Nud1p links astral microtubule organization and the control of exit from mitosis (original) (raw)
Abstract
The budding yeast spindle pole body (SPB) not only organizes the astral and nuclear microtubules but is also associated with a number of cell-cycle regulators that control mitotic exit. Here, we describe that the core SPB component Nud1p is a key protein that functions in both processes. The astral microtubule organizing function of Nud1p is mediated by its interaction with the γ-tubulin complex binding protein Spc72p. This function of Nud1p is distinct from its role in cell-cycle control: Nud1p binds the spindle checkpoint control proteins Bfa1p and Bub2p to the SPB, and is part of the mitotic exit network (MEN) in which it functions upstream of CDC15 but downstream of LTE1. In conditional lethal nud1-2 cells, the MEN component Tem1p, a GTPase, is mislocalized, whereas the kinase Cdc15p is still associated with the SPB. Thus, in nud1-2 cells the failure of Tem1p to interact with Cdc15p at the SPB probably prevents mitotic exit.
Keywords: Cdc15p/mitotic exit network/Nud1p/spindle pole body/Tem1p
Introduction
In Saccharomyces cerevisiae, nuclear and astral microtubules (AMs) are nucleated by the yeast microtubule organizing centre, the spindle pole body (SPB) (Byers and Goetsch, 1975). Nuclear microtubules segregate the duplicated chromosomes in mitosis (Jacobs et al., 1988) while the AMs play a role in orientating the spindle along the length axis of the cell and are required for the migration of the nucleus into the daughter cell body, the bud, during mitosis (Palmer et al., 1992).
At the molecular level, both sets of microtubules are organized by the yeast γ-tubulin complex (Geissler et al., 1996; Marschall et al., 1996; Spang et al., 1996). γ-tubulin complex binding proteins target the γ-tubulin complex to SPB substructures (Knop et al., 1999a) and thereby determine where microtubules become nucleated. Spc110p binds the γ-tubulin complex to the nuclear and Spc72p to the cytoplasmic side of the SPB (Knop and Schiebel, 1997, 1998).
Whereas all nuclear microtubules originate from the SPB inner plaque, AMs can be nucleated from two distinct sites at the cytoplasmic face of the SPB (Byers and Goetsch, 1975; Brachat et al., 1998): the outer plaque, which is the layer of the SPB facing the cytoplasm, and the half-bridge, which is an asymmetric extension of the SPB that projects onto the nuclear envelope. During the G1 phase of the cell cycle, the cytoplasmic microtubules were found to emanate predominantly from the half-bridge whereas in the other phases of the cell cycle they originate from the outer plaque. Finally, in mating cells, AMs originate exclusively from the half-bridge (Byers and Goetsch, 1975; Brachat et al., 1998).
Spc72p shows the same distribution between the SPB outer plaque and the half-bridge throughout the cell cycle as the AMs (Adams and Kilmartin, 1999; Pereira et al., 1999). Thus, alterations in the cell-cycle-dependent binding of Spc72p to SPB substructures are the molecular basis for the changes in AM organization by the SPB. We were recently able to demonstrate that the half-bridge protein Kar1p interacts with Spc72p during karyogamy and G1 phase, leading to nucleation of AMs from the half-bridge (Pereira et al., 1999). This leaves open the question of which SPB component interacts with Spc72p at the outer plaque in the other phases of the cell cycle.
Astral and nuclear microtubule functions are monitored by spindle checkpoints that coordinate kinetochore attachment to microtubules with the onset of anaphase (Mad2p checkpoint) (Hwang et al., 1998) and the migration of the nucleus into the bud with mitotic exit (Bub2p checkpoint) (Bardin et al., 2000; Pereira et al., 2000). Exit from mitosis requires inactivation of B-type cyclin-dependent kinase (Cdk) activity by the ubiquitin-ligase anaphase-promoting complex (APC) and the proteasome (Zachariae et al., 1996). It is not fully clear what cue finally triggers the exit from mitosis and initiates Cdk inactivation, although a network of proteins has been identified that concertedly regulates these events (reviewed by Balasubramanian et al., 2000). The components of this network are often referred to as the mitotic exit network (MEN), and include the small GTPase Tem1p (Shirayama et al., 1994b), its GDP/GTP exchange factor Lte1p (Shirayama et al., 1994a), the protein phosphatase Cdc14p (Shou et al., 1999; Visintin et al., 1999) and several kinases such as Cdc15p (Schweitzer and Philippsen, 1991) and Cdc5p (Shirayama et al., 1998).
Activation of Tem1p by Lte1p ultimately leads to activation of Cdc15p followed by the release of Cdc14p from the nucleolus, which then initiates inactivation of Cdk activity by several mechanisms (Jaspersen et al., 1998; Shou et al., 1999; Visintin et al., 1999; Bardin et al., 2000; Pereira et al., 2000). Yeast strains harbouring temperature-sensitive mutations in any of the MEN genes show a common phenotype at the restrictive temperature: they arrest in late anaphase with separated nuclei, are unable to conclude cell division and still sequester the phosphatase Cdc14p in the nucleolus (Shou et al., 1999; Visintin et al., 1999). It seems that the SPB has an important but ill-understood role in the late events of mitosis since many of the MEN components such as Tem1p (Bardin et al., 2000; Pereira et al., 2000), Cdc15p (Cenamor et al., 1999; Xu et al., 2000) and Cdc5p (Shirayama et al., 1998) localize to the SPB.
We report that the SPB component Nud1p (Adams and Kilmartin, 1999; Elliott et al., 1999) has a role in AM organization by binding Spc72p to the outer plaque in a cell-cycle-dependent manner. In addition, Nud1p binds Bfa1p and Bub2p, the GAP of Tem1p, to the SPB, and is required for mitotic exit by facilitating the binding of Tem1p to Cdc15p.
Results
Nud1p interacts with Spc72p
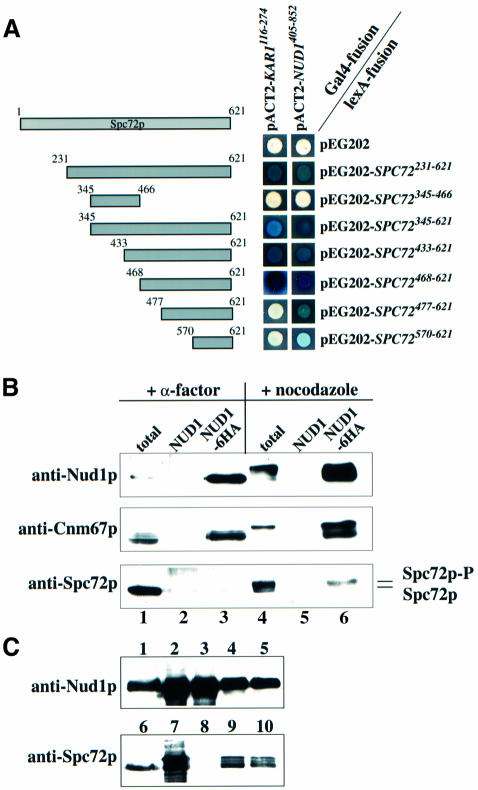
We have recently shown that Spc72p interacts with Kar1p during the G1 phase of the cell cycle and during karyogamy (Pereira et al., 1999). In order to understand how Spc72p binds to the outer plaque in other phases of the cell cycle, known outer plaque components were screened for two-hybrid interactions with Spc72p. A specific two-hybrid interaction was detected between the C-terminus of Spc72p and C-Nud1p (Figure 1A). Nud1p has been described previously as a core component of the outer plaque that interacts with Cnm67p (Brachat et al., 1998; Adams and Kilmartin, 1999; Elliott et al., 1999). As for Kar1p (Pereira et al., 1999), the interaction with Nud1p is mediated by the C-terminus of Spc72p. But since amino acids 468–477 of Spc72p are required for it to bind Kar1p, whereas this region seems to be less important for the interaction with Nud1p (Figure 1A), it is likely that the Nud1p and Kar1p binding sites of Spc72p are distinct but do overlap.
