The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions (original) (raw)
Abstract
Eukaryotic mRNAs exist in vivo as ribonucleoprotein particles (mRNPs). The protein components of mRNPs have important functions in mRNA metabolism, including effects on subcellular localization, translational efficiency and mRNA half-life. There is accumulating evidence that pre-mRNA splicing can alter mRNP structure and thereby affect downstream mRNA metabolism. Here, we report that the spliceosome stably deposits several proteins on mRNAs, probably as a single complex of ∼335 kDa. This complex protects 8 nucleotides of mRNA from complete RNase digestion at a conserved position 20–24 nucleotides upstream of exon–exon junctions. Splicing-dependent RNase protection of this region was observed in both HeLa cell nuclear extracts and Xenopus laevis oocyte nuclei. Immunoprecipitations revealed that five components of the complex are the splicing-associated factors SRm160, DEK and RNPS1, the mRNA-associated shuttling protein Y14 and the mRNA export factor REF. Possible functions for this complex in nucleocytoplasmic transport of spliced mRNA, as well as the nonsense-mediated mRNA decay pathway, are discussed.
Keywords: exon–exon junctions/nonsense-mediated decay/nucleocytoplasmic transport/mRNA/spliceosome
Introduction
Throughout their lifetimes, pre-mRNAs and their product mRNAs exist in vivo as RNA–protein particles (RNPs; Dreyfuss et al., 1993). The complement of proteins bound to these RNAs evolves significantly as nuclear pre-mRNA is processed into mRNA and nuclear mRNA is exported to the cytoplasm. For example, Pol II transcripts acquire CBC20/80, the nuclear cap-binding proteins, after extensive modifications to their 5′ ends, and these proteins are exchanged for the translation initiation factor eIF4E upon export to the cytoplasm (Izaurralde et al., 1995; Visa et al., 1996; Fortes et al., 2000). As another example, Pol II transcripts acquire PABP2, the nuclear poly(A) binding protein, as part of a series of protein–RNA interactions that direct 3′ end cleavage and polyadenylation (Wahle, 1991; Krause et al., 1994). In the cytoplasm, mRNA poly(A) tails are bound by PABP1 (Gorlach et al., 1994), which functions in the initiation of protein synthesis (reviewed by Sachs and Varani, 2000). Some heterogeneous nuclear ribonucleoprotein particle (hnRNP) proteins, which coat pre-mRNAs as they are synthesized, accompany mRNAs into the cytoplasm. Such hnRNP proteins can regulate cytoplasmic events, including mRNA localization, translation and mRNA turnover (reviewed by Shyu and Wilkinson, 2000). Therefore, the nuclear history of an mRNA can have substantial effects on its downstream metabolic fate.
Like capping and polyadenylation, it has recently become apparent that pre-mRNA splicing can significantly influence the downstream metabolism of product mRNAs. There are several examples in the literature indicating that the expression of specific genes in metazoans is enhanced by the presence of an intron (Luo and Reed, 1999 and references therein). It has also been shown that metazoan mRNAs containing an exon–exon junction >50 nucleotides (nt) downstream of the first in-frame termination codon are targeted for nonsense-mediated mRNA decay (NMD; reviewed by Maquat, 1995, 2000; Li and Wilkinson, 1998; Hentze and Kulozik, 1999). Thus, mRNAs must somehow carry a memory of the introns contained in their precursors.
The most likely mechanism mediating the downstream effects of pre-mRNA splicing on mRNA metabolism is a splicing-dependent modification of mRNP protein composition (Le Hir et al., 2000). To date, several proteins have been shown to specifically associate with spliced mRNA. Antibodies against SRm160, a co-activator of splicing (Blencowe et al., 1998), and RNPS1, a general splicing activator (Mayeda et al., 1999), specifically precipitate mRNAs produced by splicing in vitro. SRm160 and hPrp8p, a core component of the spliceosome (Luo et al., 1999), could be cross-linked to one mRNA in vitro in a splicing-dependent manner (Le Hir et al., 2000). Other proteins that are not demonstrable splicing factors have also been found associated with spliced mRNA. Antibodies against the oncoprotein DEK, which co-purifies with SRm160 (McGarvey et al., 2000), and Y14, a novel RNA-binding protein that shuttles in and out of the nucleus (Kataoka et al., 2000), preferentially precipitate spliced mRNAs. Additionally, the protein REF (also called Aly) was found to be present in spliced mRNP complexes, but absent from an analogous mRNP complex containing unspliced mRNA (Zhou et al., 2000). However, whether these proteins bind along the length of spliced mRNA or only at particular positions remained to be determined.
In a previous study, we described a novel in vitro cross-linking approach for detecting proteins that associate with mRNAs only as a consequence of splicing (Le Hir et al., 2000). While that study provided direct evidence that splicing can alter the proteins associated with mRNA, only selected positions could be assayed for an interaction with splicing-dependent proteins. Furthermore, it was not possible to determine the proportion of spliced mRNA molecules carrying these proteins. Here, we took advantage of a targeted RNase H analysis (Gunzl and Bindereif, 1999) to explore in more detail where and how the splicing machinery alters spliced mRNP structure in vitro and in vivo. Remarkably, this approach revealed that spliceosomes deposit multiple proteins, probably as a single complex, at a conserved position 20–24 nt upstream of spliced mRNA exon–exon junctions. This –20/24 complex contains SRm160, DEK, RNPS1, Y14 and REF.
