Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo (original) (raw)
Abstract
The phosphoprotein phosphatase 2A (PP2A) catalytic subunit contains a methyl ester on its C-terminus, which in mammalian cells is added by a specific carboxyl methyltransferase and removed by a specific carboxyl methylesterase. We have identified genes in yeast that show significant homology to human carboxyl methyltransferase and methylesterase. Extracts of wild-type yeast cells contain carboxyl methyltransferase activity, while extracts of strains deleted for one of the methyltransferase genes, PPM1, lack all activity. Mutation of PPM1 partially disrupts the PP2A holoenzyme in vivo and ppm1 mutations exhibit synthetic lethality with mutations in genes encoding the B or B′ regulatory subunit. Inactivation of PPM1 or overexpression of PPE1, the yeast gene homologous to bovine methylesterase, yields phenotypes similar to those observed after inactivation of either regulatory subunit. These phenotypes can be reversed by overexpression of the B regulatory subunit. These results demonstrate that Ppm1 is the sole PP2A methyltransferase in yeast and that its activity is required for the integrity of the PP2A holoenzyme.
Keywords: carboxyl methylation/catalytic subunit/phosphoprotein phosphatase 2A/Ppm1/yeast
Introduction
Reversible protein phosphorylation plays a major role in regulating most cellular processes: as many as 25% of all cellular proteins are subject to reversible phosphorylation (Chelsky et al., 1985). Much of the specificity of protein phosphorylation resides in the kinases responsible for transferring the phosphate group from ATP to the target proteins, as witnessed by the rich diversity of cellular kinases (Hunter, 1994; Plowman et al., 1999). However, specificity is also imparted by phosphoprotein phosphatases, whose complexity, achieved in part through combinatorial subunit interactions, may rival that of protein kinases (Virshup, 2000). Phosphoprotein phosphatases play critical roles in the timing, extent and localization of protein phosphorylation and in regulating those cellular processes modulated by phosphorylation.
Phosphoprotein phosphatase 2A (PP2A) is one of the four major classes of eukaryotic serine/threonine phosphoprotein phosphatases (Cohen, 1989; Wera and Hemmings, 1995). It affects such diverse cellular processes as metabolism, signal transduction, cell cycle progression, apoptosis, transcription, DNA replication and protein synthesis (Virshup, 2000). It exists predominantly as a heterotrimeric protein, consisting of a catalytic subunit (C), a regulatory subunit (B) and a scaffold protein (A) on which the catalytic and regulatory subunit sit (Cohen, 1989; Wera and Hemmings, 1995; Groves et al., 1999). Mammalian cells contain multiple isoforms of the C and A subunits. In addition, mammalian cells contain a number of regulatory subunits, which fall into three distinct and non-homologous families, B, B′ and B′′, each of which is represented by several isoforms (Kamibayashi et al., 1994; McCright and Virshup, 1995; Csortos et al., 1996; Zolnierowicz et al., 1996). Each regulatory subunit is capable of binding an AC dimer and probably imparts a particular substrate specificity to the C subunit (Hubbard and Cohen, 1993; Molloy et al., 1998). While the heterotrimeric forms of PP2A probably account for the main biological activities of the phosphatase, cell extracts contain catalytically active AC dimers and free C subunits and these may contribute to the biological function of the phosphatase (Cohen, 1989). C subunits also form heterodimers with a subunit, called α4 in mammalian cells and Tap42 in budding yeast (Saccharomyces cerevisiae), whose association with C may inhibit its catalytic activity (Di Como and Arndt, 1996; Murata et al., 1997; Chen et al., 1998; Jiang and Broach, 1999). In yeast, association of Tap42 with the C subunit is essential for viability (Di Como and Arndt, 1996; Beck and Hall, 1999). Thus, PP2A consists of a large collection of distinct entities with different ternary structures, subunit compositions and substrate recognition properties.
Genetic analysis of PP2A in S.cerevisiae has helped clarify the diverse roles PP2A that plays in the cell. Two highly homologous genes, PPH21 and PPH22, redundantly encode the C subunit of PP2A (Sneddon et al., 1990). Inactivation of both genes severely retards growth and analysis of temperature-sensitive alleles has identified a requirement for PP2A in actin cytoskeletal organization and progression through mitosis (Ronne et al., 1991; Lin and Arndt, 1995; Evans and Stark, 1997). Inactivation of PPH3, a gene with homology to PPH21/22, in a pph21 pph22 background is lethal, suggesting that Pph3 is a partially redundant isoform of C (Ronne et al., 1991). CDC55 and RTS1 encode proteins homologous to the mammalian B and B′ regulatory subunits, respectively (Healy et al., 1991; Shu et al., 1997). The yeast genome does not encode a protein homologous to the B′′ subunit. Inactivation of CDC55 renders growth cold sensitive and causes defects in cytokinesis and in the spindle checkpoint, while inactivation of RTS1 causes temperature-sensitive growth, reduced ability to use non-fermentable carbon sources and cell cycle arrest in G2. Thus, the two regulatory subunits appear to affect different cellular processes, presumably as a result of targeting the phosphatase to distinct substrates. Inactivation of the A subunit, encoded by TPD3, results in a phenotype that approximates the sum of the phenotypes induced by inactivation of the individual regulatory subunits (van Zyl et al., 1992). Tap42 competes in vivo with A and B for association with the C subunit (Jiang and Broach, 1999). The association of Tap42 with C is required for mitotic growth and this association is disrupted by nutrient starvation or treatment of cells with the macrolide drug rapamycin (Di Como and Arndt, 1996; Schmidt et al., 1998). Thus, yeast PP2A participates in a number of different cellular processes, dictated in part by the regulatory subunits with which it associates.
The C subunit of mammalian PP2A is subject to at least three post-translational modifications: methyl esterification of the C-terminal leucine, phosphorylation of a conserved tyrosine located two residues from the C-terminus and phosphorylation of an as yet unidentified threonine (Chen et al., 1992, 1994; Guo and Damuni, 1993; Lee and Stock, 1993; Xie and Clarke, 1994). Phosphorylation of either the tyrosine or the threonine site inhibits phosphatase activity in vitro. On the other hand, carboxyl methylation of the C subunit does not affect phosphatase activity in vitro (Tolstykh et al., 2000). However, purified PP2A carboxyl methyltransferase methylates the C subunit only in the context of an AC dimer and the resulting dimer containing the methylated C subunit has a higher affinity for a B regulatory subunit than does a dimer with an unmethylated C subunit (Tolstykh et al., 2000). Consistent with this observation, a tagged version of a mammalian PP2A C subunit containing a mutation of the conserved C-terminal leucine showed reduced participation in ABC heterotrimers in vivo but normal participation as an AC heterodimer (Chung et al., 1999). These observations suggest that carboxyl methylation could affect the association of the C subunit with regulatory subunits in vivo and, as a result, alter targeting of the phosphatase towards certain substrates relevant for its normal biological activity.
To examine this hypothesis, we have identified the gene encoding the budding yeast PP2A carboxyl methyltransferase and characterized strains lacking its activity. We have found that the PP2A heterotrimer is destabilized in strains lacking methyltransferase activity and that the phenotypes of such strains are quite similar to those of strains lacking the PP2A regulatory subunits. These observations confirm the biological role of PP2A carboxyl methylation and raise the possibility that this reversible modification participates in regulation of cellular growth.
