the Bassoon-Homer1 disks, on the other hand, showed substantial variations (Figure 3F). In particular, the area of synapses in the Cortex spanned over an order of magnitude, ranging from 0.01 to 0.3 µm2. The mean areas measured from different brain regions were also substantially different, with the AOB displaying the smallest and the Cortex the largest synapses (AOB: 0.046 ± 0.003 µm2, MOB: 0.058 ± 0.004 µm2, Cortex: 0.092 ± 0.003 µm2, (± SEM)) (Figure 3F). Interestingly, the overall density of excitatory synapses (Bassoon-Homer1 pairs) in the AOB (0.11 ± 0.02 synapses per µm3) was also much lower than the synapse density measured in the MOB (0.50 ± 0.02 per µm3) and Cortex (0.52 ± 0.02 per µm3).
Figure 3. Quantification of synaptic morphology in distinct regions of the brain.
(A–C) STORM images of Bassoon and Homer1 from the Cortex (A), MOB (B) and AOB (C) regions in the same mouse brain section.
(D) To determine the separation between Bassoon and Homer1 clusters, the distribution of localization points (within a ~160 nm thick region at the center of the synapse defined by the dashed lines) along the trans-synaptic axis was measured and fit with Gaussian functions, and the distance between the centroid positions of the two Gaussians were defined as Bassoon-Homer1 distances.
(E) The histogram of Bassoon-Homer1 distances of 327 synapses in mouse MOB glomeruli.
(F) Normalized distribution of the active zone area of excitatory synapses, measured from the sum of the bassoon and Homer1 signals. A total of 252 synapses in the cortex, 75 synapses in the MOB and 77 synapses in the AOB were measured.
Axial positions of synaptic proteins
The multicolor and 3D super-resolution imaging capabilities of STORM provide a means to quantitatively characterize the spatial relationship of different proteins within the synapse and to determine the relative orientation of large molecular species. Here, we present a systematic approach to map the molecular organization of synaptic proteins. In this approach, a pair of PAZ and PSD scaffolding proteins were selected as references and a third protein was placed in the reference frame using three-color, 3D STORM imaging. The procedure was then repeated to place multiple proteins in the same frame, ultimately allowing a comprehensive molecular architecture to be built.
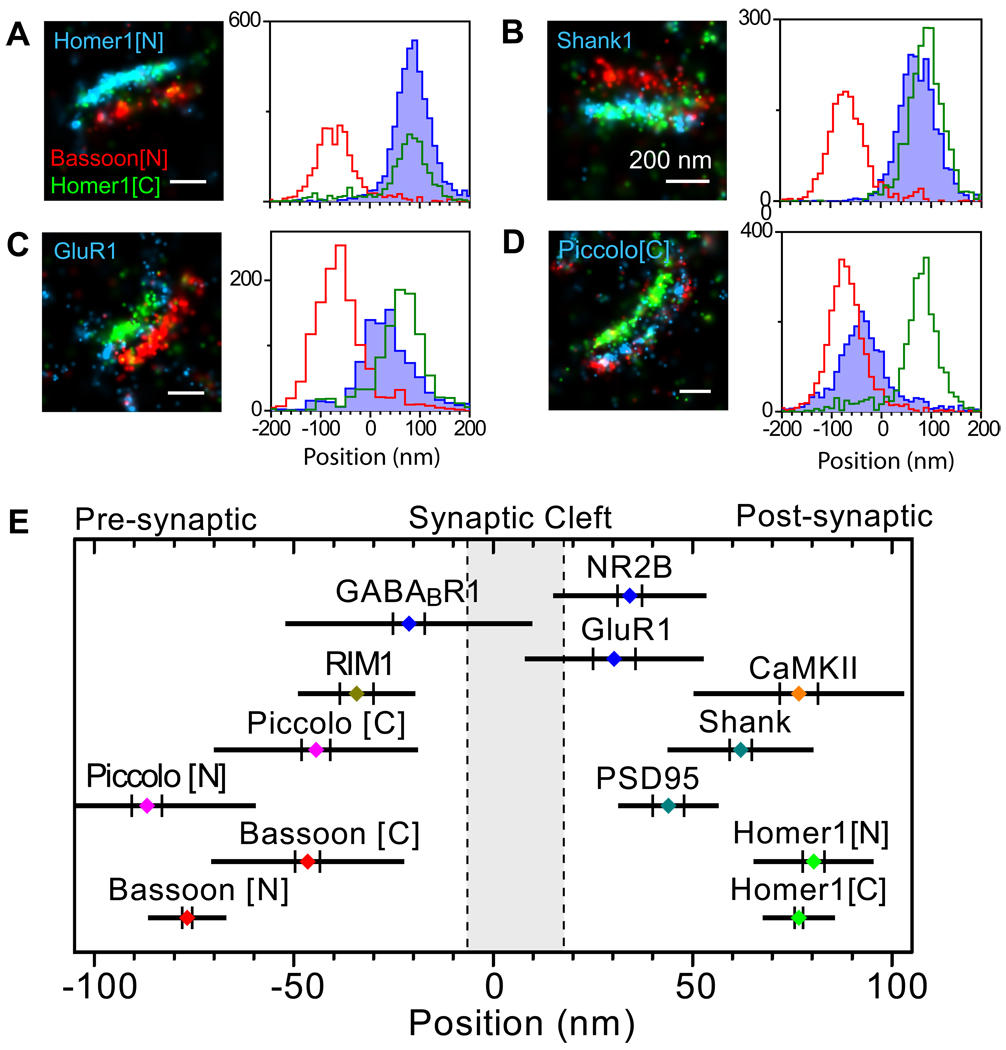
First, we focused on the axial distribution of synaptic components. The prior knowledge of some of these proteins allowed us to validate this approach. The highly uniform Bassoon-Homer1 distance across synapses makes them ideal choices as references. We thus defined a synaptic coordinate system with the origin set at the midpoint between Bassoon[N] and Homer1[C] and the positive direction pointing to the postsynaptic side. In this system, the mean positions of Bassoon[N] and Homer1[C] are −76.7 ± 1.1 nm and 76.7 ± 1.1 nm, respectively. Three-color, 3D STORM imaging of several synaptic proteins along with Bassoon[N] and Homer1[C] was then performed in the MOB to position each protein (Figure 4). Here, we note that for the axial position analysis, only synapses with the transynaptic axes close to the imaging plane were considered to take advantage of the higher localization precision in the xy plane. We also placed the synaptic cleft in our coordinate system (Figure 4E) according to the known distance between Bassoon[N] and the presynaptic membrane (Siksou et al., 2007) and width of the cleft (Zuber et al., 2005).
Figure 4. Axial positions of synaptic proteins.
(A–D) Various synaptic proteins (blue) were imaged together with Bassoon[N] (red) and Homer1[C] (green). For each protein, the left panel shows a typical three-color image of a single synapse and the right panel displays the localization distribution along the trans-synaptic axis derived from multiple synapses. The images and axial position distributions of Homer1[N], Shank1, GluR1, and Piccolo[C] are shown here. The axial distributions of PSD-95, CamKII, NR2B, GABABR1, Piccolo[N], Bassoon[C] and RIM1 are shown in Figure S3.
(E) A composite plot of the axial positions of all synaptic proteins imaged in this work. For each protein, the colored dot specifies the mean axial position, the two vertical lines represent the associated SEM, and the half length of the horizontal bar denotes the SD, primarily arising from synapse-to-synapse variation.