Fig. 1. Spc72p interacts with Nud1p. (A) Spc72p and Nud1p interact in the yeast two-hybrid system. Fragments of SPC72 subcloned into plasmid pEG202 were co-transformed into yeast strain SGY37 with pACT2 containing either KAR1 (codons 116–274) or NUD1 (codons 405–852). (B) Spc72p co-precipitates with Nud1p. NUD1 (lanes 2 and 5) and NUD1-6HA (lanes 1, 3, 4 and 6) cells were grown to early logarithmic phase and arrested with either α-factor or nocodazole. Nud1p-6HA was immunoprecipitated using anti-HA beads. Spc72p and Cnm67p in the immunoprecipitates were detected by immunoblotting. Lanes 1 and 4 show the total extract before immunoprecipitation, and lanes 2, 3, 5 and 6 the immunoprecipitates. Lanes 1–3 were loaded with four times more extract than lanes 4–6 to make up for the fact that NUD1 is expressed at much lower levels in pheromone-treated cells (Figure 2B). (C) Insect cells were transfected with either SPC72 baculovirus, _NUD1-His_6 baculovirus or both. Cells were harvested 72 h post-infection, lysed and incubated with TALON beads. Shown are yeast extracts (lanes 1 and 6), total lysates (lanes 2, 4, 7 and 9) and TALON-precipitates (lanes 3, 5, 8 and 10) of cells transfected with _NUD1-His_6 (lanes 2 and 3), SPC72 (lanes 7 and 8), and SPC72 and _NUD1-His_6 (lanes 4, 5, 9 and 10). Note that no protein was detected by the anti-Nud1p or anti-Spc72p antibodies in untransfected insect cells (not shown).
In order to corroborate the two-hybrid data showing an interaction between Spc72p and Nud1p, co-immunoprecipitation studies were carried out. A yeast strain carrying a haemagglutinin (HA)-tagged version of NUD1 (NUD1-6HA) was arrested either in G1 phase with the mating pheromone α-factor or in metaphase with the microtubule-depolymerizing drug nocodazole. Nud1p-6HA was precipitated using anti-HA beads and the precipitate analysed for the presence of Spc72p or Cnm67p, the known Nud1p interactors. Figure 1B shows that Nud1p-6HA was associated with Cnm67p in G1-arrested (+ α-factor, NUD1-6HA) and metaphase-arrested (+ nocodazole, NUD1-6HA) cells but Spc72p co-precipitated with Nud1p-6HA only in cells arrested in metaphase (compare Nud1p-6HA precipitation of cells incubated with α-factor or nocodazole). We noticed that only a slower migrating, phosphorylated form (Figure 2C) of Spc72p co-precipitated with Nud1p-6HA (Figure 1B; + nocodazole, NUD1-6HA), suggesting that Nud1p associates preferentially with phosphorylated Spc72p (see below). These precipitations were found to be specific, since Spc72p and Cnm67p were not detected in the anti-HA precipitate of the NUD1 control strain (Figure 1B; NUD1). Since Spc72p was co-precipitated less efficiently in comparison with Cnm67p we conclude that either not every molecule of Nud1p is associated with Spc72p or the interaction is unstable under our conditions.
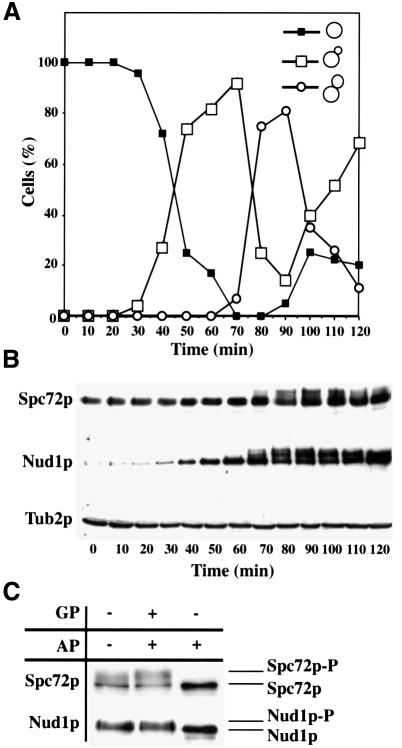
Fig. 2. Spc72p and Nud1p are phosphorylated in a cell-cycle-dependent manner. Samples of α-factor-synchronized cells were taken every 10 min and analysed for (A) budding index and (B) Spc72p, Nud1p and β-tubulin by immunoblotting. β-tubulin was used as loading control. (C) Spc72p-3HA or Nud1p-6HA was precipitated from cell extracts and treated without any addition, with alkaline phosphatase (AP) or alkaline phosphatase and β-glycerolphosphate (GP). Samples were analysed by immunoblotting.
To find out whether Nud1p and Spc72p interact directly, Spc72p and Nud1p-His6 were expressed in insect cells using the baculovirus expression system. Nud1p-His6 but not Spc72p expressed on its own could be precipitated with metal beads (Figure 1C, lanes 3 and 8). However, when both proteins were expressed together, Nud1p-His6 and Spc72p were found on the beads (lanes 5 and 10). In summary, Spc72p binds directly to Nud1p.
Spc72p and Nud1p are phosphorylated at the same point in the cell cycle
We investigated whether Nud1p and Spc72p are subject to cell-cycle-dependent modifications. Nud1p and Spc72p of a synchronized culture were analysed by immunoblotting. With the appearance of cells with buds of medium size (Figure 2A) in the S/G2 phase of the cell cycle, both proteins were separated into multiple bands (Figure 2B, 70 min). To establish that the slower migrating forms of Nud1p and Spc72p were due to phosphorylation, Nud1p-6HA or Spc72p-3HA was immunoprecipitated from the cell extracts and the precipitate treated with alkaline phosphatase (AP) with or without the phosphatase inhibitor β-glycerophosphate (GP). Upon AP treatment the bands detected by the antisera collapsed to a single species, indicating that Nud1p and Spc72p were phosphorylated (Figure 2C). However, dephosphorylation of Spc72p and Nud1p by AP was inhibited when GP was added. In conclusion, Spc72p and Nud1p are phosphorylated from S/G2 phase until the end of mitosis.
Overexpression of C-Nud1p leads to microtubule defects and cell-cycle arrest
Since the C-terminus of Nud1p is responsible for the interaction of Nud1p with Spc72p, we reasoned that overexpression of this domain of NUD1 (C-NUD1) might disrupt the interaction between endogenous Nud1p and Spc72p. An expression construct containing N-terminally HA-tagged C-NUD1 (HA-C-NUD1; codons 406–852) under the control of the galactose-inducible Gal1 promoter was generated. Expression of the protein upon addition of galactose was confirmed by anti-HA immunoblotting (Figure 3A; compare lanes 1 and 2 with 3 and 4). Continuous overexpression of HA-C-NUD1 was toxic to the cells (Figure 3B, sector 3; compare glucose and galactose plates). In contrast, overexpression of N-NUD1 (codons 1–406) had no effect on growth of cells (Figure 3B, sector 2); neither did overexpression of full-length NUD1 [Figure 3C; compare sectors 1 (wild type) and 2 (Gal1-NUD1)]. We established by immunoblotting with anti-Nud1p antibodies that N-Nud1p and Nud1p were overexpressed upon the addition of galactose (not shown).
Fig. 3. Overexpression of C-NUD1 interferes with AM organization and cell-cycle progression. (A) Cells of SPC72–GFP (UGY151, lanes 1 and 3) and NUD1–GFP (UGY153, lanes 2 and 4) with Gal1-HA-C-NUD1 were grown to early logarithmic phase in medium containing 3% raffinose. Galactose (4%) was then added to induce the Gal1 promoter. Samples were taken before (lanes 1 and 2) and 6 h after addition of galactose (lanes 3 and 4) and analysed by immunoblotting with anti-HA antibodies for the expression of HA-C-NUD1. Markers are bovine serum albumin (BSA) (66 kDa) and ovalbumin (45 kDa). (B) Yeast cells (YPH499) were transformed with vector p416-Gal1 (sector 1), p416-Gal1-N-NUD1 (sector 2) and p416-Gal1-C-NUD1 (sector 3) and grown on either glucose or galactose plates for 2 days at 30°C. (C) Diploid wild-type cells (FY1679, sector 1) or diploid cells containing one chromosomal copy of Gal1-NUD1 (UGY194, sector 2) were grown as in (B). (D) Cells of (A) were analysed for SPB-localized GFP fluorescence or (E) processed for indirect anti-tubulin immunofluorescence. DNA was stained with DAPI. In order to facilitate comparisons, control cells (–Gal) in anaphase were selected. The arrows indicate detached microtubules. (D and E) Bars, 5 µm. (F) The number of GFP-labelled SPB signals was scored before and 6 h after the addition of galactose to cells of SPC72–GFP, NUD1–GFP and CNM67–GFP with Gal1-C-NUD1 (n = 200).