Results
RNase H footprinting reveals a splicing-dependent protection of mRNA
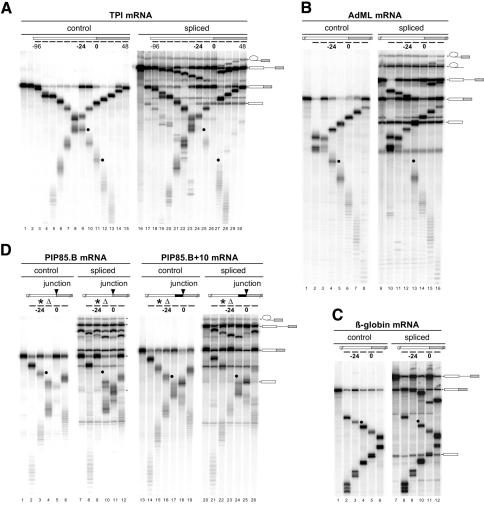
If pre-mRNA splicing stably alters mRNP structure, such alterations might be detectable as splicing-dependent protections of mRNA to RNase digestion. We initially tested this possibility by incubating a uniformly labeled pre-mRNA that derived from the human triose phosphate isomerase (TPI) gene under splicing conditions in HeLa cell nuclear extract to generate mRNA (spliced mRNA). Subsequently, the reaction was divided into aliquots and individual cDNA oligonucleotides (oligos; 12 nt each) were added to each aliquot to activate targeted RNase H cleavage (Figure 1A, right panel). Each oligo was named according to the position of its center relative to the exon–exon junction (Figure 1A, top). A series of 13 such oligos allowed for analysis across the entire TPI mRNA. To reveal any splicing-dependent changes in the mRNA cleavage pattern, an identical mRNA that had been transcribed from cDNA (control mRNA) was analyzed in parallel (Figure 1A, left panel).
Fig. 1. A splicing-dependent protection of mRNAs from targeted RNase H cleavage. (A) Uniformly labeled TPI control mRNA (lanes 1–15) or TPI pre-mRNA (lanes 16–30) was incubated under splicing conditions in HeLa cell nuclear extract for 0 (lanes 1 and 16), 45 (lanes 2–15) or 90 min (lanes 17–30). Aliquots of the 45 and 90 min reactions were further incubated with the cDNA oligos indicated (short bars underneath the mRNA schematic at the top; lanes 3–15 and 18–30). RNAs were then separated by 14% denaturing PAGE. Each oligo was named (e.g. –96, –24, 0, 48 in top panels) according to its center position relative to the exon–exon junction, which was defined as 0. Splicing substrates, intermediates and products are indicated on the right. Black dots indicate specific mRNA cleavage fragments that are markedly reduced with spliced mRNA compared with control mRNA. Similar RNase H analyses were performed with control and spliced (B) AdML mRNAs, (C) β-globin mRNAs and (D) PIP85.B (left) and PIP85.B+10 (right) mRNAs. The triangle indicates the same cDNA oligo, which was alternatively centered at –12 and –24 for PIP85.B and PIP85.B+10 mRNAs, respectively. Likewise, the asterisk indicates a single cDNA oligo centered at –24 and –36 for each mRNA, respectively. AdML, β-globin and PIP85.B RNAs were separated by 15, 8 and 15% denaturing PAGE, respectively.
We found that all regions of control mRNA were accessible to RNase H cleavage. This was shown by the appearance of two cleavage products in all digestion reactions (Figure 1A, left). Although a significant amount of uncleaved mRNA remained after digestion at some positions, this amount was not consistent in every experiment (data not shown). In reactions containing spliced mRNA, only one cleavage product specifically reflected RNase H digestion of mRNA; the other product could also be generated by digestion of pre-mRNA and/or splicing intermediates (except at the exon–exon junction). Compared with the control mRNA digestions, this mRNA-specific cleavage product was either absent or dramatically reduced at two positions in spliced mRNA: 24 nt upstream of the exon–exon junction (oligo centered at position –24; Figure 1A, lane 24), and at the junction itself (oligo centered at the exon–exon junction, position 0; Figure 1A, lane 26). This indicated that two positions in TPI mRNA were almost completely protected from RNase H cleavage after splicing.
To test the generality of the RNase H resistances observed for spliced TPI mRNA, we performed the same analysis with three other splicing substrates and their corresponding control mRNAs: AdML, β-globin and PIP85.B (Figure 1B–D). No splicing-dependent protection at the exon–exon junction was observed with any of these mRNAs (Figure 1B, lane 14; Figure 1C, lane 11; Figure 1D, lanes 11 and 25). Remarkably, however, splicing-dependent protection from cleavage by each of the oligos centered at position –24 was characteristic of all mRNAs tested (Figure 1B, lane 12; Figure 1C, lane 9; Figure 1D, lanes 9 and 23).