Results
Isolation of cDNA for the human PP2A carboxyl methyltransferase
By conventional chromatographic techniques we previously purified PP2A methyltransferase (PPMT) to homogeneity from extracts of bovine brain (Lee and Stock, 1993). We digested the resulting 47 kDa protein with chymotrypsin or endoproteinase Lys-C and obtained sequence data from the N-terminus of several of the result ing fragments. A search of the DDBJ/EMBL/GenBank expressed sequence tag (EST) database revealed nine human and one mouse entry that encoded sequences highly homologous to one or more of the peptides. From the overlap of the human cDNA entries, we assembled a 300 bp fragment with a 98 amino acid contiguous peptide sequence of the putative human methyltransferase. Using this assembled sequence, we recovered and sequenced four human cDNA clones, one of which appeared to contain the entire PPMT open reading frame. The sequence of the gene was identical to that recently reported by De Baere et al. (1999) and the predicted protein sequence of human PPMT derived from the DNA sequence contains the peptide fragments we identified.
To confirm that the recovered cDNA clone encoded human PPMT, we expressed the full-length cDNA in Escherichia coli under the control of an inducible LacZ promoter and assayed PPMT activity in extracts of cells expressing this cDNA. Extracts from induced cells but not from uninduced cells were capable of transferring [3H]methyl from _S_-adenosyl methionine to bovine PP2A AC dimer (data not shown). This confirmed that the human cDNA clone we isolated on the basis of homology to peptides derived from bovine PPMT encodes human PPMT.
Identification of yeast genes encoding PP2A carboxyl methyltransferase and methylesterase
BLAST analysis identified two genes in S.cerevisiae (YDR435c and YOL141w), two in Schizosaccharomyces pombe (emb|CAA21793| and emb|CAA19576|), one in Caenorhabditis elegans (emb|CAA84295| and WormPD B0285.4) and one in Drosophila melanogaster (gb|AAF53483|) and an additional human cDNA (dbj|BAA25473|) that encode proteins with extensive homology to human PPMT. The larger of the two genes from S.cerevisiae (YOL141w) and from S.pombe (CAA19576), as well as the second human gene, encode proteins that show homology to the entire human PPMT protein in their N-termini but extend for an additional 300 amino acids C-terminal to the region of homology. The C-terminal extensions of the three proteins show homology to each other (data not shown). The sequence relationships among these different genes are represented in Figure 1. On the basis of the homology to human PPMT and the data presented below, we have designated YDR435c and YOL141w from S.cerevisiae as the phospho protein phosphatase 2A methyltransferase genes, PPM1 and PPM2, respectively.
Fig. 1. Sequence relationship of putative human, S.cerevisiae, S.pombe, D.melanogaster and C.elegans PP2A carboxyl methyltransferases. Predicted amino acid sequences of human methyltransferases (hsmt1, AAF18267; hsmt2, BAA25473) and those of related proteins predicted from the genomic sequences of S.cerevisiae (PPM1, YDR435c; PPM2, YOL141w), S.pombe (spmt1, CAA21793; spmt2, CAA19576), D.melanogaster (dmmt, AAF53483) and C.elegans (cemt, CAA84295) are aligned using the PILEUP program from the University of Wisconsin GCG.
Lee et al. (1996) identified and purified to homogeneity a bovine methylesterase (PPME) specific for methylated PP2A C subunit. Ogris et al. (1999) subsequently found that this protein co-precipitated with a mutant form of the PP2A C subunit and proceeded to clone the gene encoding the methylesterase. By BLAST analysis, we identified a single open coding region in S.cerevisiae (YHR075C) with significant homology to the bovine enzyme. Our subsequent analysis, described below, confirmed that this gene promotes PPME activity. Accordingly, we have designated this gene PPE1.
To determine whether either of the two identified S.cerevisiae genes homologous to bovine PPMT encodes a protein with PPMT activity, we created null alleles of PPM1 and PPM2 by replacing the entire open reading frame of each gene with a selectable marker. We then generated isogenic strains in a W303 background that contained a deletion of one or the other of these genes. We tested extracts of these strains for PPMT activities by incubating them with purified bovine AC dimer and _S_-[3H]adenosyl methionine, and assessing the extent of radioactivity incorporated into the C subunit as described in Materials and methods. As shown in Figure 2A, wild-type yeast extract contains a significant level of PPMT activity, as do strains deleted for PPM2. However, extracts of ppm1 strains have no detectable PPMT activity. From these data we conclude that PPM1 encodes the entirety of yeast PPMT.
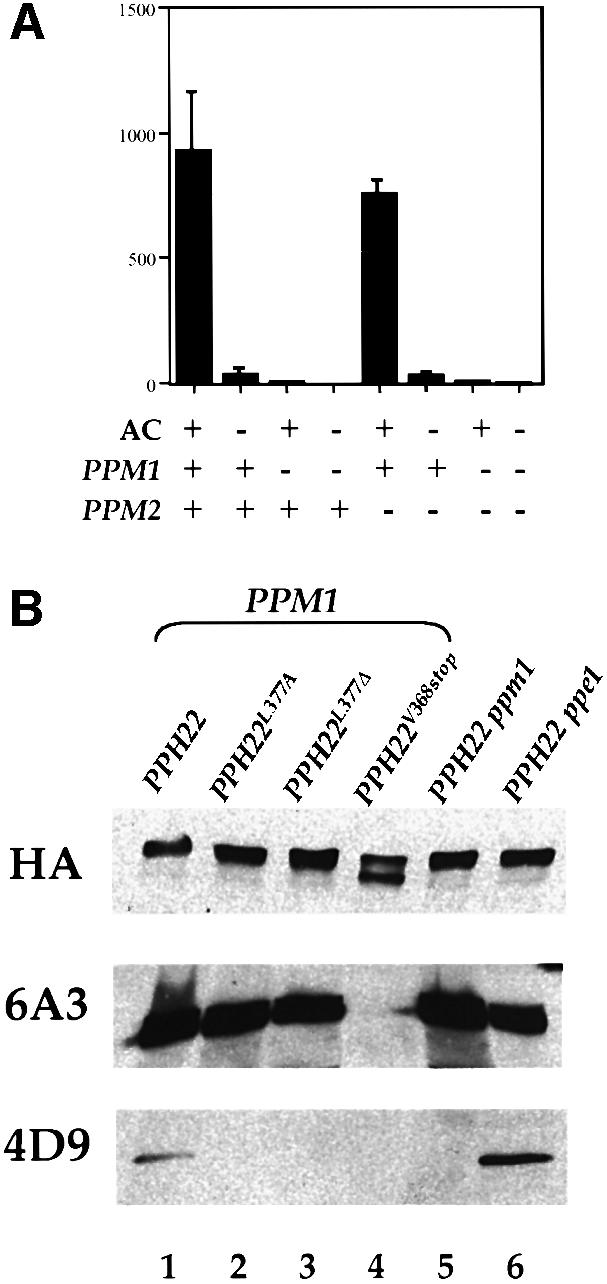
Fig. 2. PPM1 and PPE1 encode a PP2A carboxyl methyltransferase and a PP2A carboxyl methylesterase, respectively. (A) Extracts from W303-1A (PPM1 PPM2), Y2752 (ppm1), Y2739 (ppm2) and Y2740 (ppm1 ppm2) were incubated at 30°C for 30 min with or without bovine AC dimers and [3H]AdoMet, and then subjected to SDS–PAGE. The C subunit was analyzed for incorporation of [3H]methyl esters as described in Materials and methods. The data shown are means ± SD of duplicates from two separate experiments. (B) Extracts from Y2762 (pph21 pph22, lanes 1–4), Y2760 (pph21 pph22 ppm1, lane 5) or Y2761 (pph21 pph22 ppe1, lane 6) expressing triple HA-tagged Pph22 containing the indicated mutations were separated by SDS–PAGE and then transferred to PDVF membranes. The membranes were probed with a monoclonal antibody against HA, monoclonal antibody 6A3 (which recognizes both methylated and unmethylated C subunit) and monoclonal antibody 4D9 (which recognizes only methylated C subunit). Extracts were run as separate sets of tracks for each antibody.