In order to analyse the effects of overexpression of C-NUD1 on endogenous Spc72p or Nud1p, yeast strains containing either chromosomally green fluorescent protein (GFP)-tagged NUD1 (NUD1–GFP) or SPC72–GFP were transformed with Gal1-C-NUD1. After 6 h induction of Gal1-C-NUD1 with galactose, the number of SPBs displaying a GFP signal for Spc72p or Nud1p was strongly reduced (Figure 3D and F), indicating that C-Nud1p affects the SPB localization of these outer plaque components. However, the SPB localization of Cnm67p, another outer plaque component, was not influenced by the overexpression of C-NUD1 (Figure 3F), arguing against a general disruption of the outer plaque upon overexpression of Gal1-C-NUD1.
Mutations that affect the outer plaque severely impair AM organization (Brachat et al., 1998; Knop and Schiebel, 1998). Indeed, indirect tubulin immunofluorescence of Gal1-C-NUD1 cells after 6 h induction with galactose revealed that AMs were frequently detached from the SPB (Figure 3E; indicated by arrows). We also noted that upon overexpression of C-NUD1, the cells arrested with anaphase spindles with separated nuclei (Figure 3E; +Gal). This cell-cycle arrest was not due to checkpoint activation since overexpression of C-NUD1 gave similar results in cells deleted for MAD2 or BUB2 (not shown). These data indicate that overexpression of C-NUD1 interferes with both AM organization and exit out of mitosis.
Mutations in NUD1 lead to microtubule detachment and cell-cycle arrest
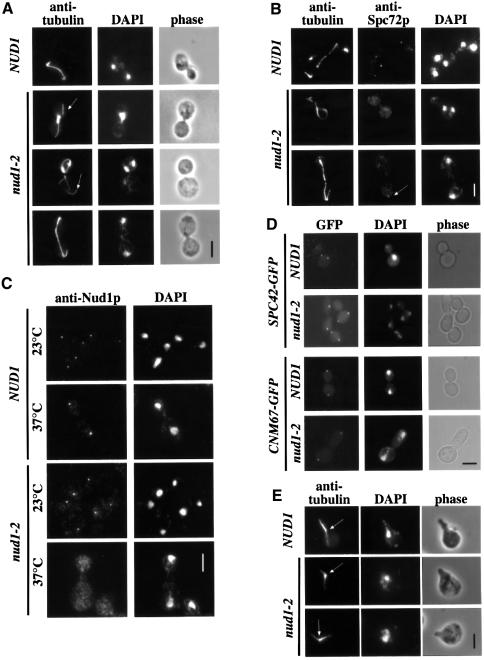
Mutations in KAR1 that affect the interaction between Spc72p and Kar1p lead to microtubule detachment from the SPB when the cells are treated with the mating pheromone α-factor (Vallen et al., 1992; Pereira et al., 1999). We wondered whether disrupting NUD1 function would result in a similar phenotype in mitotic cells. Strikingly, 90% of the conditional lethal nud1-2 cells arrested at 37°C as large-budded cells, of which ∼10–20% had an elongated bud (Figure 4D; bottom panel). The remaining 10% of cells were either small budded or without bud. This result was independent of whether the cells were synchronized with α-factor first, shifted as an asynchronous culture, or the presence of the BUB2 or MAD2 checkpoint control genes (not shown).
Fig. 4. Analysis of the phenotype of nud1-2 cells. (A–C) NUD1 and nud1-2 cells were shifted to 37°C for 3 h and then processed for (A) anti-tubulin, (B) anti-tubulin and anti-Spc72p, or (C) anti-Nud1p immunofluorescence. (A) Detached AMs are indicated by arrows. (B) Residual Spc72p-SPB staining can be observed in some nud1-2 cells (arrow). (D) Spc42p and Cnm67p localization are not affected in nud1-2 cells. SPC42–GFP NUD1 (UGY196), SPC42–GFP nud1-2 (UGY199), CNM67–GFP NUD1 (UGY198) and CNM67–GFP nud1-2 cells (UGY200) were shifted to 37°C for 3 h and inspected by phase contrast and fluorescence microscopy. (E) Pheromone-arrested nud1-2 cells have normal AM arrays at 37°C. NUD1 and nud1-2 cells were treated with α-factor for 2.5 h at 23°C and then shifted to 37°C for 30 min in the continued presence of α-factor. Microtubules were visualized using anti-tubulin antibodies. The arrows indicate the positions of the SPBs. Note also that nud1-2 cells shifted for only 30 min to 37°C showed a profound AM organization defect (not shown). (A–E) DNA was stained with DAPI. Bars, 5 µm.
Anti-tubulin immunofluorescence revealed that the AMs were severely disturbed. In ∼80% of large-budded cells, the AMs appeared to be detached from the SPB, and severed microtubule bundles were found in the cytoplasm (Figure 4A; arrows). In the residual cells, no clear AM defect was apparent (Figure 4A; bottom panel). Despite this AM defect, ∼70% of the large-budded cells had a long spindle and properly separated nuclei (Figure 4A; bottom panel). Of the remaining cells, 28% displayed a nuclear migration failure with the misoriented spindle being retained in the mother cell body (Figure 4A; nud1-2, middle panel) similar to Δkar9 or spc72(ts) cells, other mutants with AM defects (Knop and Schiebel, 1998; Miller and Rose, 1998). Approximately 2% of cells displayed a short spindle situated in the bud neck region (Figure 2A; nud1-2, top panel).
We then investigated whether the phenotype of nud1-2 cells is caused by a partial disintegration of the outer plaque. Indirect immunofluorescence with anti-Spc72p antibodies revealed that the normally focused Spc72p signal at the SPB (Figure 4B; NUD1) was strongly reduced in 80% of the nud1-2 cells incubated at 37°C, leading to high cytoplasmic staining (Figure 4B; middle panel). However, in ∼20% of nud1-2 cells a weak Spc72p signal was still observed at the SPB (Figure 4B; bottom, arrow). These results could indicate either that the interaction of the mutant Nud1p protein with Spc72p is reduced or that Nud1p-2 itself is released from the SPB. In order to investigate this point, NUD1 and nud1-2 cells were processed for indirect immunofluorescence using anti-Nud1p antibodies. In NUD1 cells (Figure 4C; NUD1) and in nud1-2 cells (Figure 4C; nud1-2) at 23°C, clear SPB staining was observed, although in the latter case a slightly higher staining in the cytoplasm was noted. In nud1-2 cells shifted to 37°C for 3 h, most cells displayed a dispersed cytoplasmic staining pattern for Nud1p (Figure 4C; bottom), and only very few cells still showed a distinct but weak SPB staining (not shown). In contrast, the SPB component Spc42p–GFP, which interacts with Cnm67p towards the centre of the SPB (Adams and Kilmartin, 1999; Elliott et al., 1999), and the outer plaque component Cnm67p–GFP still displayed normal SPB localization at the non-permissive temperature in nud1-2 cells (Figure 4D). Electron microscopy confirmed that large-budded nud1-2 cells (n = 20) (Figure 5B) contained a strongly reduced, misformed outer plaque that was not associated with AMs. This is a clear distinction from large-budded wild-type NUD1 cells (Figure 5A) in which the SPB has a dense outer plaque with attached AMs. In some nud1-2 cells (n = 5) (Figure 5B), AMs emanated from the half-bridge, an indication that the half-bridge microtubules were not affected by the nud1-2 mutation. In conclusion, the mutated Nud1p-2 probably loses interaction with Cnm67p at 37°C, resulting in the partial disintegration of the outer plaque.
Fig. 5. nud1-2 cells have a defective outer plaque. (A) NUD1 and (B) nud1-2 cells were incubated for 3 h at 37°C. Samples were prepared for thin section electron microscopy. (A) In NUD1 cells the cytoplasmic outer plaque (OP), the central plaque (CP) and AMs emanating from the outer plaque are clearly detectable. (B) In nud1-2 cells the outer plaque was misformed and reduced. AMs were associated with the half-bridge of some nud1-2 cells. AM, astral microtubules; CP, central plaque; HB, half-bridge; NE, nuclear envelope; NM, nuclear microtubules; OP, outer plaque. Bar, 120 nm.