To confirm that immuno-labeling did not add significant additional distance between molecules, we determined the average position of the N-terminus of Homer1 (Homer1[N]) in our synaptic coordinate system and compared it with that of Homer1[C]. It has been shown that Homer forms a tetramer with the dimeric coiled coils intercalating in a tail-to-tail fashion (Hayashi et al., 2009). This antiparallel structure predicts that average positions of the C and N termini of Homer1 would be nearly identical. Experimentally, using an antibody directed at the N-terminal region of Homer1, we found the mean position of Homer1[N] to be 80.4 ± 2.7 nm, which was indeed nearly identical to that of Homer1[C] (Figures 4A, 4E).
We then imaged several other major protein components of the PSD, including the scaffolding proteins, PSD-95 and Shank1, a signaling enzyme, the calcium/calmodulin-dependent protein kinase-II (CamKII), as well as subunits of the NMDA and AMPA receptors and the metabotropic GABA receptor. The average axial position of these proteins were found to be 44.0 ± 3.9 nm for PSD-95, 62.1 ± 2.8 nm for Shank1, 76.7 ± 4.8 nm for CamKII, 30.4 ± 5.3 nm for the C-terminal domain of the AMPA receptor subunit GluR1, 34.3 ± 3.1 nm for the C-terminal domain of the NMDA receptor subunit NR2B and −21.1nm ± 4.0 nm for the C-terminal domain of the GABAB receptor subunit-1 (GABABR1) (Figures 4 and S3). As expected, the neurotransmitter receptors were found in closest proximity to the synaptic cleft. The ~13 nm distance between the C-terminal domain of GluR1 and the postsynaptic membrane is in quantitative agreement with the previous estimate that the cytoplasmic domain of the AMPA receptors protrudes by 10 – 15 nm from the plasma membrane (Chen et al., 2008a; Nakagawa et al., 2005). The ~26 nm average distance between PSD-95 and the synaptic cleft is close to the 12 – 25 nm range previously determined by EM tomography and immuno-EM (Chen et al., 2008a; Petersen et al., 2003; Valtschanoff and Weinberg, 2001). The sequential distribution of the three scaffolding proteins, PSD-95, Homer1 and Shank1 with increasing distances from the synaptic cleft is also consistent with the laminar organization of postsynaptic proteins observed by EM (Valtschanoff and Weinberg, 2001). These results serve to validate our new imaging approach. Interestingly, STORM imaging of GABABR1 identified both pre- and postsynaptic localizations of this receptor, with a predominant presynaptic distribution. This presynaptic positioning may be related to the role of GABA receptors in reducing neurotransmitter release by inhibition of voltage-gated calcium channels (Ulrich and Bettler, 2007). Our data also revealed a broad distribution for CamKII, which resulted from the variation of CamKII positions from synapse to synapse. This broad variability in the CamKII position amongst synapses may be functionally significant given the dynamic role of CamKII in scaffolding and phosphorylation of a variety of molecules within the PSD (Bingol et al., 2010).
In addition to the neurotransmitter receptors and PSD components, we also determined the position of several presynaptic proteins (Figures 4 and S3). Piccolo, a scaffolding protein structurally related to Bassoon (Fenster et al., 2000), was found to have similar axial distribution as Bassoon. The N-terminal domain of Piccolo (Piccolo[N]) was located at −86.8 ± 3.8 nm, in close proximity to Bassoon[N]. RIM1, in contrast, was found to be much closer to the presynaptic membrane (−37.4 ± 4.0 nm), consistent with its role in synaptic vesicle release (Schoch et al., 2002).
Orientation of synaptic proteins
Next, we investigated the ability of the STORM-based approach to determine the orientation of synaptic proteins by taking advantage of epitopes located in different domains of the corresponding protein. We used this strategy to determine the orientation of the two large presynaptic scaffolding proteins, Bassoon and Piccolo. Three-color imaging of an antibody directed against the C-terminal region of Bassoon (Bassoon[C]) in the established Bassoon[N] - Homer1[C] reference frame revealed that Bassoon[C] was located on average at −46.5 ± 3.1 nm, about 30 nm closer to the synaptic cleft than the N terminus of Bassoon (Figure 4E). Similarly, the C-terminal region of Piccolo (Piccolo[C]) was located on average at −44.4 ± 3.6 nm, closer by more than 40 nm to the synaptic cleft than Piccolo[N] (Figure 4E). These data suggest that Bassoon and Piccolo are organized in a highly extended and oriented manner at the active zone. If these molecules had a random instead of an oriented distribution, or had any other type of central-symmetric distributions, the average positions of their N and C termini would have the same value. Indeed, for Homer1 which adopts a central-symmetric tetrameric structure (Hayashi et al., 2009), our measurements gave nearly identical Homer1[N] and Homer1[C] values with a difference of merely 3.7 nm, as described above.
Biologically, the novel finding that Bassoon and Piccolo are organized in an extended and oriented manner bears interesting functional implications for these multi-domain PAZ scaffolding proteins, which are thought to position distinct presynaptic components through interactions with their multiple domains (Jin and Garner, 2008; Schoch and Gundelfinger, 2006). The oriented organization of Bassoon and Piccolo may thus help place other presynaptic proteins in an orderly fashion. Interestingly, the Drosophila presynaptic scaffolding protein Bruchpilot, which is evolutionarily unrelated to Bassoon or Piccolo, has also been shown to exhibit an extended conformation at the neuromuscular junction, but in contrast to Bassoon and Piccolo, Bruchpilot places its N-terminal end closer to the synaptic membrane (Fouquet et al., 2009).
Technically, it is interesting to estimate a limit of the protein size for its oriented organization to be detectable. Several factors need to be considered. First, our single-molecule localization precision is ~14 nm in SD. Second, the antibody size (~15 nm) contributes additional error, which can be approximated as a Gaussian of ~15 nm in width (or equivalently ~6.5 nm in SD), though deviation may arise if the antibodies are not randomly oriented. The total convolved localization uncertainty is then ~17 nm in SD (estimated by convolving a Gaussian of 14 nm SD with two Gaussians of 6.5 nm SD each, representing the two layers of antibodies). Finally, the error is further reduced by determining the average position of an epitope in a synapse from many localizations. For example, at least 50 localizations in the center region of each synapse were used to estimate the average axial position of Homer1[C] or [N]; the standard error in the average position is thus reduced to ~17/√50 = 2.4 nm. Consistent this estimate, only a small difference of ~4 nm was observed for the positions of Homer1[C] and [N], which are predicted to be nearly identical to each other according to the tetrameric Homer structure (Hayashi et al, 2009). This estimate implies that the orientation effect of a protein less than ~5 nm in size would be difficult to detect. While this limit could be further reduced by averaging across many synapses, the existence of synapse-to-synapse variations could present additional challenges.