To confirm the assignment of PPMT activity to PPM1 and PPME activity to PPME1, we examined the methylation state of the PP2A C subunit in wild-type and various mutant yeast cells. We have previously raised monoclonal antibodies against an amidated peptide corresponding to the C-terminal domain of the C subunit of PP2A (Tolstykh et al., 2000). One of these monoclonal antibodies, 6A3, recognizes the C subunit of both the methylated and unmethylated protein. However, a second monoclonal antibody, 4D9, binds only to the carboxyl methylated C subunit. These reagents can thus be used to assess the in vivo methylation state of the PP2A C subunit. Accordingly, we constructed isogenic wild-type, _ppm1_Δ and _ppe1_Δ strains expressing a hemagglutinin (HA)-tagged version of either Pph21 or Pph22 and performed western analysis on extracts of these strains. We also performed western analysis on extracts of wild-type cells expressing HA-tagged versions of the C subunit carrying various mutations affecting the C-terminal domain of the protein. The results of this analysis are shown in Figure 2B. Both the methylation-specific and the -nonspecific monoclonal antibodies recognized the HA-tagged PP2A C subunit present in extracts of wild-type cells. The methylation-nonspecific antibody bound to mutant protein carrying a leucine to alanine substitution of the C-terminal amino acid, but the methylation-specific antibody did not. This is consistent with the apparent requirement of a terminal leucine residue in order for the C subunit to be methylated by PPMT. Furthermore, as evident from the figure, protein present in a ppm1 strain showed no cross-reactivity with the methylation-specific antibody, while protein present in a ppe1 strain showed increased cross-reactivity with the antibody relative to that seen in the PPM1 PPE1 strain. As evident from lanes reacted with anti-HA antibodies, the PP2A C subunit was expressed at equal levels in all the strains. These results indicate that methylation of the C subunit of PP2A is reduced in ppm1 strains and enhanced in ppe1 strains, consistent with the hypothesis that PPM1 encodes the yeast PPMT and PPE1 the yeast PPME. Given the degree of increased cross-reactivity of the 4D9 antibody with PP2A C in ppe1 cells compared with wild-type cells, we conclude that at least 50% of the C subunit in exponentially growing cells is unmethylated.
Methyltransferase activity is required for stable association of the PP2A C subunit with its regulatory subunits in vivo
In vitro experiments using purified PP2A components have shown that C subunit methylation enhances the binding of the B subunit to the AC dimer (Tolstykh et al., 2000). To examine in vivo the effect on heterotrimer stability of C subunit carboxyl methylation, we assessed the stability of PP2A heterotrimers in a strain lacking methyltransferase activity. We immunoprecipitated HA-tagged C subunit from extracts of PPM1 and ppm1 strains using anti-HA antibody under non-denaturing conditions. We then fractionated the immunoprecipitates by gel electrophoresis, transferred the fractionated proteins to a PVDF membrane, and probed the membrane with antibodies specific for Tpd3, Cdc55 or HA. As seen in Figure 3A and as noted previously (Jiang and Broach, 1999), a significant fraction, but by no means all, of the A and B subunits in extracts of the PPM1 strain co-precipitated with Pph21 and Pph22. In contrast, but consistent with previous in vitro results (Tolstykh et al., 2000), significantly less Cdc55 co-precipitated with Pph21 or Pph22 from extracts of the _ppm1_Δ strain. Unexpectedly, though, we observed a comparable diminution in the amount of Tpd3 that co-precipitated with Pph21 or Pph22 from extracts of the _ppm1_Δ strain. We confirmed this observation by immunoprecipitating Tpd3 from extracts of the _ppm1_Δ strain and finding less co-precipitation of Pph21 or Pph22 than obtained from extracts of the wild-type strain. Thus, we conclude that the absence of PP2A carboxyl methyltransferase activity destabilized not only the PP2A ABC heterotrimer but also the AC heterodimer in vivo.
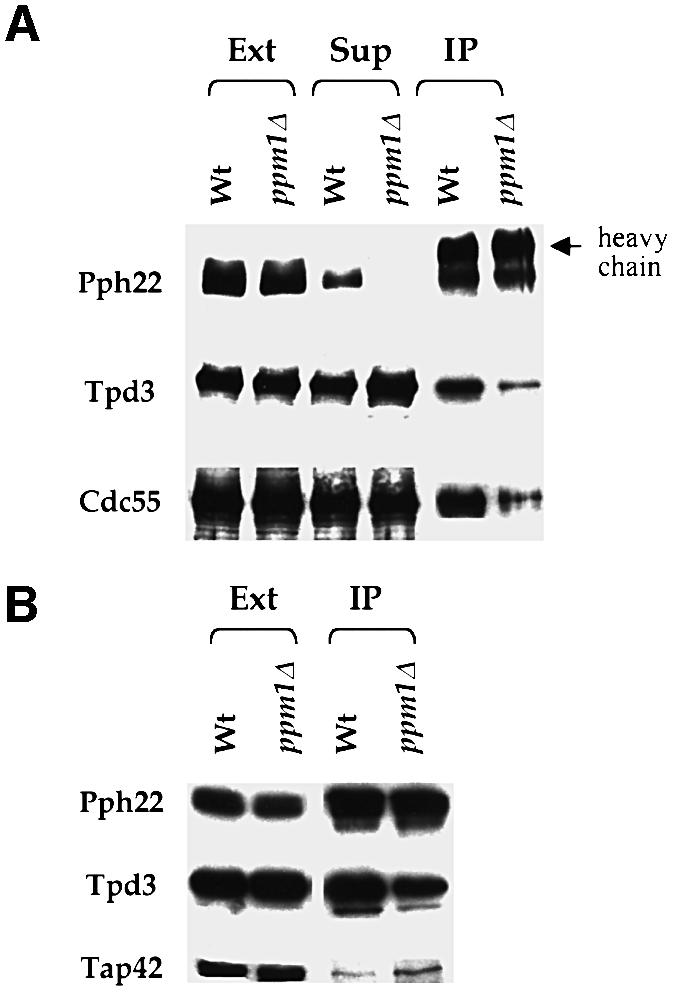
Fig. 3. Loss of Ppm1 activity affects the interaction of PP2A regulatory subunits with the C subunits. Extracts from Y2480 [_pph22:: (HA)_3_PPH22_] and Y2734 [_ppm1 pph22:: (HA)_3_PPH22_] cells were immunoprecipitated with anti-HA epitope monoclonal antibodies. Samples of the precipitates (‘IP’), extracts before precipitation (‘Ext’) and extracts after precipitation (‘Sup’) were separated by SDS–PAGE and then transferred to PVDF membranes. The membranes were probed separately with anti-HA, anti-Tpd3, anti-Cdc55 (A) and anti-Tap42 (B) polyclonal antibodies.