Spc72p is bound to the cytoplasmic side of the SPB via Nud1p from S phase until the end of mitosis, whereas it interacts with Kar1p in G1 phase and mating cells (Pereira et al., 1999). Thus, Nud1p should not have a role in AM organization in mating cells. To test this notion, nud1-2 cells were treated with α-factor at 23°C, which induces cell-cycle arrest, AM reorganization to the half-bridge and shmoo formation (Rose, 1996; Pereira et al., 1999). nud1-2 cells were then shifted to 37°C in the continued presence of α-factor. Whereas AMs detached or depolymerized under these conditions in spc72(ts) and kar1(ts) cells (Pereira et al., 1999), no such phenotype was observed in nud1-2 cells. Instead, AMs (Figure 4E; nud1-2) still originated from the SPB [Figure 4E; localized between the astral and nuclear microtubules (marked with arrow)] and contacted the shmoo tip as in wild-type cells (Figure 4E, top). This observation is in keeping with Spc72p interacting with Kar1p and not Nud1p in pheromone-treated cells.
The nuclear migration defect of nud1-2 cells is alleviated by an SPC72–CNM67 fusion protein
We investigated whether the AM organization defect and the cell-cycle arrest phenotype of nud1-2 cells were interdependent. For this study, we introduced a gene fusion of the γ-tubulin complex binding domain of Spc72p (codons 1–276) with Cnm67p (codons 1–581) (Spc72– Cnm67p) into nud1-2 cells. We reasoned that Spc72– Cnm67p will bind the γ-tubulin complex to the cytoplasmic side of the SPB without the need for Nud1p and Spc72p. Thus, Spc72–Cnm67p should bypass the requirement of Nud1p for AM organization.
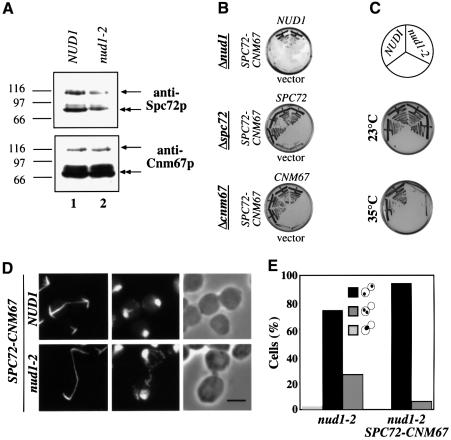
Spc72–Cnm67p was detected by the anti-Spc72p and anti-Cnm67p antibodies as a protein of ∼100 kDa (Figure 6A, arrows), in addition to the endogenous proteins (double arrows). SPC72–CNM67 complemented both the Δspc72 and Δcnm67 but not Δnud1 cells (Figure 6B). Also, the temperature-sensitive growth defect of nud1-2 cells was not rescued by SPC72–CNM67 (Figure 6C).
Fig. 6. An SPC72–CNM67 gene fusion alleviates the nuclear migration defect of nud1-2 cells. (A) Cell extracts of SPC72–CNM67 NUD1 (UGY217; lane 1) or SPC72–CNM67 nud1-2 (UGY215; lane 2) cells were prepared and analysed by immunoblotting using anti-Spc72p or anti-Cnm67p antibodies. The arrows indicate the Spc72–Cnm67p fusion protein (100 kDa), the double arrows the endogenous Spc72p or Cnm67p. Markers: β-galactosidase (116 kDa), phosphorylase b (97 kDa) and BSA (66 kDa). (B) SPC72–CNM67 complements for SPC72 or CNM67 but not for NUD1. Cells of Δnud1 pRS316-NUD1 (UGY111), Δspc72 pRS316-SPC72 (ESM448) or Δcnm67 pRS316-CNM67 (ESM431) were transformed with pRS315-NUD1, pRS315-SPC72 or pRS315-CNM67, as indicated, pRS315-SPC72–CNM67 or pRS315 (vector). Cells were grown at 23°C on 5-fluoroorotic (5-FOA) plates, which select against the _URA3_-based pRS316 plasmids. Growth indicates complementation. Transformants were tested in duplicate. (C) SPC72–CNM67 does not rescue nud1-2 cells. Cells of SPC72–CNM67 NUD1 or SPC72–CNM67 nud1-2 were grown at 23 or 35°C for 3 days. (D) SPC72–CNM67 NUD1 and SPC72–CNM67 nud1-2 cells were shifted to 37°C for 3 h and processed for indirect tubulin immunofluorescence. DNA was stained with DAPI. Bar, 5 µm. (E) Quantitative analysis of the nuclear migration defect and cell-cycle arrest of nud1-2 and SPC72–CNM67 nud1-2 cells of (D). Only large-budded cells (90% of the population) were counted. The dots within the symbolized cells indicate the position of the DAPI-staining regions.
In order to assess the effect of SPC72–CNM67 on AM organization, nud1-2 cells were shifted to the restrictive temperature and indirect anti-tubulin immunofluorescence was performed. AMs of nud1-2 SPC72–CNM67 cells (Figure 6D; nud1-2) appeared similar to wild-type (Figure 6D; NUD1), Δcnm67 or Δspc72 cells (not shown) carrying SPC72–CNM67. Consequently, the nuclear migration defect of nud1-2 cells was reduced from 30 to 6% through SPC72–CNM67 (Figure 6E). However, 90% of nud1-2 SPC72–CNM67 cells still arrested in the cell cycle in anaphase (Figure 6D and E), indicating that the cell-cycle arrest phenotype of nud1-2 cells is not a direct consequence of the AM defect. In summary, introduction of a functional SPC72–CNM67 gene fusion into nud1-2 cells alleviates the nuclear migration defect but does not rescue the mitotic exit defect.
nud1-2 cells display a MEN phenotype
The cell-cycle arrest of nud1-2 cells in late anaphase prompted us to investigate whether Nud1p could have a role in the regulation of mitotic exit. Mutants in components of the MEN are characterized by an anaphase cell-cycle arrest with 2N DNA content, stable Clb2p levels but degradation of the anaphase inhibitor Pds1p (Jaspersen et al., 1998). In addition, mutants of the MEN are suppressed by high gene dosage of the Cdk inhibitor SIC1 (Schwob et al., 1994; Jaspersen et al., 1998) and MEN mutants do not release the phosphatase Cdc14p from the nucleolus (Shou et al., 1999; Visintin et al., 1999).
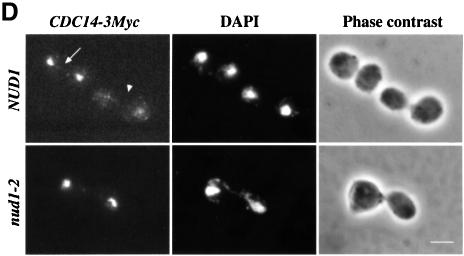
We decided to analyse whether these criteria are fulfilled by the nud1-2 cells. α-factor-synchronized nud1-2 cells replicate their DNA as wild-type NUD1 cells but then arrest with 2N DNA content (Figure 7A) as large-budded cells (Figure 7B; plateau of large-budded cells after 120 min). Analysis of the Clb2p and Pds1p protein content revealed that nud1-2 cells behaved similarly to other MEN mutants. Whereas in NUD1 cells the cycling behaviour of Clb2p was clearly evident with a peak at 90 min and another one 60 min later, Clb2p proteins started accumulating in nud1-2 cells at 90 min and remained stable until the end of the experiment (Figure 7C). Furthermore, in wild-type NUD1 cells, Pds1p was periodically degraded (Figure 7C; NUD1, 60 and 150 min) whereas in nud1-2 cells Pds1p was degraded after 60–90 min and then stayed low (Figure 7C). In addition, high gene dosage of SIC1 (2 µm plasmid) suppressed the growth defect of nud1-2 cells at 35°C (Table I). Finally, indirect anti-Myc immunofluorescence revealed that Cdc14p-3Myc remains sequestered in the nucleolus of 99% of large-budded nud1-2 cells at the restrictive temperature (Figure 7D, nud1-2) but was released into the nucleus and cytoplasm of wild-type cells at anaphase (Figure 7D; NUD1, arrowhead). These phenotypes of nud1-2 cells are typical for MEN mutants (Jaspersen et al., 1998).