Radial (lateral) distribution of synaptic proteins within individual synapses
In addition to the axial position, characterization of the radial positions of synaptic proteins is also important for understanding their functional properties. For example, detailed knowledge of the lateral distribution of neurotransmitter receptors is of considerable interest, as changes in this distribution provides a mechanism to rapidly alter the transmission properties of synapses (Adesnik et al., 2005; Ehlers et al., 2007; Groc et al., 2006; Tovar and Westbrook, 2002). However, a consensus view of how neurotransmitter receptors are distributed laterally in individual synapses is still missing, in part because the radial distribution of proteins is particularly difficult to measure at the single synapse level. Due to the low labeling efficiency of immuno-gold, typical EM imaging requires averaging of a large ensemble of synapses to determine the radial distribution of a specific protein. Freeze fracture replica labeling, in combination with SDS denaturation, can achieve higher immuno-gold labeling density and has been used to examine receptor distributions at the single synapse level (Masugi-Tokita et al., 2007). However, as fracture occurs primarily along membranes, this approach can only be applied to characterize the distributions of membrane proteins. The unpredictable nature of membrane fracture and partitioning of antigenic epitopes presents additional complication (Masugi-Tokita et al, 2007). In contrast, the multicolor, 3D imaging capability of STORM allows the radial distributions of transmembrane and cytoplasmic synaptic proteins to be determined and those of distinct proteins to be compared within individual synapses. Here we focused on the distributions of the AMPA receptor subunit GluR1 and NMDA receptor subunit NR2B, and compared them with the distributions of PSD scaffolding proteins.
We performed three-color imaging of GluR1 and NR2B together with Homer1, and Homer1 structures that showed overlap with either GluR1 or NR2B or both were used for further analysis. We observed substantial synapse-to-synapse variation in the lateral distributions of these receptor subunits in the AOB. In some synapses, NR2B and/or GluR1 were predominantly located within the PSD, as defined by the distribution of Homer1 (Figure 5A, upper panels), whereas in other synapses, the receptors were primarily distributed at the periphery of the PSD (Figure 5A, lower panels). To quantify the lateral distribution of the receptors within a synapse, we renormalized the coordinates of each receptor localization point in the plane of synaptic cleft according to the long and short axes of the PSD defined by the Homer1 distribution, and determined the normalized radial position r of the receptor molecule. The mean radial position R of all molecules of the same type within a synapse was then used as a simple measure to allow quantitative comparison between different molecules and across synapses. About half of the synapses exhibited R values for NR2B and/or GluR1 comparable to or smaller than that of Homer1, i.e. _R_receptor ≤ _R_Homer1 + 2×SD, whereas the other half exhibited _R_receptor > _R_Homer1 + 2×SD, where SD is the standard deviation of the _R_Homer1 distribution (Figure 5B). The radial distributions of the receptor subunits also show clear distinction between these two populations of synapses: the first population exhibited an overall receptor distribution nearly identical to that of the PSD scaffolding proteins Homer1 and Shank1, whereas the second population displayed a substantially broader distribution extending into the perisynaptic region (Figure 5C).
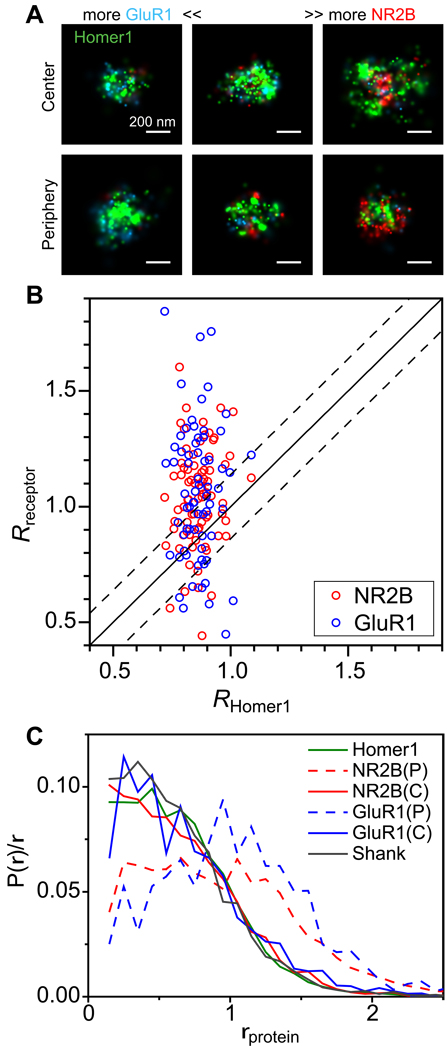
Figure 5. Radial (lateral) distribution of glutamate neurotransmitter receptors at synapses.
AMPA receptor subunit GluR1 and NMDA receptor subunit NR2B were imaged together with Homer1.
(A) “Face” views of six example synapses with the top row representing a population of synapses in which the receptor subunits take a more central distribution within the PSD and the bottom row representing a population of synapses with a more peripheral receptor distribution near the edge of the PSD. The left to right columns show synapses with mostly GluR1, both GluR1 and NR2B, and mostly NR2B, respectively.
(B) Comparison of the mean radial position R of NR2B and GluR1 in individual synapses with the R value of Homer1. The solid line is the y = x line and the dashed lines are the ± 2×SD boundary calculated from the standard deviation of _R_Homer1.
(C) Radial distribution of receptors of the two populations of synapses divided according to the upper 2×SD boundary of _R_Homer1. We refer to these two populations as the central (C) population (_R_receptor ≤ _R_Homer1 + 2×SD) and the peripheral (P) populations (_R_receptor > _R_Homer1 + 2×SD). The radial distributions of NR2B (red lines) and GluR1 (blue lines) are displayed together with those of Homer1 (green) and Shank1 (black).
The above results unambiguously demonstrate that, at the individual synapse level, both NMDA and AMPA receptors can take central or perisynaptic distribution. In fact, the division into central and perisynaptic distributions was only a coarse-grained partition. The lateral distribution of receptors showed a broad heterogeneity from synapses to synapse, with a wide variety of distributions as evidenced by the broad and continuous distribution of the receptor R value for individual synapses (Figure 5B). STORM analysis of individual synapses helps to reconcile the discrepancy found in previous descriptions of lateral receptor distributions (Antal et al., 2008; Kharazia and Weinberg, 1997; Somogyi et al., 1998), as we observed all of the previously described distribution types (central, uniform, or perisynaptic) in subgroups of synapses. Different findings reported in various studies may also in part arise from the investigation of different brain regions and receptor subunits, and future super-resolution imaging of all glutamate receptor subunits in different brain regions should help provide a complete picture on this issue.
Neurotransmitter receptor composition and plasticity within individual synapses
Finally, we demonstrate that the STORM-based super-resolution imaging approach helps provide quantitative information on the composition of receptors at individual synapses, which is a key determinant of synaptic strength and plasticity. This composition analysis takes direct advantage of the single-molecule imaging nature of STORM and derives the number of molecules from the localization points of individual molecules. To illustrate this strategy, we performed STORM imaging of NR2B and GluR1 simultaneously with Homer1 and compared the receptor composition in two brain regions, the AOB and MOB. Considerable heterogeneity in the receptor composition was observed amongst individual synapses. In both regions, subsets of synapses showed predominant GluR1 staining (Figure 5A, left panels), or predominant NR2B staining (Figure 5A, right panels), while some synapses showed significant amount of both receptor subunits (Figure 5A, middle panels).