Since no conserved sequence element was apparent in the vicinity of position –24 (data not shown), it seemed likely that the location of this splicing-dependent protection was defined mainly by its distance from the exon– exon junction rather than by RNA sequence. To test this hypothesis, we inserted 10 nt at the 3′ end of PIP85.B exon 1 (Figure 1D). Although this insertion moved the sequence surrounding position –24 in the PIP85.B construct to the vicinity of position –34 in the PIP85.B+10 construct, the protected region in the new construct remained 24 nt upstream of the exon–exon junction. Thus, a general consequence of pre-mRNA splicing is the deposition of some stably associated species a fixed distance 24 nt upstream of mRNA exon–exon junctions.
Splicing-dependent protection of mRNA is conserved in Xenopus oocytes
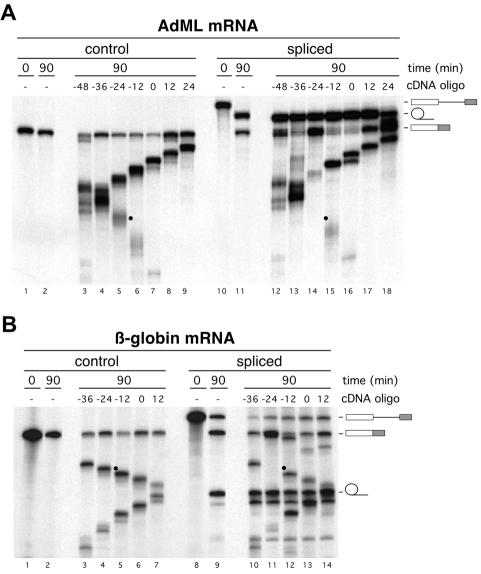
To test whether the splicing-dependent protection of mRNA observed in HeLa cell nuclear extract could be reproduced in vivo, we performed a similar RNase H analysis in Xenopus laevis oocytes (Figure 2). The same AdML (Figure 2A) and β-globin (Figure 2B) pre-mRNAs and control mRNAs as above were injected into oocyte nuclei. After 90 min, more than half of each pre-mRNA had been converted to spliced products. Following oocyte dissection, isolated nuclei were further incubated in the presence of the indicated cDNA oligos to activate endogenous RNase H.
Fig. 2. A splicing-dependent protection of AdML (A) and β-globin (B) mRNAs from RNase H cleavage in X.laevis oocytes. Control mRNA [(A) lanes 1–9; (B) lanes 1–7] or pre-mRNA [(A) lanes 10–18; (B) lanes 8–14] was injected into oocyte nuclei. Nuclear RNAs were collected immediately [(A) lanes 1 and 10; (B) lanes 1 and 8] or 90 min after injection [(A) lanes 2–9 and 11–18; (B) lanes 2–7 and 9–14]. Where indicated, nuclear RNA samples were incubated with individual cDNA oligos to activate endogenous RNAse H [(A) lanes 3–9 and 12–18; (B) lanes 3–7 and 10–14]. AdML and β-globin RNAs were separated by 15 and 10% denaturing PAGE, respectively. For uncleaved and cleaved samples, one and four oocyte equivalents of RNA, respectively, from pools of 10 oocytes were loaded per lane. Splicing substrates and products are indicated on the right. Black dots indicate specific mRNA cleavage fragments that are markedly reduced with spliced mRNA compared with control mRNA.
Remarkably, we observed exactly the same cleavage and protection patterns for control and spliced mRNAs in Xenopus oocytes in vivo as were observed in HeLa cell nuclear extract in vitro. These results indicate that the splicing-dependent protection at –24 (Figure 2A, lane 14; Figure 2B, lane 11) is conserved from amphibians to humans.
Identification of proteins associated with the exon–exon junction region of spliced mRNA
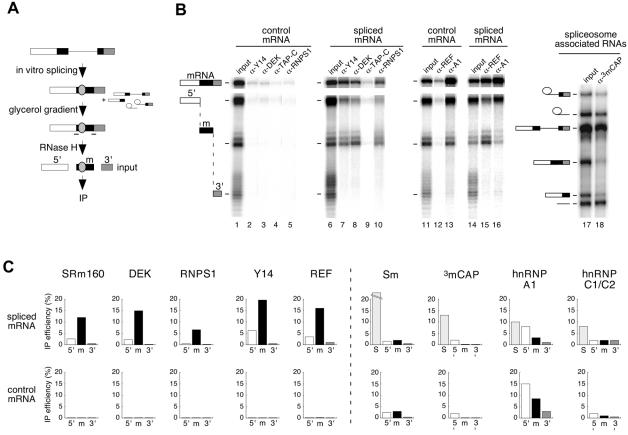
To identify factors that associate with the region around the exon–exon junction in a splicing-dependent manner, we undertook a selective co-immunoprecipitation (co-IP) strategy (Figure 3A). Briefly, uniformly labeled AdML pre-mRNA was spliced in HeLa cell nuclear extract. To ensure that only released mRNPs were analyzed, spliced mRNPs were separated from spliceosomes by glycerol gradient fractionation (Le Hir et al., 2000). The mRNA in these isolated mRNPs was then subjected to RNase H cleavage by a pair of oligos, one annealing upstream and the other annealing downstream of the exon–exon junction, to generate three separate mRNA fragments: 5′ (83 nt), middle (m; 64 nt) and 3′ (36 nt), where the –24 region and the exon–exon junction were contained in the middle fragment. Reactions were then subjected to IP with different antibodies, and the co-IP of mRNA fragments was analyzed by denaturing PAGE. In parallel, control mRNA was incubated under splicing conditions, cleaved with the identical oligos, and subjected to IP directly (Figure 3B).