Fig. 7. nud1-2 cells exhibit a phenotype similar to MEN mutants. α-factor-synchronized NUD1 and nud1-2 cells with CLB2-6HA or PDS1-6HA were released into 37°C pre-warmed medium. Samples were taken every 30 min and (A) analysed for DNA content by FACS analysis, (B) budding index or (C) Pds1p-6HA and Clb2p-6HA by immunoblotting. Tub2p was detected as loading control. (D) NUD1 and nud1-2 cells carrying CDC14-3Myc were shifted to 37°C for 2 h and processed for indirect immunofluorescence using anti-Myc antibodies. DNA was stained with DAPI. The upper panel shows two large-budded wild-type cells, one of which has already released Cdc14p into the nucleus and cytoplasm (arrowhead) and the other one in which Cdc14p is still held in the nucleolus (arrow). Out of 200 large-budded nud1-2 CDC14-3Myc cells, only three were found in which Cdc14p had been released from the nucleolus. Cdc14p-3Myc was still retained in the nucleolus when nud1-2 cells were incubated for 3 or 4 h at 37°C. Bar, 5 µm.
Table I. Suppression of nud1-2 and Δlte1 cells by components of the MEN and SIC1.
| | Growth at the restrictive temperature | | | | | | | | | | ---------------------------------------- | ----------- | ----------- | ----------- | ------------ | ----------- | --------- | ----------- | -- | | | Control vector | Gal1-NUD1 | Gal1-LTE1 | Gal1-TEM1 | Gal1-CDC15 | Gal1-CDC5 | 2µ-SIC1 | CEN-CDC14 | | | nud1-2 | – | +++ | – | – | + | +/– | +++ | + | | Δlte1 | – | +++ | +++ | +++ | +++ | ND | ND | ND |
Genetic interaction of NUD1 with CDC14, CDC15, LTE1, SPO12 and TEM1
Components of the MEN show multiple genetic interactions including high copy suppression of the conditional lethal growth defect (Jaspersen et al., 1998). To establish further that NUD1 is part of the MEN, we tested for suppression of the nud1-2 phenotype by other components of the MEN. The data are summarized in Table I. Most efficient suppression of the nud1-2 mutation was achieved by Gal1-SPO12. SPO12 is a gene with an unknown function in mitosis. It has been described previously as a suppressor of MEN mutants (Jaspersen et al., 1998). In addition, a weak suppression of nud1-2 cells was observed by Gal1-CDC15 and Gal1-CDC5. In contrast, TEM1, which functions upstream of CDC15 (Shirayama et al., 1994b; Jaspersen et al., 1998), did not relieve the growth defect of nud1-2 cells upon overexpression. We also noticed that CDC14 on a centromere-based plasmid suppressed the growth defect of nud1-2 cells. Stronger expression of CDC14 (2 µm plasmid) was toxic for nud1-2 cells, explaining why 2 µm-CDC14 did not allow growth of nud1-2 cells at the restrictive temperature (not shown).
Further, we discovered overexpression of Gal1-NUD1 did not suppress the growth defect of cdc15-1 or tem1-3 cells (not shown). However, the cold sensitivity of Δlte1 cells was suppressed by Gal-TEM1 (Shirayama et al., 1994b; Table I), Gal1-CDC15 and Gal1-NUD1 (Table I), indicating that NUD1 functions downstream of LTE1. When taken together, our data suggest that NUD1 is a component of the MEN that functions upstream of CDC15 but downstream of LTE1.
Components of the MEN are mislocalized in nud1-2 cells
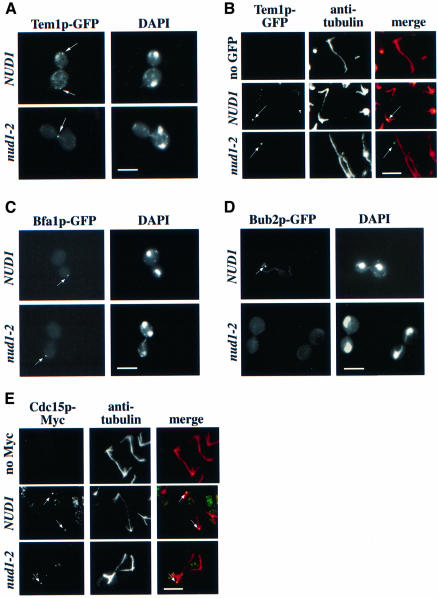
In order to assess the cell-cycle arrest of nud1-2 cells more closely we analysed the localization of the MEN component Tem1p and Cdc15p as well as of the mitotic checkpoint components Bfa1p and Bub2p in nud1-2 cells at 37°C. In wild-type cells, Tem1p is associated with the SPB that enters the bud throughout most of the cell cycle in a manner dependent on Bfa1p and Bub2p, the two components of the GAP of Tem1p. However, in late anaphase or telophase, Tem1p is bound to the SPB independently of Bfa1p and Bub2p (Pereira et al., 2000). Cdc15p is probably found at one SPB in early anaphase but at both in late anaphase (Cenamor et al., 1999; Xu et al., 2000).
Tem1p–GFP of nud1-2 cells was still dot-like but was not localized with the DAPI-staining region (Figure 8A; nud1-2, arrow), as was the case in NUD1 wild-type cells (Figure 8A; NUD1, arrow). Labelling with anti-tubulin antibodies and confocal microscopy confirmed that the Tem1p–GFP signal was not SPB associated (Figure 8B; compare NUD1 with nud1-2 cells); rather, it resided at or close to the cell cortex (Figure 8B; nud1-2). Similarly, we observed mislocalization of the Bfa1p–GFP signal (Figure 8C; nud1-2), whereas the Bub2p–GFP signal was undetectable in 94% of the nud1-2 cells at the restrictive temperature (Figure 8D). We generated a BFA1–GFP TEM1-9Myc nud1-2 strain in order to prove co-localization of Bfa1p and Tem1p to the same dot-like structure at the non-permissive temperature. We observed co-localization of Bfa1p and Tem1p in nud1-2 cells incubated at the non-permissive temperature. However, the BFA1–GFP TEM1-9Myc nud1-2 cells were already sick at the permissive temperature, presumably caused by the additional disturbance of the protein complex by the double tag (not shown). Taken together, these results suggest that in nud1-2 cells the interaction between Bub2p, Bfa1p and Tem1p and the SPB is disturbed. The detached Bfa1p–Tem1p complex seems to remain stable under these conditions whereas Bub2p becomes dispersed.
Fig. 8. Localization of MEN components in nud1-2 cells. (A) Tem1p–GFP is mislocalized in nud1-2 mutants. TEM1–GFP NUD1 (UGY218) or TEM1–GFP nud1-2 cells (UGY201) were synchronized with α-factor and then shifted to 37°C. The cells were fixed after 105 min when most of the TEM1–GFP cells reached anaphase. The Tem1p–GFP signal was inspected by fluorescence microscopy. (B) Cells of (A) were analysed by anti-tubulin staining and confocal microscopy. In NUD1 cells, Tem1p–GFP co-localized with the SPB whereas in nud1-2 cells the Tem1p–GFP signal (green) was distinct from the tubulin staining (red). Note that the overlap of the green and red signals results in a yellow dot. (C) The Bfa1p–GFP signal is mislocalized in nud1-2 cells. Cells of BFA1–GFP NUD1 and BFA1–GFP nud1-2 were shifted to 37°C for 3 h and then analysed by fluorescence microscopy. (D) The Bub2p–GFP signal is lost in nud1-2 cells. Cells of BUB2–GFP NUD1 and BUB2–GFP nud1-2 were treated as in (C). (E) nud1-2 cells in late anaphase exhibit only one SPB-associated Cdc15p-9Myc signal. NUD1 and nud1-2 cells carrying CDC15–9Myc were grown to early logarithmic phase. Cells were then shifted to 37°C for 3 h and processed for tubulin immunofluorescence. (A–E) The arrows indicate the GFP and 9Myc signals. When indicated, DNA was stained with DAPI. Bars, 5 µm.