A portion of the PP2A C subunit is associated with Tap42 in exponentially growing yeast cells. This association is diminished in stationary phase cells and in cells treated with rapamycin, and is increased in cells deleted for Tpd3 (Di Como and Arndt, 1996; Jiang and Broach, 1999). Accordingly, we investigated whether loss of Ppm1 activity affected the extent of association of Tap42 with the PP2A C subunit. As shown in Figure 3B, a slightly larger amount of Tap42 co-precipitates with PP2A C from extracts of the _ppm1_Δ strain than from an isogenic wild-type strain. Quantification of signals on the western blot suggests that the increase is ∼50%. While this increase is small, we obtained essentially the same results with different extracts of several different strains, so we can conclude that the interaction of Tap42 with PP2A C does not depend on Ppm1 activity and may even be enhanced in its absence.
Phenotypes of ppm1 strains are similar to those of cdc55 and rts1 strains
Because of the reduced association of the PP2A regulatory subunits with the C subunits in ppm1 strains in vivo, we examined whether such strains were phenotypically similar to those lacking these regulatory subunits. cdc55 mutants were originally identified as isolates exhibiting cytokinesis and morphological defects at low temperature, and subsequently as mutants defective in cell cycle arrest in response to spindle damage (Healy et al., 1991; Minshull et al., 1996; Wang and Burke, 1997). These two defects may both derive from a role of Cdc55 in promoting dephosphorylation of Cdc28, since certain cdc20 and cdc28 alleles suppress both the checkpoint and morphological defects caused by cdc55 mutations (Minshull et al., 1996; Wang and Burke, 1997). More recently, we have shown that Cdc55 antagonizes the Tor signaling pathway, in that cdc55 mutants exhibit rapamycin resistance as well as an enhanced association in vivo between the Tap42 and the C subunit of PP2A (Jiang and Broach, 1999). RTS1 was initially identified by homology to a mammalian B′ subunit and subsequent characterization revealed deletion mutants to be temperature sensitive for growth, deficient for growth on non-fermentable carbon sources and defective in the G2–M transition in the cell cycle (Shu et al., 1997). Accordingly, we tested ppm1 strains for many of these phenotypes associated with inactivation of CDC55 or RTS1.
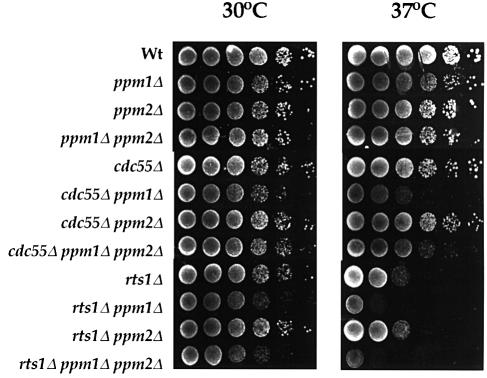
As shown in Figure 4, _ppm1_Δ strains showed no growth defects on rich media. While cdc55 strains do not exhibit a growth defect at reduced temperatures, as measured by plating efficiency, they are morphologically distinctive, mainly because of a defect in cytokinesis. We saw no such morphological defect for _ppm1_Δ strains in the W303 background or in the S288C background, in which the cdc55 morphological phenotype is more pronounced (data not shown). As noted above, rts1 strains were reported to be deficient in growth on non-fermentable carbon sources. However, we observed no growth defect of any of the mutants on YEP media with glycerol, ethanol or glycerol plus ethanol as carbon source (data not shown).
Fig. 4. Genetic interactions between Ppm1 and PP2A B regulatory subunit genes. Exponentially growing cultures of W303-1A (wild type), Y2752 (ppm1), Y2739 (ppm2), Y2740 (ppm1 ppm2), Y2483 (cdc55), Y2745 (ppm1 cdc55), Y2741 (ppm2 cdc55), Y2742 (ppm1 ppm2 cdc55), Y2736 (rts1), Y2737 (ppm1 rts1), Y2738 (ppm2 rts1) and Y2744 (ppm1 ppm2 rts1) were 10-fold serially diluted and spotted on to YEPD plates. Growth is shown after 2 days at 30 or 37°C.
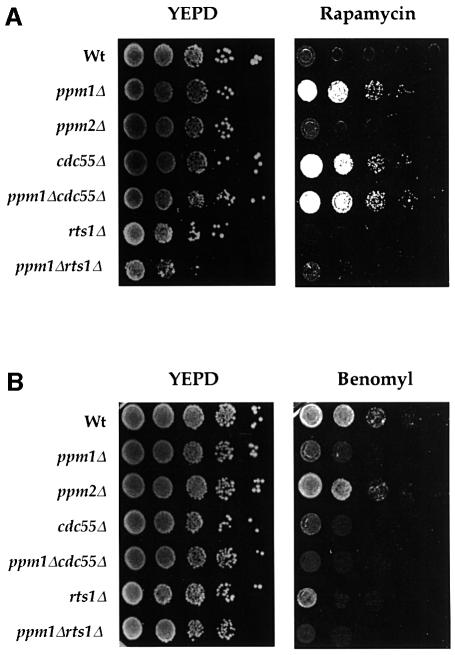
The macrolide drug rapamycin inhibits yeast growth by inhibiting those essential activities of Tor kinase mediated at least in part through Tap42. cdc55 strains are resistant to rapamycin, since Cdc55 is required to reverse Tor-mediated phosphorylation of Tap42. As shown in Figure 5A, the ppm1 strain can grow in the presence of 100 nM rapamycin, a concentration to which the isogenic wild-type and ppm2 strains are sensitive but on which cdc55 strains can grow. Thus, ppm1 strains mirror this phenotype of cdc55 strains.
Fig. 5. ppm1 cells are resistant to and supersensitive to benomyl. Exponentially growing cultures of wild-type cells, Y2752 (ppm1), Y2739 (ppm2), Y2483 (cdc55), Y2736 (rts1), Y2745 (ppm1 cdc55) and Y2737 (ppm1 rts1) were 10-fold serially diluted and spotted on to YEPD plates containing 100 nM rapamycin (A) or 12 µg/ml benomyl (B). Growth is shown after incubation at 30°C for 2 days without drug or 4 days in the presence of rapamycin and benomyl.
To test whether ppm1 strains exhibit a checkpoint defect similar to that caused by loss of CDC55, we examined the benomyl sensitivity of such strains. Strains defective in the spindle checkpoint pathway are more sensitive to low levels of benomyl, a microtubule-depolymerizing agent, probably due to an increased proportion of cells that transit mitosis in the absence of an intact spindle (Hoyt et al., 1991; Li and Murray, 1991). As shown in Figure 5B, both cdc55 and ppm1 strains show substantially lower plating efficiency on benomyl than does the wild-type strain. rts1 strains were also more sensitive to benomyl, which was surprising since Rts1 had not previously been implicated in spindle checkpoint function. However, in a systematic study of phenotypes of yeast strains carrying insertions in individual genes, K.H.Cheung, A.Kumar, X.Liu and P.Ross-Macdonald (http://ygac.med.yale.edu/) list rts1::Tn strains as benomyl supersensitive, confirming our results.