Fig. 3. Identification of proteins associated with the exon–exon junction region by RNA co-IP. (A) Scheme of co-IP strategy. AdML pre-mRNA was spliced for 2 h, and mRNP was separated from spliceosomes by glycerol gradient fractionation. Spliced mRNA was subsequently cleaved using RNase H and two oligos (–) to produce three fragments: 5′ (white), m (black) and 3′ (shaded). The fragment ‘m’ contains the –20/24 protected region (gray oval). This reaction mixture was then subjected to IP with different antibodies. Control mRNA was analyzed in parallel reactions except that it was not fractionated on a glycerol gradient. (B) Representative RNA co-IP experiments performed with antibodies against Y14, DEK, TAP-C, RNPS1, REF, hnRNPA1 (A1) and 3mCAP. Lanes 1, 6, 11, 14 and 17 correspond to one-fifteenth of input RNA. Lane 18 corresponds to co-IP of spliceosome-associated RNAs. AdML RNAs were analyzed as indicated in Figure 1. Structures of splicing substrates, intermediates and products as well as mRNA fragments are diagrammed. (C) Co-IP efficiencies of mRNA fragments (5′, m and 3′) and spliceosomes (S, taken as splicing intermediates) with antibodies against the species indicated. Upper and lower rows of histograms correspond to fragments of spliced and control mRNAs, respectively. Co-IP efficiencies are averages of 2–5 independent experiments.
As expected from previous studies (see Introduction), antibodies against SRm160, DEK, RNPS1, Y14 and REF all precipitated full-length spliced mRNA to a much greater extent than control mRNA (Figure 3B and data not shown). Remarkably, all of these antibodies also selectively co-immunoprecipitated the middle (m) fragment from spliced mRNA, but not the same fragment from control mRNA (Figure 3B and C). All other fragments, whether arising from the spliced or control mRNA, were either precipitated with greatly reduced efficiency (e.g. the 5′ fragment from spliced mRNA) or were observed at background levels only. These differences in IP efficiencies were readily quantifiable, as the histograms shown in Figure 3C represent averages of 2–5 independent experiments. The generality of the splicing-dependent association of SRm160, DEK, RNPS1, Y14 and REF with the region around the exon–exon junction of spliced mRNA was assessed by performing similar analyses with TPI, β-globin and PIP85.B mRNAs. In those experiments, antisera against these proteins invariably co-immunoprecipitated RNA fragments containing the –24 region of spliced mRNA more efficiently than any other fragment (data not shown). Thus, the association of SRm160, DEK, RNPS1, Y14 and REF with this region is a general feature of spliced mRNA.
Selective co-IP of the middle fragment from spliced mRNA was not a general property of all antibodies tested. Antibodies against the mRNA export factor TAP (Gruter et al., 1998; Braun et al., 1999; Katahira et al., 1999) did not co-immunoprecipitate any RNA we tested (Figure 3B and data not shown). Anti-trimethyl cap antibodies, which recognize U1, U2, U4 and U5 spliceosomal snRNAs, and Y12 antibodies, which recognize the core Sm snRNP proteins, failed to co-immunoprecipitate full-length spliced mRNA, control mRNA, or any RNase H-generated fragment thereof. However, both antibodies did effectively co-immunoprecipitate RNAs contained in spliceosomes (S) purified from the same glycerol gradients as spliced mRNP (Figure 3B and C). Antisera against the hnRNP A1 protein, which actively shuttles between the nucleus and cytoplasm and is thought to be associated with mRNAs in vivo (Nakielny and Dreyfuss, 1999), co-immunoprecipitated both spliced and control mRNAs (and their respective RNase H digestion fragments) with comparable efficiencies (Figure 3B and C). In this case, co-IP efficiency correlated more with RNA length than any other attribute, consistent with the minimal RNA recognition requirements of hnRNP A1 (see below). Antisera recognizing hnRNP C1/C2, a non-shuttling protein (Nakielny and Dreyfuss, 1996), co-immunoprecipitated RNAs contained in spliceosomes, but neither spliced nor control mRNA or their fragments. Similar results were obtained with an antiserum directed against the C-terminus of hPrp8p (Le Hir et al., 2000). Thus, the epitopes recognized by these antisera are either not present or are inaccessible in spliced mRNP.