Finally, we investigated the localization of chromosomally Myc-tagged CDC15-9Myc in nud1-2 cells. As reported before for Cdc15p–GFP (Cenamor et al., 1999; Xu et al., 2000), endogenous Cdc15p-9Myc was associated with one SPB in early anaphase (not shown) but with both SPBs in anaphase of wild-type cells (Figure 8E; NUD1). In contrast, in 80% of nud1-2 cells the Cdc15p-9Myc signal was associated with the SPB in the bud (Figure 8E) as indicated by the co-localization of the anti-tubulin and the Cdc15p-9Myc signal (Figure 8E, the merged signals appear yellow). In the residual 20% of cells, the Cdc15p-9Myc signal was mislocalized (not shown). In summary, Cdc15p but not Bfa1p, Bub2p or Tem1p is still associated with the SPB in the majority of nud1-2 cells.
Nud1p interacts with Bub2p
The loss of Bub2p–GFP signal and the mislocalization of Bfa1p and Tem1p in nud1-2 cells led us to test whether any of these components interact with Nud1p in the yeast two-hybrid system. A strong two-hybrid interaction was found between Nud1p405–852 and Bub2p (Figure 9A). This result was confirmed by co-immunoprecipitation of Nud1p with Bub2p-3HA (Figure 9B, lane 6). In addition, co-immunoprecipitation of Nud1p with Bfa1p-3HA (lane 8) but not with Tem1p-3HA (lane 7) was observed, suggesting that the Nud1p–Bfa1p–Bub2p complex was precipitated. Tem1p probably dissociated from the Nud1p–Bfa1p–Bub2p complex during the extraction of SPBs before the immunoprecipitation experiment. We also noticed that the phosphorylated forms of Nud1p preferentially co-immunoprecipitated with Bfa1p and Bub2p (Figure 9B, lanes 6 and 8). Thus, the Bfa1p– Bub2p complex interacts with Nud1p.
Fig. 9. Bub2p interacts with Nud1p. (A) Two-hybrid interaction of Bub2p and Nud1p. (B) Wild-type (lanes 1 and 5), BUB2-3HA (lanes 2 and 6), TEM1-3HA (lanes 3 and 7) and BFA1-3HA (lanes 4 and 8) cells were arrested with nocodazole. Cell extracts were incubated with anti-HA antibodies bound to protein G beads. The total extracts (lanes 1–4) and immunoprecipitates (lanes 5–8) were tested for Nud1p (anti-Nud1p) and HA-tagged proteins (anti-HA) by immunoblotting.
Discussion
Spatially distinct nucleation of AMs is achieved by interaction of Spc72p with Nud1p and Kar1p
The AMs in budding yeast originate from either the half-bridge or the outer plaque depending on the phase of the cell cycle (Byers and Goetsch, 1975; Pereira et al., 1999). We now provide evidence suggesting that the formation of the two microtubule arrays might be mediated by the differential interaction of Spc72p with either Kar1p at the half-bridge (Pereira et al., 1999) or the core SPB component Nud1p at the outer plaque (Adams and Kilmartin, 1999). Several lines of evidence indicate that Nud1p is the direct binding partner for Spc72p at the outer plaque. First, Spc72p and Nud1p interact in yeast two-hybrid assays. Secondly, Spc72p can be co-precipitated with Nud1p from yeast cells. Thirdly, both proteins interact when expressed in insect cells. Finally, overexpression of the C-NUD1 or temperature-sensitive mutations in NUD1 leads to delocalization of Spc72p from the SPB and misorganized AMs.
How the differential binding of Spc72p to either Kar1p or Nud1p is regulated is still unclear and we are currently investigating this point in more detail. Because Spc72p and Nud1p are phosphorylated in a cell-cycle-dependent manner, we speculate that the phosphorylation status of Spc72p and/or Nud1p influences its affinity for its respective binding partners. This is consistent with the finding that only phosphorylated Spc72p was precipitated with Nud1p. Considering the phosphorylation pattern of Spc72p and its cell-cycle-dependent distribution (Adams and Kilmartin, 1999; Pereira et al., 1999), unphosphorylated Spc72p probably binds to Kar1p in the G1 phase of the cell cycle, whereas upon phosphorylation Spc72p interacts preferentially with Nud1p in S/G2 phase and mitosis.
The functional consequences of the cell-cycle-dependent AM fluctuation between the half-bridge and the outer plaque are not understood. AM fluctuations are probably not essential during vegetative growth, since the Spc72–Kar1p or Spc72–Cnm67p fusion proteins, which tether the AMs constitutively either to the half-bridge (Pereira et al., 1999) or the outer plaque, are fully functional based on the structure of the AMs and the migration of the nucleus into the bud (Pereira et al., 1999). AM organization by the half-bridge in the G1 phase of the cell cycle may prepare cells for the differentiation process induced by mating pheromone since in mating cells all AMs originate from the half-bridge (Byers and Goetsch, 1975; Pereira et al., 1999). Alternatively, AM’s attachment to SPB substructures may influence their number and properties. An indication for this comes from studying AM’s properties throughout the cell cycle (Carminati and Stearns, 1997; Shaw et al., 1997).
Our finding extends the previous model of microtubule organization by the SPB (Knop et al., 1999a). γ-tubulin complex binding proteins (GTBPs) like Spc72p may be dynamic entities that change their position within a microtubule organizing centre such as the SPB, with dependence on the cell cycle and upon external stimuli. Key factors in regulating microtubule organization besides GTBPs are interacting components such as Kar1p and Nud1p.
Nud1p has two distinct functions
Based on the phenotype of nud1-44 cells, it has been proposed that Nud1p may regulate events in mitosis (Adams and Kilmartin, 1999). However, whether the cell-cycle arrest of nud1-44 cells was caused by spindle checkpoint activation, a defective outer plaque or a direct involvement of Nud1p in the MEN, and whether Nud1p has an additional function in AM organization remained unclear.
We report here that nud1-2 cells arrest in late anaphase not in response to the Bub2p or Mad2p spindle checkpoint but because Nud1p is a component of the MEN. Furthermore, nud1-2 cells display an AM defect, caused by mislocalized Nud1p and Spc72p, which results in nuclear migration failure in ∼30% of nud1-2 cells. The AM defect, however, was not responsible for the cell-cycle arrest since constitutive nucleation of AMs mediated by an Spc72–Cnm67p protein ‘cured’ the nuclear migration failure but did not suppress the cell-cycle defect. These observations imply that Nud1p fulfils two distinct functions during the cell cycle: a non-essential attachment of cytoplasmic microtubules to the outer plaque during S, G2 and M phase, and an essential function in controlling mitotic exit.
In contrast to other mutations that affect AM organization, such as spc72-10 (Pereira et al., 2000), cell-cycle arrest of nud1-2 cells was not dependent on the presence of the Bub2p checkpoint. Our interpretation of these data is that even though the cells have a clear AM defect, deletion of the checkpoint did not affect cell-cycle arrest because the downstream event of the Bub2p checkpoint, inhibition of mitotic exit (Pereira et al., 2000), is already blocked through the nud1-2 mutation.
Whereas the majority of Δcnm67 or spc72(ts) cells have a nuclear migration defect (Brachat et al., 1998; Knop and Schiebel, 1998), this phenotype was not very pronounced in nud1-2 cells. It is likely that the AMs organized via the Kar1p–Spc72p complex at the half-bridge contribute to nuclear migration in nud1-2 cells. Consistent with this notion, we observed AMs originating from the half-bridge in nud1-2 cells incubated at the restrictive temperature. In addition, bundles of AMs may only detach from the outer plaque during nuclear migration into the bud, probably because of forces applied onto the outer plaque.
Nud1p is a core SPB component with a function in mitotic exit
Genetic interaction of NUD1 with components of the MEN and SIC1, as well as the cell-cycle arrest phenotype of nud1-2 cells in respect of Clb2p stabilization and Pds1p degradation, clearly demonstrate that Nud1p is a component of the MEN. What is the role of Nud1p in the MEN? An indication for the defect of nud1-2 cells came from studying the localization of MEN components. In nud1-2 cells incubated at the restrictive temperature, Bfa1p, Bub2p and Tem1p detach from the SPB. Whereas Tem1p and Bfa1p appeared as a dot-like signal frequently associated with the cell cortex, Bub2p was not detected in a defined structure. These findings are most easily reconciled by assuming that upon binding of Bub2p to Nud1p at the SPB a Bfa1p–Tem1p complex assembles that is then mislocalized upon loss of Bub2p from the SPB when nud1-2 cells are shifted to the restrictive temperature. This model is consistent with our finding that Bub2p interacts with Nud1p.