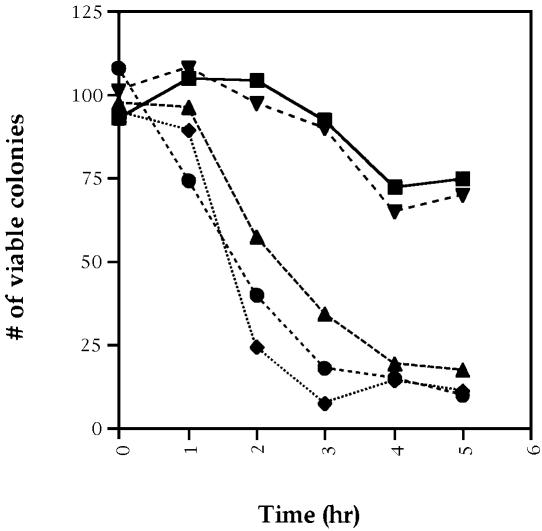
To confirm the checkpoint deficiency of ppm1 strains, we examined the loss of viability of a ppm1 strain as a function of cell cycle progression in the presence of nocodazole, another microtubule-depolymerizing agent. Cells of the test strain were arrested at G1 by treatment with α-factor and then released from the G1 block in the presence of nocodazole. Samples were removed at various times after release and analyzed for position in the cell cycle and for viability. Cells with an intact spindle checkpoint arrest in mitosis and retain viability, whereas cells defective for the spindle checkpoint proceed through mitosis in the absence of a spindle and lose viability. Accordingly, loss of viability coincides with transit through mitosis. As evident from Figure 6, both the wild-type strain and the ppm2 strain arrest with a G2 content of DNA and retain viability during the course of the experiment. In contrast, the viability of cdc55, rts1 and ppm1 strains declines steeply, coincident with transit through mitosis. From these results we conclude that the ppm1 and rts1 mutants show a checkpoint defect similar to that of cdc55 mutants.
Fig. 6. ppm1, cdc55 and rts1 are required for spindle assembly checkpoint activity. Exponentially growing cultures of W303-1A (wild type), Y2752 (ppm1), Y2739 (ppm2), Y2483 (cdc55) and Y2736 (rts1) were incubated in the presence of α-factor for 3 h at 30°C and then transferred into fresh YEPD medium containing 12 µg/ml nocodazole. At different times after release from cell cycle arrest, cells were removed, sonicated, counted and then plated on to YEPD plates. Viable colonies were counted after 2 days. Squares, wild type; diamonds, cdc55; circles, rts1; upward pointing triangles, ppm1; downward pointing triangles, ppm2.
PPE1 overexpression causes phenotypes similar to those of ppm1 strains
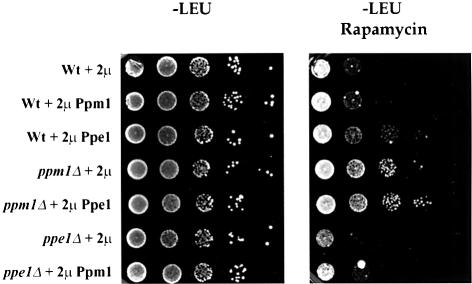
If the in vivo level of methylation of PP2A results from a dynamic equilibrium between the activities of PPMT and PPME, we would expect that overexpression of PPE1 should induce phenotypes similar to those associated with loss of PPM1. As shown in Figure 7, this is the case. A strain containing PPE1 on a high-copy-number vector exhibits rapamycin resistance equivalent to that of a ppm1 strain. This strain is also supersensitive to benomyl (data not shown). We have not seen any obvious phenotypes associated with loss of Ppe1 despite the fact that such strains exhibit enhanced methylation of PP2A (Figure 2B). ppe1 strains grow normally at all temperatures on fermentable and non-fermentable carbon sources, and show wild-type sensitivities to nocodazole, benomyl and rapamycin (data not shown). Thus, while reduced methylation of PP2A, whether achieved by reduced PPMT activity or increased PPME activity, yields observable phenotypes, we have not yet identified phenotypes associated with increased methylation.
Fig. 7. Overexpression of PPE1 induces rapamycin resistance. Exponentially growing cultures of strain W303-1A containing plasmid pRS425 (2µ), pJW131 (2µ PPM1) or pJW141 (2µ PPE1) and strain Y2752 (ppm1) containing plasmid pRS425 or pJW141 and strain Y2746 (ppe1) containing plasmid pRS425 or pJW131 were 10-fold serially diluted and spotted on to SD – Leu plates without rapamycin or with 100 nM rapamycin. Growth is shown after 2 days at 30°C in the absence of rapamycin or after 4 days in the presence of rapamycin.
Overexpression of CDC55 reverses the rapamycin resistance of ppm1
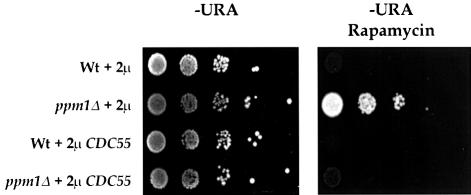
We hypothesize that the phenotypic alterations seen in the ppm1 strain derive from a reduced association between the regulatory and C subunits rather than a diminished activity of the C subunit. If our hypothesis were correct, then we would anticipate that increased expression of CDC55 might increase the concentration of PP2A heterotrimer by mass action and reverse the phenotypes of the ppm1 strain. To test this, we introduced CDC55 under its own promoter on a high-copy 2µ plasmid into a ppm1 strain and then examined the rapamycin sensitivity of the resulting strain. As is evident from Figure 8, overexpression of CDC55 restored rapamycin sensitivity to the ppm1 strain.
Fig. 8. Overexpression of Cdc55 restores rapamycin sensitivity to ppm1 strains. Exponentially growing cultures of strains W303-1A and Y2752 (ppm1) containing either pRS426 (2µ) or YEpCDC55 (2µ CDC55) were 10-fold serially diluted and spotted on to SD – Ura plates containing no rapamycin or 100 nM rapamycin. Growth is shown after incubation at 30°C for 2 days in the absence of rapamycin or after 4 days in the presence of rapamycin.
Discussion
Yeast cells contain a number of proteins modified by carboxyl methylation (Hrycyna et al., 1994). The identities of most of these proteins are unknown, as are the identities of the methyltransferases responsible for their modification. However, three such proteins, Ras, a-mating factor and Ste18, the γ subunit of the heterotrimeric G protein, are substrates of a single methyltransferase encoded by STE14. Loss of Ste14 activity completely eliminates a-mating factor activity, has no apparent affect on Ras activity and reduces basal, but not induced, pheromone signaling mediated by Ste18 (Hrycyna et al., 1991; Sapperstein et al., 1994). Thus, carboxyl methylation can have varied, and sometimes substantial, effects on the activities of proteins.