Taken together, the above results indicate that the SRm160, DEK, RNPS1, Y14 and REF proteins all become associated with the exon–exon junction region of mRNA as a consequence of splicing. Furthermore, their association with spliced mRNA is sufficiently stable to withstand glycerol gradient fractionation followed by IP. Finally, in contrast to hnRNP A1, which apparently binds at multiple points along the length of mRNA, selective co-IP of the exon–exon junction region by antibodies against SRm160, DEK, RNPS1, Y14 and REF suggests that this region is their primary point of association with spliced mRNA.
The splicing-dependent protection spans 8 nt, including positions –20 and –24
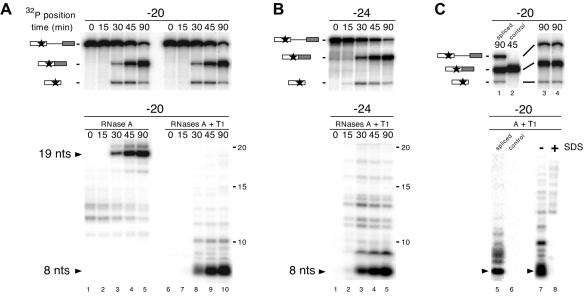
The above experiments indicated that antibodies against the SRm160, DEK, RNPS1, Y14 and REF proteins all selectively co-immunoprecipitate a 64 nt fragment containing position –24 and the exon–exon junction of spliced mRNA. We next investigated whether the association point of these proteins corresponded exactly to the region protected by splicing from RNase H cleavage (i.e. the nucleotides immediately flanking position –24). To do so, we first determined whether the splicing-dependent protection of the –24 region could withstand more stringent RNase digestion. AdML pre-mRNA containing a single labeled phosphate located 20 nt upstream of the 5′ splice site was incubated under splicing conditions for various times and then each reaction was divided in half. One half was analyzed directly by denaturing PAGE to follow the evolution of splicing intermediates and products (Figure 4A, upper panels). The other half was treated with RNase A or RNases A and T1 before PAGE to detect any protected fragments (Figure 4A, lower panels).
Fig. 4. Complete RNase digestion of singly labeled RNAs. (A) Upper panel: splicing time courses for AdML pre-mRNAs containing a single labeled phosphate (star) positioned 20 nt upstream of the 5′ splice site. Splicing substrates, intermediates and products are indicated on the left. RNAs were analyzed as in Figure 1. Lower panel: complete RNase A (left) or RNase A + T1 (right) digestion of RNAs at each time point yielded 19 and 8 nt protected fragments (arrows). Protected fragments were separated by 20% denaturing PAGE. (B) Same as (A) with AdML pre-mRNAs containing a single labeled phosphate positioned 24 nt upstream of the 5′ splice site. (C) Upper panels: AdML pre-mRNA (lanes 1, 3 and 4) or control mRNA (lane 2) containing a single labeled phosphate positioned 20 nt upstream of the 5′ splice site was incubated under splicing conditions for the time indicated and separated by 20% (lanes 1 and 2) or 15% (lanes 3 and 4) denaturing PAGE. Lower panels: aliquots of each splicing reaction were subjected to complete RNase A + T1 digestion with (lane 8) or without (lanes 5–7) prior SDS treatment. Protected fragments were separated as in (A).
Prior to the formation of splicing products, no major protected fragments were observed (Figure 4A, lanes 1, 2, 6 and 7). However, as spliced mRNA appeared, a 19 nt protected fragment appeared with identical kinetics in the RNase A-digested samples (lanes 3–5). Similarly, an 8 nt protected fragment appeared in the RNase A- and T1-digested samples (lanes 8–10). An 8 nt protected fragment was also observed when the single label was incorporated into pre-mRNA at position –24 (Figure 4B). In contrast, no protected fragments were observed when control mRNA containing a single labeled phosphate at position –20 was incubated under splicing conditions and then digested with RNases A and T1 (Figure 4C, lane 6). Neither were any protected fragments observed when a sample containing spliced mRNA was treated by SDS prior to RNase addition (Figure 4C, lane 8). Thus, the splicing-dependent protection of AdML mRNA spans 8 nt, including positions –24 and –20 relative to the exon–exon junction. Furthermore, whereas the species responsible for this protection is resistant to extensive RNase degradation, its sensitivity to SDS treatment suggests that it is not covalently attached to mRNA.