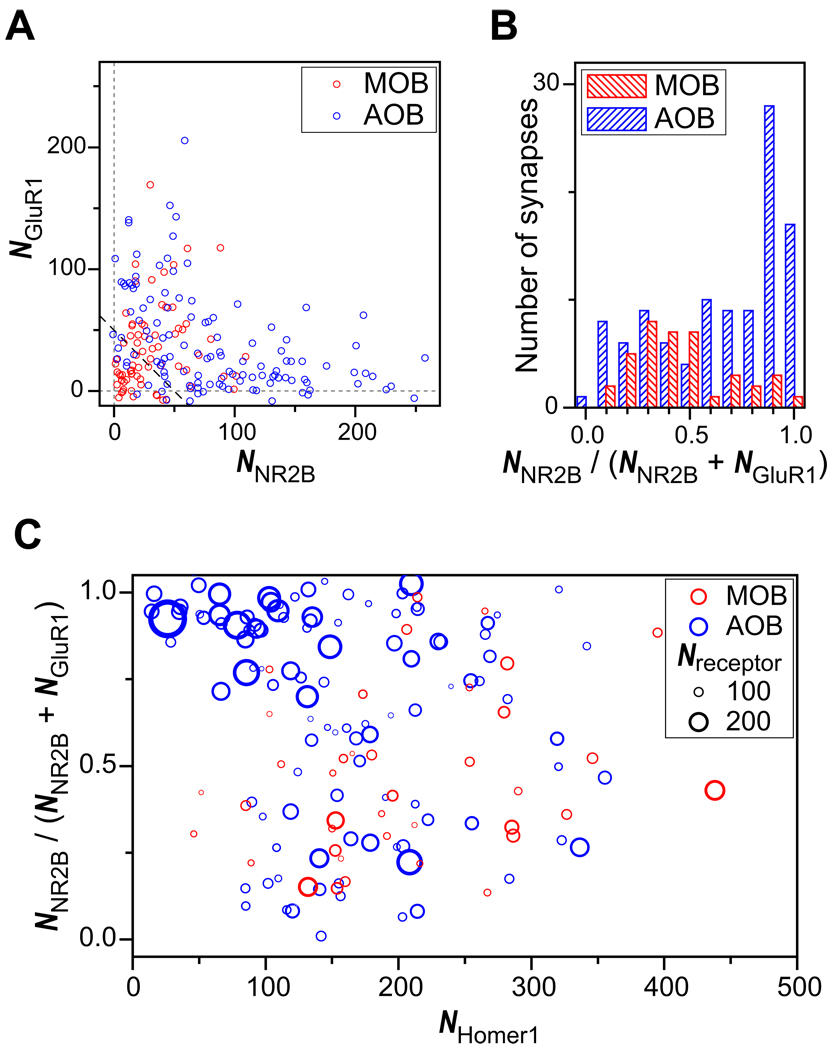
To characterize the synapse-to-synapse variability in receptor composition, we quantified the number of NR2B and GluR1 localization points (_N_NR2B and _N_GluR1) per synapse. We note that _N_NR2B and _N_GluR1 in each synapse are not exactly equal to the number of receptors for two reasons: (1) antibody-epitope binding is not necessarily 100% efficient; (2) each antibody molecule can yield more than one localization points. However, the epitope binding efficiency of a given antibody should be comparable across synapses. Moreover, we labeled all antibody types with similar dye composition and further adjusted activation laser intensities to obtain on average identical numbers of localization points per NR2B and GluR1 antibodies. Therefore, _N_NR2B and _N_GluR1 should scale with the number of receptor molecules and can be used as an indication of relative receptor abundance across synapses. Our analysis showed that while synapses in both MOB and AOB regions displayed heterogeneous NR2B-to-GluR1 ratio, the heterogeneity appeared much larger in the AOB. This latter region contained a large fraction of synapses with a large _N_NR2B value but a very small _N_GluR1 value (Figure 6A, B). The synaptic content of NR2B and GluR1 appeared anti-correlated in the AOB, but not in the MOB (Figure 6A). Interestingly, an anti-correlation was also observed between NR2B and Homer1 in AOB, but not in MOB (Figure S4A), whereas GluR1 and Homer1 showed positive correlations in both regions (Figure S4B). As a consequence, most of the AOB synapses with very large NR2B-to-GluR1 ratios also contain low amounts of Homer1, likely representing “immature” synapses, while such synapses were nearly absent in the MOB (Figure 6B, C). Notably, the content of different scaffolding proteins (e.g. Bassoon, Homer1 and Shank1) all appeared to be positively correlated in the AOB and MOB in a similar manner (Figure S4C, D). Thus, the anti-correlations observed between NR2B and GluR1 and between NR2B and Homer1in the AOB, but not in the MOB, as well as the differences in the NR2B-to-GluR1 ratio distributions observed between AOB and MOB synapses were unlikely to result from experimental artifacts.
Figure 6. Composition of glutamate neurotransmitter receptors at synapses.
(A) Number of NR2B localization points (_N_NR2B) versus number of GluR1 localization points (_N_GluR1) for each synapse in the AOB and MOB. The correlation coefficient between _N_NR2B and _N_GluR1 is −0.42 in the AOB and 0.23 in the MOB.
(B) The histograms of NR2B fraction (_N_NR2B / (_N_NR2B + _N_GluR1)) for synapses in the MOB and AOB regions.
(C) A scatter plot of the NR2B fraction in relation with the number of Homer1 localization points (_N_Homer1) for each synapse in the AOB and MOB. The circle diameters are proportional to the total number of receptor localizations.
Structures with the total number of receptor localization points smaller than 50 (data points below the dashed line in (A)) are not included in (B) and (C), as a low total number of receptor localizations introduces a large error in the _N_NR2B / (_N_NR2B + _N_GluR1) calculation.
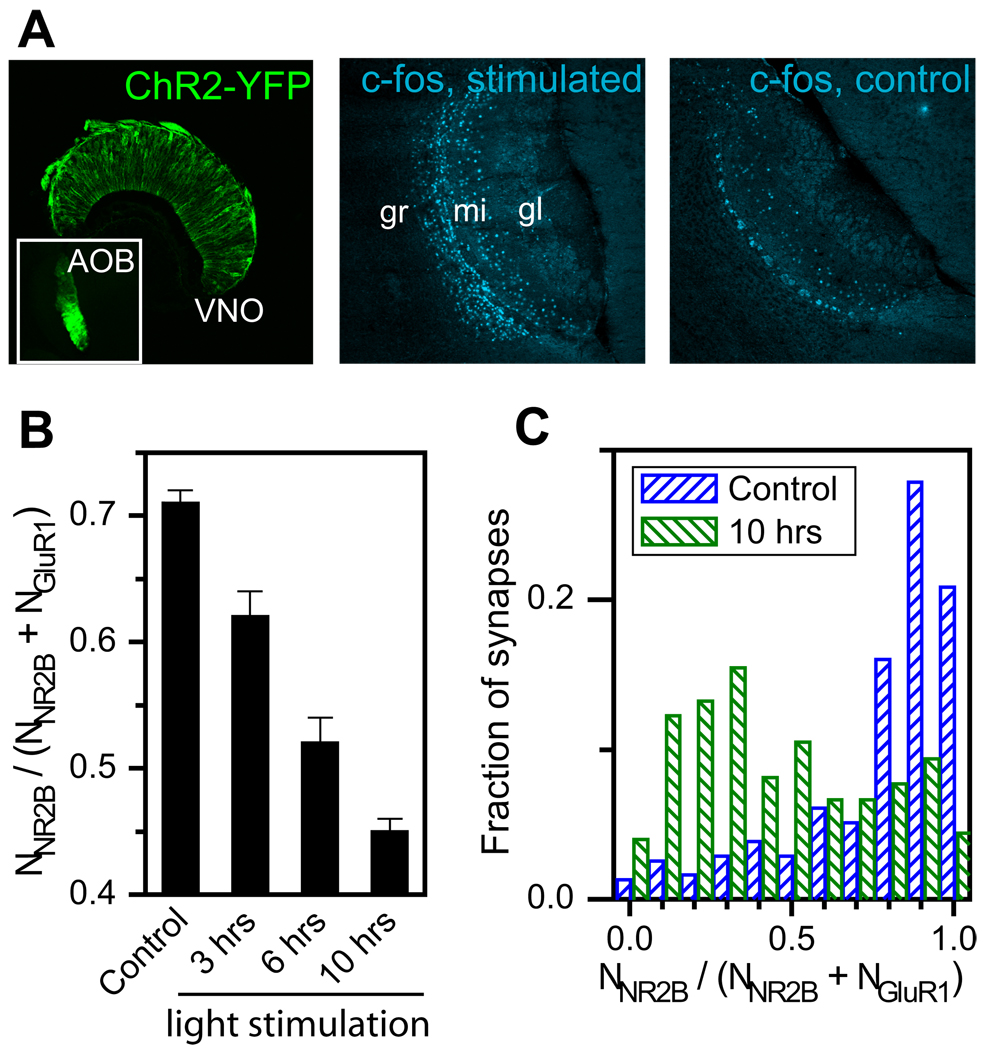
We then tested whether the receptor composition in AOB synapses could be modulated by neural activity using an optogenetic approach (Boyden et al., 2005). Considering that the mitral cells in the AOB receive synaptic inputs from sensory neurons in the vomeronasal organ (VNO), we took advantage of a transgenic mouse line that specifically expresses channelrhodopsin-2 fused with yellow fluorescent protein (ChR2-YFP) in the VNO neurons (Figure 7A, left panel). Stimulation of the VNO neurons in the ChR2-YFP expressing mice with blue light caused induction of the immediate early gene, c-fos, within the mitral and granule cells of the AOB (Figure 7A, middle panel), whereas identical light stimulation of control ChR2-YFP null mice did not show such c-fos induction (Figure 7A, right panel), suggesting specific activation of the ChR2-positive VNO neurons. Robust activation of the VNO neurons by light was further confirmed by electrophysiological recording in the AOB (Figure S5). Using this ChR2-YFP mouse line as a tool, we analyzed the response of AOB synapses to increased neuronal activity in the VNO. Statistics on the STORM images of individual AOB synapses showed a significant increase in the synaptic content of GluR1 and Homer1 and a moderate decrease in NR2B upon light stimulation. Ten hours after stimulation, the average value of _N_GluR1 derived from many synapses increased by 2 fold, the average value of _N_Homer1 increased by 50%, and that of _N_NR2B decreased by 25%. The average NR2B-to-GluR1 ratio decreased consistent over time after stimulation (Figure 7B). Analysis of synapse-to-synapse variations further showed that the fraction of synapses with very large NR2B-to-GluR1 ratios was reduced substantially (Figure 7C). Taken together, these data suggest a significant synapse maturation effect due to the VNO stimulation. This shift was not observed in control ChR2-YFP mice that did not receive light stimulation. These results suggest that the first order VNO sensory synapses in the AOB are capable of rapid transformation in their molecular properties in response to neuronal activity.