We have identified two genes in Saccharomyces that show homology to bovine PPMT and provided evidence that inactivation of one of these genes, PPM1, eliminates in vivo PPMT activity. Despite the high level of homology between the two yeast genes, mutation of the second gene, PPM2, does not reduce PPMT activity in vitro or lead to any of the phenotypes associated with altered PP2A function in vivo. PPM2 encodes a protein with a 300 amino acid N-terminal domain homologous to yeast and human PPMT but also includes a 300 amino acid C-terminal extension region. A gene fully homologous to both domains of this extended form is present in S.pombe and in the human genome, although not in C.elegans or D.melanogaster, suggesting a conserved function for this protein. The high degree of sequence conservation with yeast and mammalian PPMT argues for a functional conservation too. Accordingly, we are examining yeast cells for their spectrum of carboxyl-methylated proteins in isogenic _ppm2_– and PPM2+ strains in an attempt to identify a substrate for this likely carboxyl methyltransferase.
In vitro analysis has shown that methylation of the C subunit of PP2A by bovine PPMT increases the affinity of the C subunit for at least one B regulatory subunit (Tolstykh et al., 2000). The effect of methylation on other regulatory subunits was not tested, so we do not know whether methylation diminishes the affinity of all regulatory subunits equally or whether it affects some subunits more than others. These results are consistent with previous analyses of C subunit mutants. Chung et al. (1999) recently showed that a human PP2A C subunit mutated at the carboxyl methylation site (L309Q) exhibited reduced association with B subunits in extracts of COS-7 cells but was still able to associate with α4. Evans et al. (1999) showed that deletion of the C-terminal leucine did not impair the ability of human PP2A C subunit to complement a pph21 pph22 yeast strain, although more substantial alterations of the C-terminus eliminated complementing activity. The results presented in this report indicate that loss of methylation even of the wild-type C subunit abrogates its association in vivo with the B subunit. We have also shown that mutation of the terminal leucine to alanine completely eliminates carboxyl methylation. However, the loss of methylation only diminishes, rather than fully abrogating, PP2A function. Thus, our results are consistent with the previous observation that mutation of the C-terminal leucine does not render the gene inactive.
The in vitro observations with bovine PP2A prompted us to test whether inactivation of yeast PPMT would have effects similar to those seen in mutants lacking one or the other B regulatory subunit. We indeed found that ppm1 strains mirror many, albeit not all, of the phenotypes of cdc55 (lacking the B subunit) or rts1 (lacking the B′ subunit) strains. In particular, ppm1 strains show the same rapamycin resistance as cdc55 strains, the same temperature sensitivity as the rts1, and the same sensitivity to benomyl and nocodazole as both rts1 and cdc55 strains. This latter observation indicates that ppm1 strains probably have the same spindle checkpoint defect as cdc55 strains. In cdc55 strains, the checkpoint defect results from premature inactivation of Cdc28 in arrested cells, probably due to persistent phosphorylation of Cdc28 Tyr15 (Minshull et al., 1996; Wang and Burke, 1997). PP2A does not directly dephosphorylate Cdc28 Tyr15, so the precise protein substrate for Cdc55-targeted PP2A activity that maintains Tyr15 in the phosphorylated state under arrest conditions is not known. The results presented in this report revealed that Rts1 activity is also required for spindle checkpoint integrity. We do not know from these observations whether the substrate for Rts1-directed PP2A activity in maintaining checkpoint integrity is the same as that of Cdc55-directed activity. In sum, though, our results indicate that PPMT activity is required for full PP2A activity in vivo.
The phenotypes of cdc55 or rts1 are not identical to those of a ppm1 strain. For instance, ppm1 strains do not show the morphological abnormalities manifested by cdc55 strains. This probably reflects the fact that the affinity of the C subunit for its regulatory partners is diminished but not eliminated, so that ppm1 strains retain some Cdc55- and Rts1-directed PP2A activity. This interpretation is strengthened by the observation that ppm1 deletion shows synergistic effects with cdc55 or rts1 deletion (Figure 4), indicating that loss of Ppm1 activity is not completely equivalent to loss of Cdc55 or Rts1. Furthermore, overexpression of Cdc55 reversed at least one of the phenotypes of ppm1 strains, suggesting that the C subunit in this strain is capable of binding the B subunit but does so with reduced affinity.
In contrast to its effect on heterotrimer formation, loss of PPMT activity does not diminish the affinity of the C subunit for another regulatory partner in the cell, Tap42. The retention of interaction of the unmethylated C subunit with Tap42 is consistent with the rapamycin resistance of ppm1 strains, since resistance to rapamycin depends on retaining association between Tap42 and the C subunit (Di Como and Arndt, 1996; Jiang and Broach, 1999). This difference in the effect of carboxyl methylation on affinity of the C subunit for B regulatory subunits compared with Tap42/α4 raises the possibility that methylation could provide a mechanism for modulating the distribution of the C subunit among these different regulatory elements. Our results indicate that methylation of the PP2A C subunits is in dynamic equilibrium through the competing reactions catalyzed by Ppm1 and Ppe1. Accordingly, inhibition of PPMT or activation of PPME would tend to diminish PP2A activity while strengthening the Tor-mediated pathway. Thus, PPMT or PPME could serve as a locus through which internal or external signals impinged on cellular proliferation. To investigate this possibility, we are currently exploring whether C subunit methylation responds to internal or external signals.
Materials and methods
Yeast strains, plasmids and media
The strains used in this study (listed in Table I) were all derived from W303-1A or W303-1B. Cells were grown at 30°C in YEPD or synthetic (SD) medium (Kaiser et al., 1994). Rapamycin was added to solid medium to a final concentration of 100 nM from a stock of 1 mM in 100% ethanol. Benomyl was added to solid medium to a final concentration of 15 µg/ml from a stock of 3 mg/ml in dimethylsulfoxide (DMSO). Nocadazole was added to liquid YEPD to a final concentration of 12 µg/ml from a stock of 2 mg/ml in DMSO. α-factor was added to liquid YEPD to a final concentration of 8 µg/ml from a stock of 2 mg/ml. The plasmids YEpCDC55 (Jiang and Broach, 1999), pRS (Christianson et al., 1992) and pCRtm2.1 (Invitrogen) have been described previously. pJW131 was constructed by inserting a 1.5 kb _Bam_HI–_Sal_I PPM1 PCR fragment from W303-1A genomic DNA into pRS426. pJW142 was constructed by inserting a 1.6 kb _Bam_HI–_Sal_I PPE1 PCR fragment from W303-1A genomic DNA into pRS426.