In most nud1-2 cells, the effector of Tem1p, Cdc15p, was associated with the SPB that entered the bud, a localization that seems to reflect the situation in early anaphase of wild-type cells (Cenamor et al., 1999). Assuming that the Tem1p and Cdc15p signals in nud1-2 cells are a reflection of the normal programme at the end of anaphase, nud1-2 cells are defective in the transition to the state where both SPBs carry Cdc15p and Tem1p and where Tem1p is bound to the SPB independently of Bfa1p and Bub2p (Cenamor et al., 1999; Pereira et al., 2000; Xu et al., 2000). It is also important to note that in nud1-2 cells Cdc15p and Tem1p fail to interact at the SPB, a step that is probably crucial for mitotic exit (Bardin et al., 2000).
How Nud1p functions at the molecular level is less clear. Our genetic data order NUD1 between LTE1 and CDC15. This, together with the failure of Tem1p to interact with Cdc15p at the SPB of nud1-2 cells, raises the possibility that Nud1p regulates or facilitates the binding of Tem1p to Cdc15p. Although we were unable to co-immunoprecipitate Tem1p and Nud1p, the possibility of a transient, cell-cycle-dependent interaction between both proteins cannot be excluded. Nud1p may assist the transfer of Bfa1p–Bub2p-bound Tem1p to Cdc15p at the end of anaphase or it may allow the direct binding of cytoplasmic Tem1p. In this respect it is interesting that Nud1p is an atypical SPB component that does not contain coiled-coil regions but instead has six leucine-rich repeats as a prominent feature (Adams and Kilmartin, 1999). Another protein with leucine-rich repeats that is involved in signal transduction is SUR-8. SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf (Li et al., 2000). Thus, like SUR-8, Nud1p may function as a scaffold that enhances Tem1p–Cdc15p signal transduction.
Recently, evidence was obtained that mammalian centrosomes regulate exit of mitosis (Piel et al., 2000). It is therefore likely that also in mammalian cells core centrosomal components may play a role in controlling mitotic exit.
Materials and methods
General yeast methods
Yeast cells were grown and manipulated according to standard protocols (Sherman, 1991). Strains and plasmids are listed in Table II. Yeast strains were constructed and manipulated using PCR-based cassettes (Knop et al., 1999b). Temperature-sensitive mutants in NUD1 were generated by a PCR-based strategy (Geissler et al., 1996). The nud1-2 allele carries the mutations Q6E, S224I, G316C, L548H and L683F.
Table II. Yeast strains and plasmids.
| Name | Genotype/construction | Source or reference |
|---|---|---|
| Yeast strains | ||
| ESM431 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δcnm67 pSM431 | Elliott et al. (1999) |
| ESM448-1 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δspc72::kanMX pSM447 | Knop and Schiebel (1998) |
| ESM943 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CLB2-6HA::klTRP1 | this study |
| ESM1192 | MATa ura3-52 his3Δ200 leu2Δ1 Δlte1::kanMX6 | this study |
| FY1679 | _MATa/_α ura3-52/ura3-52 TRP1/trp1Δ63 LEU2/leu2Δ1 HIS3/his3Δ200 | Eurofan |
| GPY60-3 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1 SPC72-3HA::kanMX4 | this study |
| GPY69 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1 BUB2-3HA::kanMX4 | Pereira et al. (2000) |
| GPY130 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1 TEM1-3HA::kanMX4 | Pereira et al. (2000) |
| GPY190 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1 BFA1-3HA::kanMX4 | Pereira et al. (2000) |
| GPY193 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δbub2::klTRP1 TEM1-GFP::kanMX4 | Pereira et al. (2000) |
| GPY195 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1 CDC15-9Myc::klTRP1 | this study |
| GPY235 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 PDS1-6HA::klTRP1 | this study |
| KCY2 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 | this study |
| KCY7 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 CLB2-6HA:klTRP1 | this study |
| KCY12 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 PDS1-6HA::klTRP1 | this study |
| SGY37 | MATa ura3-52::URA3-lexA-op-LacZ trp1 his3 leu2 | Geissler et al. (1996) |
| UGY111 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δnud1::kanMX4 pSM783 | this study |
| UGY121 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63::NUD1-6HA::TRP1 his3Δ200 leu2Δ1 Δsst1 | this study |
| UGY151 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC72-GFP:kanMX4 pUG95 | this study |
| UGY153 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NUD1-GFP::kanMX4 pUG95 | this study |
| UGY166 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NUD1-GFP::kanMX4 p416-Gal1 | this study |
| UGY181 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NUD1-GFP::kanMX4 pUG113 | this study |
| UGY184 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CNM67-GFP::kanMX4 pUG95 | this study |
| UGY194 | _MATa/_α ura3-52/ura3-52 TRP1/trp1Δ63 LEU2/leu2Δ1 HIS3/his3Δ200 NUD1/Gal1-GFP-NUD1::kanMX4 | this study |
| UGY196 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC42-GFP::HIS3 | this study |
| UGY198 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CNM67-GFP::HIS3 | this study |
| UGY199 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 SPC42-GFP::HIS3 | this study |
| UGY200 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 CNM67-GFP::HIS3 | this study |
| UGY201 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 TEM1-GFP::klTRP1 | this study |
| UGY215 | _MATa ura3-52::pSM798::URA3 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 | this study |
| UGY217 | MATa ura3-52::pSM798::URA3 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | this study |
| UGY218 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 TEM1-GFP::HIS3 | this study |
| UGY238 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 Δbub2::HIS3 TEM1-GFP::klTRP1 | this study |
| UGY248 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 CDC14-3Myc::HIS3 | this study |
| UGY255 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CDC14-3Myc::HIS3 | this study |
| UGY262 | _MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::_pKC8 Δnud1::kanMX4 CDC15-9Myc::klTRP1 | this study |
| YAS8 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1 | Spang et al. (1996) |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | Sikorski and Hieter (1989) |
| Plasmids | ||
| p414-Gal1 | pRS414 containing the Gal1 promoter | Mumberg et al. (1995) |
| p416-Gal1 | pRS416 containing the Gal1 promoter | Mumberg et al. (1995) |
| pACT2 | 2 µm, _LEU2_-based vector carrying the GAL4 activator domain | Durfee et al. (1993) |
| pEG202 | 2 µm, _HIS3_-based vector carrying the lexA DNA-binding domain | Gyuris et al. (1993) |
| pGP67 | pEG202 containing _SPC72_231–621 | Knop and Schiebel (1998) |
| pGP69 | pEG202 containing BUB2 | this study |
| pGP98 | pRS316 containing Gal1-GFP-CDC15 | this study |
| pKC8 | pRS305 containing nud1-2 | this study |
| pMK209 | pACT2 containing _KAR1_116–274 | Pereira et al. (1999) |
| pRS305 | _LEU2_-based yeast integration vector | Sikorski and Hieter (1989) |
| pRS306 | _URA3_-based yeast integration vector | Sikorski and Hieter (1989) |
| pRS314 | CEN6, TRP1_-based yeast–_E.coli shuttle vector | Sikorski and Hieter (1989) |
| pRS315 | CEN6, LEU2_-based yeast–_E.coli shuttle vector | Sikorski and Hieter (1989) |
| pRS316 | CEN6, URA3_-based yeast–_E.coli shuttle vector | Sikorski and Hieter (1989) |
| pRS414 | CEN6, TRP1_-based yeast–_E.coli shuttle vector | Stratagene |
| pRS416 | CEN6, URA3_-based yeast–_E.coli shuttle vector | Stratagene |
| pSM262 | pYES2 containing TEM1 | this study |
| pSM263 | pYES2 containing tem1-100 | this study |
| pSM431 | pRS316 containing CNM67 | Knop and Schiebel (1998) |
| pSM447 | pRS316 containing SPC72 | Knop and Schiebel (1998) |
| pSM613 | pACT2 containing _NUD1_405–852 | Elliott et al. (1999) |
| pSM653 | p416-Gal1 containing NUD1 | this study |
| pSM716 | pRS315 containing _pSPC72-SPC72_1–276-_CNM67_1–581 | this study |
| pSM783 | pRS316 containing NUD1 | this study |
| pSM798 | pRS306 containing _pSPC72_-_SPC72_1–276-_CNM67_1–581 | this study |
| pSM897 | p414-Gal1 containing SPO12 | this study |
| pUG9 | pR315 containing _pSPC72_-_SPC72_1–276-_KAR1_192–433 | Pereira et al. (1999) |
| pUG44 | pEG202 containing _SPC72_433–621 | this study |
| pUG56 | pEG202 containing _SPC72_345–466 | this study |
| pUG95 | p416-Gal1 containing _HA-NUD1_405–852 | this study |
| pUG97 | pEG202 containing _SPC72_468–621 | this study |
| pUG101 | pEG202 containing _SPC72_570–621 | this study |
| pUG105 | pEG202 containing _SPC72_477–621 | this study |
| pUG113 | p416-Gal1 containing _NUD1_1–406 | this study |
| pUG115 | pRS314 containing NUD1 | this study |
| pUG120 | pRS316 containing CDC14 | this study |
| pYES2 | 2 µm, _URA3_-based vector carrying the Gal1 promoter | Invitrogen |
Two-hybrid interactions
Two-hybrid interactions were determined in yeast strain SGY37 (Geissler et al., 1996) with derivatives of pACT2 (Durfee et al., 1993) and pEG202 (Gyuris et al., 1993). β-galactosidase activity was measured using a semi-quantitative overlay assay (Geissler et al., 1996).