Figure 7. Changes in the receptor composition of AOB synapses in response to VNO stimulation.
(A) Light-activation of VNO neurons expressing ChR2-YFP in ORC-V mice. (left panel) YFP image of the VNO neurons. Inset, YFP image of the AOB showing ChR2-YFP expression in the glomerular region. (Middle panel) Immunofluorescence image of c-fos in the AOB two hours after light stimulation. gr: granule cell; mi: mitral cell; gl: glomerular layers. (right panel) c-fos image in the AOB of a control mouse that was similarly light stimulated but does not express ChR2-YFP in the VNO.
(B) Average value of _N_NR2B / (_N_NR2B + _N_GluR1) derived from many AOB synapses sampled at various times after light stimulation of the ChR2-YFP expressing VNO neurons in ORC-V mice and in control ChR2-YFP expressing ORC-V littermates without light stimulation.
(C) Distribution of _N_NR2B / (_N_NR2B + _N_GluR1) constructed from individual AOB synapse in ChR2-YFP expressing mice 10 hours after light stimulation and in control ChR2-YFP expressing mice without light stimulation.
The presence of such a large fraction of NR2B-rich and GluR1-poor synapses with substantial activity-dependent plasticity in the adult AOB is highly surprising. It is widely recognized that, at birth, most central synapses are strongly NMDA-dependent and that early postnatal maturation coincides with a major shift in synapse receptor composition, with substantial increase in AMPA receptor-mediated transmission (Malenka and Bear, 2004; Malinow and Malenka, 2002). Maturation of the NMDA receptor-dominant synapses by gaining fast transmitting AMPA receptors provides a postsynaptic paradigm for long-term potentiation during postnatal development (Kerchner and Nicoll, 2008). Such NMDA receptor-dominant synapses have been found in early postnatal visual, somatosensory, and olfactory cortical areas, and are thought to play a key role in experience-dependant modifications of synaptic inputs during critical periods of sensory development (Franks and Isaacson, 2005; Isaac et al., 1997; Rumpel et al., 1998). In the olfactory cortex, this mechanism has been proposed to mediate early olfactory experience-dependent plasticity leading to the behavioral “imprinting” of pups to maternal odors (Franks and Isaacson, 2005). While our observation that most adult MOB synapses exhibit a large amount of GluR1 is consistent with the postnatal switch to AMPA-receptor enriched synapses, the observed prevalence of NR2B-rich/GluR1-poor synapses with substantial plasticity in the adult AOB is extremely intriguing, especially considering that the AOB is thought to relay chemosensory input from the VNO to higher order brain centers involved in innate behavioral responses (Dulac and Torello, 2003). Despite existing electrophysiology and imaging studies of synapses in the AOB region (Castro et al., 2007; Urban and Castro, 2005), the prevalence of such synapses had not been observed previously. While future studies are needed to further elucidate the composition and function of these synapses in AOB circuits, their discovery demonstrates the ability of the STORM-based super-resolution imaging approach to reveal novel properties of synapses and circuits.
DISCUSSION
A chemical synapse is a complex structure composed of thousands of proteins working in concert to mediate neurotransmitter-based inter-neuron communication. Mechanistic and structural insights into this basic functional unit of the brain are likely to have far reaching consequences for the understanding of normal and pathological neuronal circuits. Because of its small size, the characterization of the synaptic ultrastructure has primarily relied on EM, which offers exquisite spatial resolution but has limitations in multi-component 3D imaging. Fluorescence microscopy overcomes these limitations, but its diffraction-limited resolution prohibits accurate position analysis of sub-synaptic components.
In this work, we established a super-resolution fluorescence imaging approach to determine the protein architecture of brain sub-cellular structures, such as the excitatory chemical synapses. The sub-diffraction-limit resolution offered by STORM, together with the multicolor and 3D imaging capabilities, allowed us to resolve the molecular organization of the presynaptic active zone and postsynatic density in dense scattering brain tissues with nanometer-scale precision. The relatively fast volumetric imaging process further allowed recording of large sets high-resolution images of individual synapses to facilitate comparative analysis amongst synapses and across different brain regions. Using this approach, we determined the organization of ten pre-and postsynaptic proteins. On the presynaptic side, we observed a novel, highly oriented organization of the scaffolding proteins, Bassoon and Piccolo. On the postsynaptic side, a laminar organization of PSD proteins and substantial synapse-to-synapse variability in the lateral distributions of neurotransmitter receptors were observed. Moreover, quantitative receptor composition analysis led to the discovery of a surprisingly large fraction of immature synapses with activity-dependent plasticity in the adult AOB that conventional imaging and electrophysiological strategies had so far failed to identify.