All five spliced mRNA-specific proteins are associated with the 8 nt protected fragment
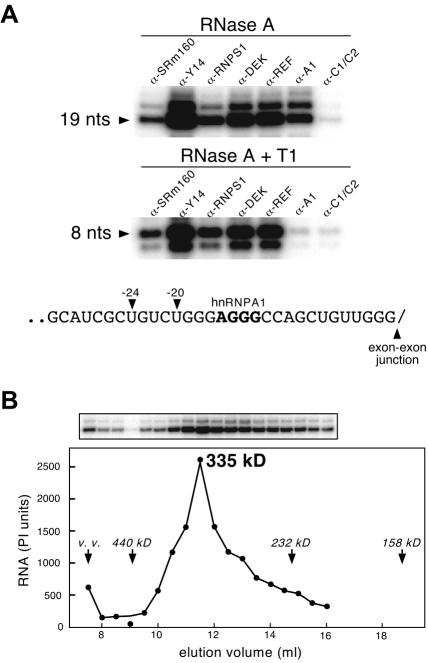
To examine which proteins remained associated with the protected region of spliced mRNA after stringent RNase digestion, we next performed IPs from digestion reactions similar to those above with antibodies against SRm160, DEK, RNPS1, Y14, REF, hnRNP A1 and hnRNP C1/C2 (Figure 5A). Notably, antibodies against all five of the spliced mRNA-specific proteins (SRm160, DEK, RNPS1, Y14 and REF) precipitated both the 19 and 8 nt protected fragments from spliced mRNA. Each of these antibodies precipitated both fragments with equal efficiencies, and these efficiencies were similar to those observed for the larger RNase H fragments in Figure 3. In contrast, antibodies against hnRNP A1 precipitated the 19 nt fragment, but not the 8 nt fragment. This latter result may be explained by the existence of a nearly consensus binding site for hnRNP A1 spanning nt –16 to –13 [AGGG (Figure 5A, bottom); the hnRNP A1 consensus is uAGGGa/u, where upper case letters indicate the most important positions for efficient binding (Burd and Dreyfuss, 1994)]. As expected, antibodies against hnRNP C1/C2 failed to precipitate either fragment. These results indicate that SRm160, DEK, RNPS1, Y14 and REF are all part of the species responsible for the splicing-dependent RNase protection of 8 nt. However, consistent with the results from Figure 3, neither hnRNP A1 nor C1/C2 is specifically associated with this region.
Fig. 5. Immunoprecipitation of the protected fragments from spliced mRNA and molecular mass determination of the –20/24 complex. (A) Co-IP of 19 nt (upper panel) or 8 nt (middle panel) protected fragments generated by RNase A or RNase A + T1 digestion, respectively, of spliced AdML mRNA containing a single labeled phosphate at position –20. Antibodies are as indicated. Lower panel: sequence of AdML mRNA exon 1 between position –31 and the exon–exon junction. Arrows and bold letters indicate –20 and –24 positions and the likely binding site of hnRNP A1, respectively. (B) Superose 6 gel filtration profile of the complex associated with the RNase A-resistant 19 nt fragment of spliced mRNA labeled at position –20 (top). The amount of labeled fragment in each fraction (bottom) was determined by PhosphorImaging (PI units). Elution positions of molecular weight standards and the void volume (v.v.) are indicated.
The –20/24 complex has an apparent molecular mass of 335 kDa
SRm160, DEK, RNPS1, Y14 and REF have a combined molecular mass of at least 222 kDa [individual molecular masses (in kDa) based on sequence calculations are: SRm160, 93.5; DEK, 42.7; RNPS1, 34.2; REF, 32; and Y14, 20; although some of these proteins are likely to be subject to post-translational modification]. To determine whether the –20/24 complex was large enough to accommodate all of these proteins, we measured its approximate mass by gel filtration. To do so, AdML pre-mRNA containing a single labeled phosphate at position –20 was incubated under splicing conditions to generate mRNA; the reaction was treated with RNase A and then loaded onto a Superose 6 gel filtration column. Compared with molecular weight standards (data not shown), the 19 nt protected fragment eluted as a single peak with an apparent molecular mass of 335 kDa (Figure 5B). Even when hnRNP A1 (38.5 kDa), which is associated with this particular RNA fragment (see above), and the RNA fragment itself (6 kDa) are taken into account, the measured mass is large enough to accommodate all of the factors we have described here. Taken together with the IP results, the observation of a single gel filtration peak suggests that SRm160, DEK, RNPS1, Y14 and REF associate with this region of spliced mRNA as a single multi-protein complex.
Discussion
In this study, we demonstrate that pre-mRNA splicing stably alters mRNP structure at a conserved position, 20–24 nt upstream of mRNA exon–exon junctions, both in vitro and in vivo. At least five proteins become tightly associated with this region, probably as part of a single complex (Figure 6). The diverse attributes of these proteins may explain previously reported effects of pre-mRNA splicing on downstream mRNA metabolism (see below).