Table I. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| Y2480 | MATa _pph22::(HA)3_PPH22 | laboratory stock |
| Y2483 | MATa cdc55::LEU2 | laboratory stock |
| Y2734 | _MAT_α _pph22::(HA)3_PPH22, ppm1::HIS3 | laboratory stock |
| Y2736 | MATa rts1::URA3 | this study |
| Y2737 | MATa rts1::URA3, ppm1::HIS3 | this study |
| Y2738 | _MAT_α rts1::URA3, ppm2::TRP1 | this study |
| Y2739 | MATa ppm2::TRP1 | this study |
| Y2740 | MATa ppm1::HIS3, ppm2::TRP1 | this study |
| Y2741 | MATa cdc55::LEU2, ppm2::TRP1 | this study |
| Y2742 | MATa cdc55::LEU2, ppm1::HIS3, ppm2::TRP1 | this study |
| Y2744 | MATa rts1::URA3, ppm1::HIS3, ppm2::TRP1 | this study |
| Y2745 | MATa cdc55::LEU2, ppm1::HIS3 | this study |
| Y2746 | MATa ppe1::TRP1 | this study |
| Y2752 | MATa ppm1::HIS3 | this study |
| Y2754 | MATa ppm1::TRP1 | this study |
| Y2765 | _MAT_α pph21::LEU2, pph22::HIS3, ppm1::TRP1 + [pRS416_–(HA)_3_PPH22_] | this study |
| Y2766 | _MAT_α pph21::LEU2, pph22::HIS3, ppe1::TRP1 + [pRS416_–(HA)_3_PPH22_] | this study |
| Y2767 | _MAT_α pph21::LEU2, pph22::HIS3 + [pRS416_–(HA)_3_PPH22_] | this study |
| Y2768 | _MAT_α pph21::LEU2, pph22::HIS3 + [pRS416_–(HA)_3_PPH22_L377A] | this study |
| Y2769 | _MAT_α pph21::LEU2, pph22::HIS3 + [pRS416_–(HA)_3_PPH22_L377Δ] | this study |
| Y2770 | _MAT_α pph21::LEU2, pph22::HIS3 + [pRS416_–(HA)_3_PPH22_V368stop] | this study |
Site-directed mutagenesis
Site-directed mutagenesis of PPH22 was performed using a QuickChange Site-Directed Mutagenesis Kit from Stratagene. All reactions were carried out using plasmid pJ295 (Jiang and Broach, 1999).
Gene deletion and epitope tagging
PPM1 was amplified by PCR from Y2098 genomic DNA as a 1.7 kb fragment, including 430 bp upstream and 300 bp downstream from the open reading frame. The fragment was cloned into the pCRtm2.1 vector, resulting in the plasmid pJW100. PPM1 was excised from pJW100 using _Eco_RI, then ligated to _Eco_RI-digested pUC18, resulting in pJW103. HIS3 was amplified from pRS413 and cloned into pCRtm2.1 (giving pJW105A). The ppm1::HIS3 allele was constructed by replacing the 690 bp _Eco_RV–_Nco_I fragment in pJW103 with the _HIS3 Eco_RV–_Nco_I fragment from pJW105A, resulting in pJW106. Yeast ppm1::HIS3 strains were obtained by transforming cells with _Bam_HI-digested pJW106 DNA. The ppmt2::TRP1 allele was constructed using a PCR-based method that has been described previously (Lorenz et al., 1995). The TRP1 gene was amplified from pRS414 using a pair of primers, one of which included 50 bp of sequence immediately 5′ of the PPM2 coding sequence and one of which included 50 bp of sequence immediately 3′ of PPM2. The PCR product was used directly for transformation into yeast. The rts1::URA3 allele was made by the same method. All marker integrations were verified by PCR. CDC55 was deleted as described previously (Jiang and Broach, 1999). The double and triple deletion strains were made by mating the single allele strains and isolating the desired haploid strains after sporulation and dissection. Constructs designed to yield HA-tagged Pph21 and Pph22 have been described previously (Jiang and Broach, 1999).
Assay of methyltransferase activity
To prepare yeast extracts, cells were grown overnight in YEPD to ∼5 × 107 cells/ml. Cells were washed once with double-distilled water and once with buffer B [20 mM MOPS pH 7.2, 1 mM EDTA, 1 mM dithiothreitol (DTT)]. Cells were resuspended in 250 µl of buffer B plus protease inhibitors, and glass beads were added to ∼50% final volume. Cells were then lysed by vortexing at 4°C for 7 min. Beads and cellular debris were removed by microcentrifugation. Methyltransferase activity in crude extracts of E.coli or yeast strains was determined by incubating ∼40 µg protein at 30°C for 30 min in 40 µl of 50 mM MOPS pH 7.2, 1.0 mM EDTA, 5.0 mM DTT, 0.7 µM [3H]AdoMet (80 Ci/mmol) and 1 pmol of purified bovine AC dimer (Tolstykh et al., 2000). Reactions were terminated by addition of TCA to 10% and the extent of 3H incorporation into the C subunit was determined as previously described (Stock et al., 1984).
Immunoprecipitation
Yeast cells were grown in YEPD to ∼2 × 107 cells/ml. Cells were harvested and washed once with double-distilled water and once with cold 10 mM NaN3. Spheroplasts were then prepared by incubating cells for 1 h at 37°C in spheroplast buffer (1.4 M sorbitol, 50 mM K2HPO4 pH 7.5, 5 mM NaN3, 40 mM β-mercaptoethanol) with 10 µg of Zymolyase 100T (ICN) per OD unit of cells. Spheroplasts were harvested and disrupted in lysis buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 2% Triton X-100, protease inhibitors). The lysate was diluted 4-fold with buffer A [20 mM HEPES pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, protease inhibitors (Complete Protease Inhibitors Set, Roche Molecular Biochemicals)]. Insoluble debris was removed by centrifugation. Supernatant containing 0.7 mg of protein was incubated with 5 µl of anti-HA antibody on a nutator at 4°C for 2.5 h. Protein A beads were added to precipitate the immunocomplex for 90 min at 4°C. Beads were washed three times with buffer A. Beads were boiled for 5 min in 300 µl of 1× SDS loading buffer. Various volumes (4–16 µl) of each sample were loaded on to 10% SDS–polyacrylamide gels and subjected to electrophoresis. Samples were then transferred on to PVDF membranes. The membranes were probed with antibodies against HA epitope (1:3000), Tpd3 (1:2000) and Cdc55 (1:2000). Anti-HA (12CA5) cell culture supernatants were obtained from the Princeton University Monoclonal Antibody Facility. Anti-Tpd3 and anti-Cdc55 poly clonal antibodies were raised in rabbits against recombinant glutathione _S_-transferase (GST)–Tpd3 and GST–Cdc55, respectively (Jiang and Broach, 1999).
Acknowledgments
Acknowledgements
We would like to thank Dr Jiang Yu for thoughtful comments and suggestions. This work was supported by NIH grant CA41086 to J.R.B. J.W. was supported by training grant GM07388.