The rapid multicolor and 3D imaging, combined with the use of standard immunohistochemical labeling of synapses, should allow a straightforward extension of this approach to a larger list of synaptic proteins, synaptic types and ultimately a full characterization of the molecular architecture of synapses. By exploiting transgenic animals expressing photoswitchable fluorescent proteins fused to synaptic components, we expect this super-resolution imaging approach to further provide a dynamic picture of synapses and additional insights into synaptic function and plasticity. In addition to STORM, super-resolution STED microscopy has also been used for imaging brain structures. These approaches have distinct advantages and limitations. When applied to imaging a small area, STED has achieved impressive video-rate imaging, and thus is particularly powerful for probing fast dynamics in small brain structures, such as a dendritic spine (Nagerl et al., 2008; Westphal et al., 2008). However, as a point-scanning approach, the image acquisition time of STED increases linearly with the sample volume, resulting in relatively low time resolution for large volume imaging. The large spectral coverage involved in the STED scheme has also limited its multi-color imaging capability and to date, only two-color imaging has been reported. On the other hand, the single-molecule-localization based STORM/(F)PALM is a wide-field imaging method that does not require sample scanning for even 3D imaging. This allows a large volume to be imaged in a relatively short amount of time and enables high-throughput analysis, though the imaging speed of STORM does not increase with decreasing sample volume as rapidly as STED. Furthermore, the multi-color imaging capability of STORM greatly facilitates multi-component analysis and allows the molecular architecture of complex structures to be constructed with a systematic three-color imaging strategy demonstrated in this work. Together, super-resolution fluorescence approaches provide powerful options for imaging specific protein structures in the brain with high resolution. However, unlike EM, they do not provide intrinsic contrast of membranes, which is particularly powerful for indentifying structures such as the synaptic cleft, vesicles, and other organelles near the synaptic active zone. Future development of membrane markers for super-resolution imaging and correlative EM / fluorescence microscopy approaches will help provide an even more complete ultrastructural characterization of synapses and other brain sub-cellular structures.
EXPERIMENTAL PROCEDURES
Immunohistochemistry
Two different types of mouse brain tissue sections, thick vibratome sections (~70 µm) and thin cryostat sections (~10 µm), were initially tested, yielding no appreciable difference in the observed synapse ultrastructure. For all experiments reported here, we used cryosections to provide a higher yield of sections and more consistent antibody penetration. Brains from adult C57Bl/6 mice were dissected out and freshly frozen by embedding into tissue-tek OCT medium and placing tissue blocks on dry ice. Sections of 10–12 µm thickness were cut on a cryostat and collected on glass slides. Sections were fixed for 10 minutes with 4% Paraformaldehyde (PFA, Electron Microscopy Sciences), followed by washing off excess PFA and quenching with 20mM Glycine in phosphate buffered saline (DPBS), pH 7.4. Sections were then blocked and permeabilized using 3% BSA in DPBS with 0.1% Triton X-100, followed by incubation with primary antibody (3 hours at room temperature or overnight at 4°C) and secondary antibodies (2 hours at room temperature), and finally fixed for 5 minutes in 3% PFA with 0.05% glutaraldehyde after antibody labeling. Secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA) and labeled with photoswitchable dye pairs as described before (Bates et al., 2007), with 2 activator dyes and 0.6–0.7 reporter dyes per antibody molecule on average. In the case where two monoclonal mouse primary antibodies for different synaptic proteins were used simultaneously, isotype specific secondary antibodies were used to distinguish the primary antibodies. Table S1 shows the primary antibodies used in this work.
STORM setup
The STORM microscope was constructed around an Olympus IX-71 inverted microscope operating in wide-field epi-fluorescence mode. Four activation/imaging lasers (405 nm from CUBE 405-50C, Coherent, 460 nm from Sapphire 460-10, Coherent, 532 nm from GCL-200-L, Crystalaser, and 657 nm from RCL-200-656, Crystalaser) were individually shuttered and then coupled into a single-mode optical fiber (OZ Optics). The fiber output was collimated and focused to the back focal plane of the oil immersion objective (100× UPlanSApo NA 1.4, Olympus) on the microscope. A dichroic mirror (T660LPXR, Chroma) and a band pass filter (ET705/70m, Chroma) separates the fluorescence signal collected by the same objective from the excitation light. A Roper Scientific DualView system were inserted between the microscope and the camera as the image relay, of which the splitting cassette was replaced with a cylindrical lens (focal length = 1 m) to create the astigmatism for 3D imaging. The images were recorded with an EMCCD camera (Ixon DV897DCS-BV, Andor). To stabilize the focusing of the microscope, a near infra-red laser (850 nm) was send into the edge of the microscope objective back aperture, and its reflection from the glass/sample interface was imaged onto a quadrant photodiode. The quadrant photodiode output drives a piezo objective positioner (Nano F-100, Madcity Labs) to compensate for the sample drift in the z-direction during the experiment. All instrument control, data acquisition and data analysis was performed using custom written software.
STORM image acquisition
Prior to STORM imaging, a low magnification fluorescence image (using a 10× objective) was acquired to identify the region of interest. After switching to the 100× objective, conventional fluorescence images for different color channels were first acquired. STORM data acquisition was then started with imaging cycles containing one frame of activation laser illumination (405 nm, 460 nm or 532 nm) followed by three frames of imaging laser illumination (657 nm) at 60 frames per second. The power of the activation lasers were typically 1–5 µW entering the back port of the microscope and that of the imaging laser was 35 mW. To reduce the out-of-focus fluorescence background, samples were first illuminated with the imaging/deactivation laser at a low incidence angle to deactivate fluorophores above and below the focal plane, then a highly oblique incidence geometry with incidence angle only slightly smaller than the critical angle was used for activation and excitation, restricting illumination to a 2–3 µm depth into the tissue sample. Typically, one STORM image acquired in several minutes covers an imaging volume of 30 µm × 30 µm × 650 nm without the need of sample scanning.
For STORM experiments, all sections were mounted in imaging buffer made by mixing the following four solutions with a volume ratio of 80:10:10:1 immediately before applying to the sections: DPBS, 1 M mercaptoethylamine with pH adjusted to 8.5 using HCl, 50% glucose solution in water, and an anti-beaching oxygen scavenger system (10 mg of glucose oxidase + 25 µL of catalase and 100 µL of DPBS, mix well and centrifuge for 1 min). Clean coverglass was placed on top of the sections and excess imaging buffer was removed followed by sealing the edges with nail polish.
Multicolor 3D imaging and crosstalk subtraction
Photoswitchable dye pairs, Cy3-A647, Cy2-A647, and A405-A647, were used for multicolor STORM imaging. The different probes are distinguished by the wavelength of light used to activate them (Figure S1A). During the imaging process, the corresponding activation laser (405 nm for A405, 460 nm for Cy2, and 532 nm for Cy3) was used to activate a small fraction of the A647 reporters at a time, and individual activated fluorophores were imaged and localized with a 657 nm laser. Specifically, in a three-color imaging experiment, the illumination sequence is 532 nm × 1 (1 frame of 532 nm laser), 657 nm × 3, 460 nm × 1, 657 nm × 3, 405 nm × 1, and finally 657 nm × 3 before repeating. Photoactivation events detected in the first imaging frames following the 532 nm, 460 nm and 405 nm activation frames were assigned to the Cy3-A647, Cy2-A647, and A405-A647 pairs, respectively. To determine the 3D position of the activated probes, a cylindrical lens was inserted into the imaging path to render the image of individual molecules in an elliptical shape. The image from each activated molecule was then fit to an elliptical Gaussian function, the centroid position of which was assigned as the x and y coordinates of the molecule and the ellipticity of which was used to determine the z coordinate (Huang et al., 2008b). The numerous localizations of the molecules determined in this manner were plotted according to their coordinates and color to construct the multi-color 3D image.