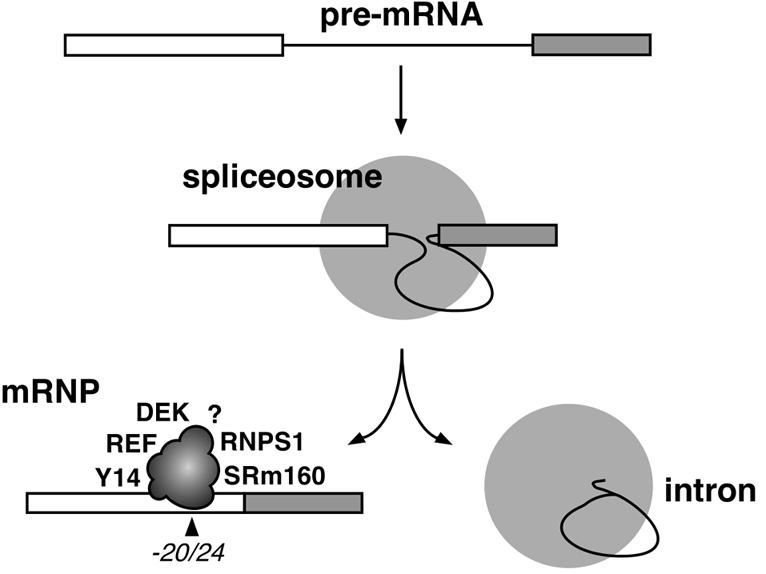
Fig. 6. Schematic illustration of the –20/24 complex deposited on mRNA as a consequence of pre-mRNA splicing. Identified proteins are indicated at arbitrarily chosen positions. The question mark symbolizes potential unidentified component(s).
Pre-mRNA splicing deposits multiple proteins at a conserved position upstream of exon–exon junctions independent of mRNA sequence
The RNase H analysis performed here is a very useful and straightforward assay for detecting stable structural changes in RNA–protein complexes (Gunzl and Bindereif, 1999). In this study, splicing-dependent changes in mRNP structure were readily detected by comparing the cleavage patterns of spliced and control mRNAs (Figure 1). When the entire length of TPI mRNA was evaluated, two regions were sufficiently altered by splicing to produce significant resistance to RNase H: the exon–exon junction and a region centered 24 nt upstream of the junction. Surprisingly, the latter protection proved characteristic of every mRNA tested. This protection exists in vivo and is conserved between amphibians and humans (Figure 2). The protection can be attributed to the splicing-dependent deposition of at least five proteins (Figure 3), probably as a single complex (Figure 5). The binding site for this complex spans 8 nt, including positions –20 and –24 relative to the exon–exon junction (Figure 4).
That pre-mRNA splicing can alter mRNP structure was expected from several earlier studies (Thermann et al., 1998; Zhang et al., 1998a,b;Luo and Reed, 1999; Le Hir et al., 2000; Zhou et al., 2000). However, the exact location(s) on mRNA where such changes occur had not been previously mapped in any systematic way. Our finding that the spliceosome deposits a multi-protein complex 20–24 nt upstream of exon–exon junctions on multiple mRNAs was unexpected. Not only is there is no consensus sequence in this region, but exons as short as 12 nt can support both steps of splicing in vitro (Duchene et al., 1988). In fact, previous studies analyzing protein–RNA interactions in splicing complexes have almost entirely ignored this region. In one study, Chiara et al. (1996) did report that the SR protein splicing factors SRp20 and SRp30 could be cross-linked in the early (E) splicing complex at positions –31 and –26 relative to the 3′ end of exon 1 of an AdML derivative. However, neither the generality of those cross-links nor whether they were detectable in later splicing complexes was examined. Our results indicate that contact between the spliceosome and the –20 to –24 region of the 5′ exon is a general occurrence, at least late in the splicing reaction.
The –20/24 proteins remain stably bound to mRNA without apparent preference for a particular RNA sequence. In this regard, the –20/24 proteins show some similarity to the protein–RNA interactions observed at a site in the intron that helps anchor U2 snRNP during assembly of splicing complex A. This ‘anchoring site’ corresponds to the 20 nt region immediately 5′ of the branch site. Although components of U2 snRNP make stable interactions with the anchoring site, this region contains no obvious conserved sequence elements (Gozani et al., 1996). Consistent with there being no consensus sequence in the –24 region of the 5′ exon, we demonstrated that the position of –20/24 protection is determined exclusively by its distance from the exon–exon junction and not by exon sequence (Figure 1D). Furthermore, –20/24 protein association with spliced mRNA is stable enough to survive complete RNase digestion or glycerol gradient fractionation followed by IP (Figures 3–5). It will be of interest to determine which of the protein(s) in this region interacts directly with mRNA and how these proteins maintain their grip without substantial base-specific contacts.
The –20/24 proteins include both pre-mRNA splicing and mRNA export factors
Three identified components of the splicing-dependent –20/24 complex are directly or indirectly related to the splicing process. SRm160, originally defined as a nuclear matrix component (Blencowe et al., 1994), is a member of the SR splicing factor family (Blencowe et al., 1998). A complex formed by SRm160 and SRm300, another nuclear matrix component, activates splicing of specific pre-mRNAs by interacting with SR proteins and U1 snRNP (Eldridge et al., 1999; Blencowe et al., 2000). Like SRm160, the phosphoprotein DEK associates with some pre-mRNAs and splicing complexes in vitro through interactions mediated by SR proteins (McGarvey et al., 2000). Interestingly, DEK also has chromatin remodeling activity (Alexiadis et al., 2000), but the relationship of this function to pre-mRNA splicing, if any, is unknown. RNPS1 contains an RNA recognition motif (RRM) and has been shown to generally activate splicing of both constitutively and alternatively spliced pre-mRNAs in synergy with SR proteins (Mayeda et al., 1999). Thus, the association of SRm160, DEK and RNPS1 with spliced mRNA may result from their associations with pre-mRNA during the early stages of spliceosome assembly.