Immunological techniques, phosphatase treatment, flow cytometry and electron microscopy
Indirect immunofluorescence was performed as described (Jacobs et al., 1988). Microtubules were stained with the mouse monoclonal antibody WA3 and HA- or Myc-tagged proteins with mouse monoclonal 12CA5 (anti-HA; Hiss Diagnostics) or 9E10 (anti-Myc; Boehringer Mannheim) antibodies. Spc72p and Cnm67p were detected with affinity-purified antibodies (Knop and Schiebel, 1998; Elliott et al., 1999). Samples were inspected with an Axiophot fluorescence microscope (Zeiss) or an MRC-600 confocal microscope (Bio-Rad). Affinity-purified antibodies against the N-terminus (amino acids 1–230) of Nud1p were used to detect Nud1p. Nud1p-6HA was precipitated using 12CA5 covalently coupled to protein G–Sepharose (Geissler et al., 1996). Immunoprecipitations were carried out in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 100 mM β-glycerophosphate, 5% glycerol, 0.5% Triton X-100, 50 mM NaF, 0.2 mM sodium orthovanadate, 1 µM okadaic acid, 0.5 µM staurosporine and protease inhibitors. Phosphatase treatment of samples was as described (Pereira et al., 1998). Yeast cells were prepared for flow cytometry as described (Hutter and Eipel, 1979). Samples for electron microscopy were prepared as described (Byers and Goetsch, 1975).
Binding of Nud1p to Spc72p in insect cell extracts
_NUD1-His_6 and SPC72 were expressed in insect cells (Sf9) using the baculovirus expression system (Pharmingen). Infected insect cells were washed with cold phosphate-buffered saline (PBS) and then incubated in lysis buffer [10 mM Tris–HCl pH 8, 10 mM sodium phosphate pH 8, 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM sodium pyrophosphate, complete EDTA-free protease inhibitors (Boehringer Mannheim)]. The cleared lysate was incubated for 30 min at 4°C with 50 µl of TALON resin (Clontech). The resin was washed three times with PBS, 1% Triton X-100 and PBS, 1% Trition X-100, 15 mM imidazole. Proteins were eluted by heating the beads for 15 min at 65°C with sample buffer.
Acknowledgments
Acknowledgements
We thank Drs F.Barr for valuable advice, discussions and help with the figures, G.Pereira for discussions, the construction of plasmids and yeast strains, and W.Kolch for helpful comments. This work was carried out with support by a project grant (Ref. SP 2469/0101) from the Cancer Research Campaign.
References
- Adams I.R. and Kilmartin,J.V. (1999) Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol., 145, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum,D. and Surana,U. (2000) Tying the knot: linking cytokinesis to the nuclear cycle. J. Cell Sci., 113, 1503–1513. [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Visintin,R. and Amon,A. (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell, 102, 21–31. [DOI] [PubMed] [Google Scholar]
- Brachat A., Kilmartin,J.V., Wach,A. and Philippsen,P. (1998) Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol. Biol. Cell, 9, 977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. and Goetsch,L. (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol., 124, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J.L. and Stearns,T. (1997) Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol., 138, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenamor R., Jimenez,J., Cid,V.J., Nombela,C. and Sanchez,M. (1999) The budding yeast Cdc15 localizes to the spindle pole body in a cell-cycle-dependent manner. Mol. Cell. Biol. Res. Commun., 2, 178–184. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A.E., Lee,W.H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Elliott S., Knop,M., Schlenstedt,G. and Schiebel,E. (1999) Spc29p is a component of the Spc110p-subcomplex and is essential for spindle pole body duplication. Proc. Natl Acad. Sci. USA, 96, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S., Pereira,G., Spang,A., Knop,M., Souès,S., Kilmartin,J. and Schiebel,E. (1996) The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J., 15, 3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hutter K.J. and Eipel,H.E. (1979) Microbial determination by flow cytometry. J. Gen. Microbiol., 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Jacobs C.W., Adams,A.E.M., Szaniszlo,P.J. and Pringle,J.R. (1988) Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol., 107, 1409–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles,J.F., Tinker-Kulberg,R.L. and Morgan,D.O. (1998) A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell, 9, 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Schiebel,E. (1997) Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J., 16, 6985–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Schiebel,E. (1998) Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J., 17, 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Pereira,G. and Schiebel,E. (1999a) Microtubule organization by the budding yeast spindle pole body. Biol. Cell, 91, 291–304. [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999b) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Li W., Han,M. and Guan,K.-L. (2000) The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev., 14, 895–900. [PMC free article] [PubMed] [Google Scholar]
- Marschall L.G., Jeng,R.L., Mulholland,J. and Stearns,T. (1996) Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule-organizing center function. J. Cell Biol., 134, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K. and Rose,M.D. (1998) Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol., 140, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Palmer R.E., Sullivan,D.S., Huffaker,T. and Koshland,D. (1992) Role of astral microtubules and actin in spindle orientation and migration in the budding yeast Saccharomyces cerevisiae. J. Cell Biol., 119, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Knop,M. and Schiebel,E. (1998) Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell, 9, 775–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Gruneberg,U., Knop,M. and Schiebel,E. (1999) Interaction of the yeast γ-tubulin complex binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J., 18, 4180–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Höfken,T., Grindlay,J., Manson,C. and Schiebel,E. (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell, 6, 1–10. [PubMed] [Google Scholar]
- Piel M., Meyer,P., Khodjakow,A., Rieder,C.L. and Bornens,M. (2000) The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol., 149, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D. (1996) Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Dev. Biol., 12, 663–695. [DOI] [PubMed] [Google Scholar]
- Schweitzer B.I. and Philippsen,P. (1991) CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast, 7, 265–273. [DOI] [PubMed] [Google Scholar]
- Schwob E., Bohm,T., Mendenhall,M.D. and Nasmyth,K. (1994) The B-type cyclin kinase inhibitor p40 (Sic1) controls the G1 to S transition in Saccharomyces cerevisiae. Cell, 79, 233–244. [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Yeh,E.Y., Maddox,P., Salmon,E.D. and Bloom,K. (1997) Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol., 139, 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui,Y., Tanaka,K. and Toh-e,A. (1994a) Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast, 10, 451–461. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui,Y. and Toh-e,A. (1994b) The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M-phase. Mol. Cell. Biol., 14, 7476–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Zachariae,W., Ciosk,R. and Nasmyth,K. (1998) The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J., 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W. et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Geissler,S., Grein,K. and Schiebel,E. (1996) γ-tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol., 134, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Hiller,M.A., Scherson,T.Y. and Rose,M.D. (1992) Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J. Cell Biol., 117, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Hwang,E.S. and Amon,A. (1999) Cif prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature, 398, 818–823. [DOI] [PubMed] [Google Scholar]
- Xu S., Huang,H.-K., Kaiser,P., Latterich,M. and Hunter,T. (2000) Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr. Biol., 10, 329–332. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shin,T.H., Galova,M., Obermaier,B. and Nasmyth,K. (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science, 274, 1201–1204. [DOI] [PubMed] [Google Scholar]