In this multicolor imaging scheme, the crosstalk between color channels mainly comes from nonspecific activation of the A647 reporter by the 657 nm imaging laser in an activator-independent way. In addition, the 460 nm activation laser also leads to small cross activation of the Cy3-A647 pair, albeit 10 times less efficient than by the 532 nm laser (Bates et al., 2007). Here we present a method to determine the nonspecific activation in situ and subtract color crosstalk using an automated algorithm. In this approach, each activation laser pulse was followed by three imaging frames where the 657 nm imaging laser was on, as described above (Figure S1B). We define the numbers of specific activation events contained in the imaging frames immediately following the 405 nm, 460 nm and 532 nm activation pulses as _N_A405, _N_Cy2 and _N_Cy3, respectively, and the number of nonspecific activation events contained in these frames as _n_0. Thus, the total numbers of activation events in these frames are given by (Figure S1B):
nA405=NA405+n0nCy2=NCy2+n0+I46010I532NCy3nCy3=NCy3+n0
where _I_460 and _I_532 are the intensity of the 460 nm and 532 nm lasers at the sample, respectively. Since the probability of nonspecific activation due to the imaging laser should remain constant across frames regardless of whether the frame is preceded by an activation pulse, we can determine the value of _n_0 from the frames not immediately following the activation pulses. Note that if a fluorophore remained fluorescent for more than one consecutive frames, it is considered as an activation event only in the imaging frame during which it initially appeared fluorescent. The number of specific activation events _N_A405, _N_Cy2 and _N_Cy3 can then be solved from the above equations using the measured _n_A405, _n_Cy2, _n_Cy3, and _n_0 values. To be statistically sound, the values of _n_A405, _n_Cy2, _n_Cy3, and _n_0 were determined from the sums of all imaging cycles. These numbers then allowed us to statistically subtract crosstalk in the reconstructed image: for each localization point in a specific color channel A of the raw image, we determined the “local density” of _n_A and _N_A within a certain radius of the localization (typically 50 nm) using the above equations and then removed this localization with a probability of P = (_n_A – _N_A) / _n_A. Independent check on isolated probes, which should appear as a single-color object, showed that this cross-talk subtraction method can reduce the crosstalk to 1–3% between channels.
Quantification of the synapse size and the axial and radial positions of synaptic proteins
The image of each excitatory synapse, as identified by a juxtaposed Bassoon-Homer1 pairs, was rotated in 3D such that its trans-synaptic axis, and the long and short axes of the Bassoon-Homer1 disks were aligned along the z, x, and y axes, respectively. The axial distance between the Bassoon and Homer1 clusters, and the axial position of the other synaptic proteins, were calculated as described in the main text. Because some synapses were curved, we only used localization points in a narrow window about 160 nm wide in the direction of the long axis to construct the localization histograms (e.g. region between the dashed lines in Figure 3D), so that synaptic curvature is insignificant within this window. To measure the area of a synapse, a one-dimensional histogram was built for each synapse by projecting its localization points to its long axis, and a second histogram built along its short axis. The dimensions of a synapse along its long and short axes were then estimated by fitting the two histograms with a rectangular function convolved with a Gaussian function representing the localization precision. The area of the synapse was then estimated from these two lengths defined by the rectangular functions, assuming an elliptical shape.
To analyze the radial distribution of receptors, the entire three-color image of NR2B and GluR1 with Homer1 was used. For any localization point at the position (x, y) in the NR2B, GluR1 or Homer1 channel, its normalized radial position r was defined as r=[(x−x0)/Rx]2+[(y−y0)/Ry]2, where Rx and Ry are the radii of the Homer1 distribution along the long and short axes calculated as described above and (_x_0, _y_0) is the center position of the Homer1 distribution. The radial distribution of r for NR2B, GluR1 and Homer1 in each synapse was the calculated. The mean value of r, defined to be R, was computed for each synapse from the radial distribution. Considering the relatively small distances in the axial dimension of synapses, we used synapses with trans-synaptic axis close to the imaging plan for analysis of the axial positions, so as to take advantage of the better image resolution in the xy plane. For the analyses of the active-zone area, radial distribution of receptors, and receptor composition, synapses with trans-synaptic axis in all orientations were considered.
Vomeronasal stimulation of mice expressing ChR2-YFP in the VNO neurons
ORC-V mice with ChR2-YFP expressed in the VNO neurons (Dhawale et al., 2010) were used. To probe the change in receptor composition upon vomeronasal stimulation, 3 weeks old ORC-V mice were anesthetized by injection of a mixture of ketamine and xylazine, and then stimulated by implanting a blue LED light (Philips luxeon rebel blue) in the oral cavity pointing dorsally towards the VNO. Light stimulation was performed using a train of 5 light pulses (100 Hz; 50% duty cycle) occurring every 2 seconds, for a total duration of 40 minutes. As a negative control, ORC-V littermates were anesthetized but left unstimulated for the same duration. Animals were then allowed to recover from the effects of anesthesia and were euthanized at different times post stimulation to examine the effects of the stimulation on the AOB.
To detect the expression of the immediate early gene, c-fos, upon stimulation, 8 weeks old ORC-V mice were stimulated and euthanized after 2 hours. As a negative control, C57Bl/6 mice that do not express ChR2-YFP were treated in a similar manner and stimulated in parallel. To detect activation with electrophysiological recording, 10 weeks old ORC-V mice were anesthetized with a mixture of ketamine and xylazine, and maintained on 1–2% isoflurane in oxygen. Light stimulation was performed with 50 ms duration every 1 second inter-stimulus interval. A small craniotomy was opened dorsal to the AOB. Multichannel electrophysiological recordings were targeted to the AOB based on stereotaxic coordinates using 4 shank silicon probes (Neuronexus; A4x8-5mm 100-200-177). Full electrophysiological waveforms were continuously sampled at 25 kHz using custom software (Ben-Shaul et al., 2010) to allow offline spike sorting analysis using Klusters (Hazan et al., 2006). Transthoracic electrocardiographic signals were monitored to ensure stable anesthesia. Electrophysiological probes were treated with DiI prior to experiments to allow visualization of the probe position, and accurate electrode targeting was verified using histological electrode tract tracing.
Supplementary Material
01
02
Acknowledgments
We thank Prof. Venkatesh Murthy for helpful discussions and for providing the ORC-V mouse line. This work was in part supported by the Howard Hughes Medical Institute (C.D. and X.Z.), NIH NIDCD (C.D.), and Gatsby Foundation (for X.Z.). X.Z. and C.D. are Howard Hughes Medical Institute investigators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include five supplemental figures, one supplemental table, and one supplemental movie.