References
- Beck T. and Hall,M.N. (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature, 402, 689–692. [DOI] [PubMed] [Google Scholar]
- Chelsky D., Ruskin,B. and Koshland,D.E.,Jr (1985) Methyl-esterified proteins in a mammalian cell line. Biochemistry, 24, 6651–6658. [DOI] [PubMed] [Google Scholar]
- Chen J., Martin,B.L. and Brautigan,D.L. (1992) Regulation of protein serine–threonine phosphatase type-2A by tyrosine phosphorylation. Science, 257, 1261–1264. [DOI] [PubMed] [Google Scholar]
- Chen J., Parsons,S. and Brautigan,D.L. (1994) Tyrosine phosphorylation of protein phosphatase 2A in response to growth stimulation and v-src transformation of fibroblasts. J. Biol. Chem., 269, 7957–7962. [PubMed] [Google Scholar]
- Chen J., Peterson,R.T. and Schreiber,S.L. (1998) α4 associates with protein phosphatases 2A, 4 and 6. Biochem. Biophys. Res. Commun., 247, 827–832. [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski,R.S., Dante,M., Shero,J.H. and Hieter,P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene, 110, 119–122. [DOI] [PubMed] [Google Scholar]
- Chung H., Nairn,A.C., Murata,K. and Brautigan,D.L. (1999) Mutation of Tyr307 and Leu309 in the protein phosphatase 2A catalytic subunit favors association with the α4 subunit which promotes dephosphorylation of elongation factor-2. Biochemistry, 38, 10371–10376. [DOI] [PubMed] [Google Scholar]
- Cohen P. (1989) The structure and regulation of protein phosphatases. Annu. Rev. Biochem., 58, 453–508. [DOI] [PubMed] [Google Scholar]
- Csortos C., Zolnierowicz,S., Bako,E., Durbin,S.D. and DePaoli-Roach,A.A. (1996) High complexity in the expression of the B′ subunit of protein phosphatase 2A0. Evidence for the existence of at least seven novel isoforms. J. Biol. Chem., 271, 2578–2588. [DOI] [PubMed] [Google Scholar]
- De Baere I., Derua,R., Janssens,V., Van Hoof,C., Waelkens,E., Merlevede,W. and Goris,J. (1999) Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the human homologue. Biochemistry, 38, 16539–16547. [DOI] [PubMed] [Google Scholar]
- Di Como C.J. and Arndt,K.T. (1996) Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev., 10, 1904–1916. [DOI] [PubMed] [Google Scholar]
- Evans D.R. and Stark,M.J. (1997) Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics, 145, 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.R., Myles,T., Hofsteenge,J. and Hemmings,B.A. (1999) Functional expression of human PP2Ac in yeast permits the identification of novel C-terminal and dominant-negative mutant forms. J. Biol. Chem., 274, 24038–24046. [DOI] [PubMed] [Google Scholar]
- Groves M.R., Hanlon,N., Turowski,P., Hemmings,B.A. and Barford,D. (1999) The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell, 96, 99–110. [DOI] [PubMed] [Google Scholar]
- Guo H. and Damuni,Z. (1993) Autophosphorylation-activated protein kinase phosphorylates and inactivates protein phosphatase 2A. Proc. Natl Acad. Sci. USA, 90, 2500–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy A.M., Zolnierowicz,S., Stapleton,A.E., Goebl,M., DePaoli-Roach,A.A. and Pringle,J.R. (1991) CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol., 11, 5767–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis,L. and Roberts,B.T. (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Hrycyna C.A., Sapperstein,S.K., Clarke,S. and Michaelis,S. (1991) The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J., 10, 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna C.A., Yang,M.C. and Clarke,S. (1994) Protein carboxyl methylation in Saccharomyces cerevisiae: evidence for STE14-dependent and STE14-independent pathways. Biochemistry, 33, 9806–9812. [DOI] [PubMed] [Google Scholar]
- Hubbard M.J. and Cohen,P. (1993) On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci., 18, 172–177. [DOI] [PubMed] [Google Scholar]
- Hunter T. (1994) 1001 protein kinases redux—towards 2000. Semin. Cell Biol., 5, 367–376. [DOI] [PubMed] [Google Scholar]
- Jiang Y. and Broach,J.R. (1999) Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J., 18, 2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kamibayashi C., Estes,R., Lickteig,R.L., Yang,S.I., Craft,C. and Mumby,M.C. (1994) Comparison of heterotrimeric protein phos phatase 2A containing different B subunits. J. Biol. Chem., 269, 20139–20148. [PubMed] [Google Scholar]
- Lee J. and Stock,J. (1993) Protein phosphatase 2A catalytic subunit is methyl-esterified at its carboxyl terminus by a novel methyltransferase. J. Biol. Chem., 268, 19192–19195. [PubMed] [Google Scholar]
- Lee J., Chen,Y., Tolstykh,T. and Stock,J. (1996) A specific protein carboxyl methylesterase that demethylates phosphoprotein phosphatase 2A in bovine brain. Proc. Natl Acad. Sci. USA, 93, 6043–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. and Murray,A.W. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Lin F.C. and Arndt,K.T. (1995) The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J., 14, 2745–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.C., Muir,R.S., Lim,E., McElver,J., Weber,S.C. and Heitman,J. (1995) Gene disruption with PCR products in Saccharomyces cerevisiae. Gene, 158, 113–117. [DOI] [PubMed] [Google Scholar]
- McCright B. and Virshup,D.M. (1995) Identification of a new family of protein phosphatase 2A regulatory subunits. J. Biol. Chem., 270, 26123–26128. [DOI] [PubMed] [Google Scholar]
- Minshull J., Straight,A., Rudner,A.D., Dernburg,A.F., Belmont,A. and Murray,A.W. (1996) Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol., 6, 1609–1620. [DOI] [PubMed] [Google Scholar]
- Molloy S.S., Thomas,L., Kamibayashi,C., Mumby,M.C. and Thomas,G. (1998) Regulation of endosome sorting by a specific PP2A isoform. J. Cell Biol., 142, 1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K., Wu,J. and Brautigan,D.L. (1997) B cell receptor-associated protein α4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl Acad. Sci. USA, 94, 10624–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris E., Du,X., Nelson,K.C., Mak,E.K., Yu,X.X., Lane,W.S. and Pallas,D.C. (1999) A protein phosphatase methylesterase (PME-1) is one of several novel proteins stably associating with two inactive mutants of protein phosphatase 2A. J. Biol. Chem., 274, 14382–14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman G.D., Sudarsanam,S., Bingham,J., Whyte,D. and Hunter,T. (1999) The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc. Natl Acad. Sci. USA, 96, 13603–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne H., Carlberg,M., Hu,G.Z. and Nehlin,J.O. (1991) Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol., 11, 4876–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein S., Berkower,C. and Michaelis,S. (1994) Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase and demonstration of its essential role in a-factor export. Mol. Cell. Biol., 14, 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Beck,T., Koller,A., Kunz,J. and Hall,M.N. (1998) The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J., 17, 6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Yang,H., Hallberg,E. and Hallberg,R. (1997) Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol., 17, 3242–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon A.A., Cohen,P.T. and Stark,M.J. (1990) Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J., 9, 4339–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J.B., Clarke,S. and Koshland,D.E.,Jr (1984) The protein carboxylmethyltransferase involved in Escherichia coli and Salmonella typhimurium chemotaxis. Methods Enzymol., 106, 310–321. [DOI] [PubMed] [Google Scholar]
- Tolstykh T., Lee,J., Vafai,S. and Stock,J. (2000) Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. EMBO J., 19, 5682–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl W., Huang,W., Sneddon,A.A., Stark,M., Camier,S., Werner,M., Marck,C., Sentenac,A. and Broach,J.R. (1992) Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 4946–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup D.M. (2000) Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol., 12, 180–185. [DOI] [PubMed] [Google Scholar]
- Wang Y. and Burke,D.J. (1997) Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 620–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wera S. and Hemmings,B.A. (1995) Serine/threonine protein phosphatases. Biochem. J., 311, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H. and Clarke,S. (1994) Protein phosphatase 2A is reversibly modified by methyl esterification at its C-terminal leucine residue in bovine brain. J. Biol. Chem., 269, 1981–1984. [PubMed] [Google Scholar]
- Zolnierowicz S., Van Hoof,C., Andjelkovic,N., Cron,P., Stevens,I., Merlevede,W., Goris,J. and Hemmings,B.A. (1996) The variable subunit associated with protein phosphatase 2A0 defines a novel multimember family of regulatory subunits. Biochem. J., 317, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]