References
- Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. doi: 10.1016/j.neuron.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Antal M, et al. Numbers, densities, and colocalization of AMPA- and NMDA-type glutamate receptors at individual synapses in the superficial spinal dorsal horn of rats. J Neurosci. 2008;28:9692–9701. doi: 10.1523/JNEUROSCI.1551-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur CP, Serrell DB, Pagratis M, Potter DL, Finch DS, Stowell MH. Electron tomographic methods for studying the chemical synapse. Methods Cell Biol. 2007;79:241–257. doi: 10.1016/S0091-679X(06)79010-0. [DOI] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang XW. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shaul Y, Katz LC, Mooney R, Dulac C. In vivo vomeronasal stimulation reveals sensory encoding of conspecific and allospecific cues by the mouse accessory olfactory bulb. Proc Natl Acad Sci U S A. 2010;107:5172–5177. doi: 10.1073/pnas.0915147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Castro JB, Hovis KR, Urban NN. Recurrent dendrodendritic inhibition of accessory olfactory bulb mitral cells requires activation of group I metabotropic glutamate receptors. J Neurosci. 2007;27:5664–5671. doi: 10.1523/JNEUROSCI.0613-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. Organization of the core structure of the postsynaptic density. Proc Natl Acad Sci U S A. 2008a;105:4453–4458. doi: 10.1073/pnas.0800897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Winters CA, Reese TS. Life inside a thin section: tomography. J Neurosci. 2008b;28:9321–9327. doi: 10.1523/JNEUROSCI.2992-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97 Suppl 1:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, Albeanu DF. Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse. Nature Neurosci. doi: 10.1038/nn.2673. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JB, Takasaki KT, Sabatini BL. Supraresolution imaging in brain slices using stimulated-emission depletion two-photon laser scanning microscopy. Neuron. 2009;63:429–437. doi: 10.1016/j.neuron.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- Ehlers MD, Heine M, Groc L, Lee MC, Choquet D. Diffusional trapping of GluR1 AMPA receptors by input-specific synaptic activity. Neuron. 2007;54:447–460. doi: 10.1016/j.neuron.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster SD, et al. Piccolo, a presynaptic zinc finger protein structurally related to bassoon. Neuron. 2000;25:203–214. doi: 10.1016/s0896-6273(00)80883-1. [DOI] [PubMed] [Google Scholar]
- Fernandez E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet W, et al. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks KM, Isaacson JS. Synapse-specific downregulation of NMDA receptors by early experience: a critical period for plasticity of sensory input to olfactory cortex. Neuron. 2005;47:101–114. doi: 10.1016/j.neuron.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-Molecule Discrimination of Discrete Perisynaptic and Distributed Sites of Actin Filament Assembly within Dendritic Spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groc L, et al. NMDA receptor surface mobility depends on NR2A-2B subunits. Proc Natl Acad Sci U S A. 2006;103:18769–18774. doi: 10.1073/pnas.0605238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, et al. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan L, Zugaro M, Buzsaki G. Klusters, NeuroScope, NDManager: a free software suite for neurophysiological data processing and visualization. J Neurosci Methods. 2006;155:207–216. doi: 10.1016/j.jneumeth.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Heintzmann R, Gustafsson MGL. Subdiffraction resolution in continuous samples. Nature Photonics. 2009;3:362–364. [Google Scholar]
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical Journal. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008a;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang WQ, Bates M, Zhuang XW. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008b;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac JT, Crair MC, Nicoll RA, Malenka RC. Silent synapses during development of thalamocortical inputs. Neuron. 1997;18:269–280. doi: 10.1016/s0896-6273(00)80267-6. [DOI] [PubMed] [Google Scholar]
- Jin Y, Garner CC. Molecular mechanisms of presynaptic differentiation. Annu Rev Cell Dev Biol. 2008;24:237–262. doi: 10.1146/annurev.cellbio.23.090506.123417. [DOI] [PubMed] [Google Scholar]
- Kerchner GA, Nicoll RA. Silent synapses and the emergence of a postsynaptic mechanism for LTP. Nat Rev Neurosci. 2008;9:813–825. doi: 10.1038/nrn2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharazia VN, Weinberg RJ. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci Lett. 1997;238:41–44. doi: 10.1016/s0304-3940(97)00846-x. [DOI] [PubMed] [Google Scholar]
- Kittel RJ, et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Klar TA, Hell SW. Subdiffraction resolution in far-field fluorescence microscopy. Opt Lett. 1999;24:954–956. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Masugi-Tokita M, Tarusawa E, Watanabe M, Molnar E, Fujimoto K, Shigemoto R. Number and density of AMPA receptors in individual synapses in the rat cerebellum as revealed by SDS-digested freeze-fracture replica labeling. J Neurosci. 2007;27:2135–2144. doi: 10.1523/JNEUROSCI.2861-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AC, et al. Tuning of synapse number, structure and function in the cochlea. Nat Neurosci. 2009;12:444–453. doi: 10.1038/nn.2293. [DOI] [PubMed] [Google Scholar]
- Nagerl UV, Willig KI, Hein B, Hell SW, Bonhoeffer T. Live-cell imaging of dendritic spines by STED microscopy. Proc Natl Acad Sci U S A. 2008;105:18982–18987. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Okabe S. Molecular anatomy of the postsynaptic density. Mol Cell Neurosci. 2007;34:503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Peters A, Palay SL, Webster Hd. The fine structure of the nervous system : neurons and their supporting cells. New York: Oxford University Press; 1991. [Google Scholar]
- Petersen JD, Chen X, Vinade L, Dosemeci A, Lisman JE, Reese TS. Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J Neurosci. 2003;23:11270–11278. doi: 10.1523/JNEUROSCI.23-35-11270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpel S, Hatt H, Gottmann K. Silent synapses in the developing rat visual cortex: evidence for postsynaptic expression of synaptic plasticity. J Neurosci. 1998;18:8863–8874. doi: 10.1523/JNEUROSCI.18-21-08863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, et al. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- Schoch S, Gundelfinger ED. Molecular organization of the presynaptic active zone. Cell Tissue Res. 2006;326:379–391. doi: 10.1007/s00441-006-0244-y. [DOI] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. The synaptic organization of the brain. 5th edn. Oxford ; New York: Oxford University Press; 2004. [Google Scholar]
- Siksou L, et al. Three-dimensional architecture of presynaptic terminal cytomatrix. J Neurosci. 2007;27:6868–6877. doi: 10.1523/JNEUROSCI.1773-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Tamas G, Lujan R, Buhl EH. Salient features of synaptic organisation in the cerebral cortex. Brain Res Brain Res Rev. 1998;26:113–135. doi: 10.1016/s0165-0173(97)00061-1. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. Mobile NMDA receptors at hippocampal synapses. Neuron. 2002;34:255–264. doi: 10.1016/s0896-6273(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Ulrich D, Bettler B. GABA(B) receptors: synaptic functions and mechanisms of diversity. Curr Opin Neurobiol. 2007;17:298–303. doi: 10.1016/j.conb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Urban NN, Castro JB. Tuft calcium spikes in accessory olfactory bulb mitral cells. J Neurosci. 2005;25:5024–5028. doi: 10.1523/JNEUROSCI.0297-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtschanoff JG, Weinberg RJ. Laminar organization of the NMDA receptor complex within the postsynaptic density. J Neurosci. 2001;21:1211–1217. doi: 10.1523/JNEUROSCI.21-04-01211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- Zuber B, Nikonenko I, Klauser P, Muller D, Dubochet J. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc Natl Acad Sci U S A. 2005;102:19192–19197. doi: 10.1073/pnas.0509527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02