The Y14 and REF proteins have no known functions in the splicing process per se, but they have both been linked to mRNA export (Kataoka et al., 2000; Stutz et al., 2000; Zhou et al., 2000; E.Izaurralde, unpublished results). Like RNPS1, both proteins contain RRMs, and REF can bind RNA in vitro (E.Izaurralde, unpublished results). Thus, RNPS1, Y14 and REF are all good candidates for being the –20/24 complex components that interact directly with spliced mRNA.
Another splicing factor known to interact directly with the 5′ exon during splicing is hPrp8p, a core component of U5 snRNP (Teigelkamp et al., 1995; Collins and Guthrie, 1999; Luo et al., 1999; Siatecka et al., 1999). We have previously shown that, like SRm160, hPrp8p can be cross-linked to the PIP85.B exon–exon junction specifically as a consequence of splicing (Le Hir et al., 2000). However, our current data suggest that hPrp8p is unlikely to be a general mRNP component. Not only is the mass of the –20/24 mRNP complex too small to accommodate a 270 kDa protein, but anti-hPrp8p antibodies fail to co-immunoprecipitate any mRNA that has been purified away from spliceosomes (data not shown). Furthermore, although hPrp8p could also be cross-linked to the exon– exon junction of AdML and TPI mRNAs in a splicing-dependent manner, not all such cross-linked species were released from spliceosomes (data not shown). This suggests that, while hPrp8p does generally interact with mRNA exon–exon junctions prior to their release from spliceosomes, this interaction does not persist in most mRNP complexes.
Functional implications of the –20/24 complex for mRNA export
It was recently demonstrated that some mRNAs produced by splicing are exported from the nucleus more efficiently than those not produced by splicing, suggesting that spliceosomes alter the mRNP so as to enhance its productive interaction with the nucleocytoplasmic transport machinery (Luo and Reed, 1999; Zhou et al., 2000). Therefore, it is notable that two of the –20/24 complex components defined here have been directly or indirectly linked to mRNA export.
REF, like its yeast ortholog Yra1p, is functionally required for mRNA export (E.Izaurralde, unpublished results; Strasser and Hurt, 2000; Stutz et al., 2000; Zhou et al., 2000). REF/Yra1p can bind RNA and simultaneously contact the mRNA export factor TAP (Mex67p in yeast; E.Izaurralde, unpublished results; Strasser and Hurt, 2000; Stutz et al., 2000). TAP/Mex67p is a shuttling protein that interacts with components of the nuclear pore complex, and Mex67p is essential for viability in yeast (Katahira et al., 1999; Bachi et al., 2000). Although TAP can bind directly to certain viral RNA export elements (Gruter et al., 1998; Bear et al., 1999), it does not have high affinity for RNA in general (Braun et al., 1999; Stutz et al., 2000). It has been suggested that the principal means of TAP association with cellular mRNA is through protein–protein interactions (Bachi et al., 2000; Strasser and Hurt, 2000; Stutz et al., 2000; E.Izaurralde, unpublished results). In addition to REF, TAP can associate with other RNA binding proteins in vitro and in vivo. One of these is Y14, which is found complexed in vivo with TAP, spliced mRNA and hnRNP proteins (Kataoka et al., 2000). Our findings that both REF and Y14 are contained in the –20/24 complex suggest that these proteins may mediate the preferential export of spliced mRNA by recruiting export factors, such as TAP, to spliced mRNPs. Since we could not detect TAP in the –20/24 complex (Figure 3B), it is possible that TAP is recruited at an intermediate stage between splicing and export that is not replicated under our in vitro conditions.
The conserved position of the –20/24 complex suggests a role in NMD
In addition to a role in mRNA export, it is possible that components of the –20/24 complex may ‘mark’ the positions of exon–exon junctions for the process of NMD (reviewed in Li and Wilkinson, 1998;Nagy and Maquat, 1998; Hentze and Kulozik, 1999; Maquat, 2000). In particular, the location of this complex at a fixed position upstream of the exon–exon junction could explain the previous observations that in-frame termination codons are generally not recognized as premature unless they are located >50–55 nt upstream of the 3′-most exon–exon junction of an mRNA. It is now known that introns and, by implication, a splicing-dependent modification of mRNP, are required for both nucleus-associated and cytoplasmic NMD (Sun et al., 2000). While it is debatable whether nucleus-associated NMD involves cytoplasmic ribosomes, the involvement of cytoplasmic ribosomes in cytoplasmic NMD indicates that at least some components of the splicing-dependent modification to mRNP accompany mRNA into the cytoplasm. Since REF and Y14 are known shuttling proteins (Kataoka et al., 2000; Zhou et al., 2000; E.Izaurralde, unpublished results), they are candidates for such a role. Future studies will explore possible functional interactions between components of the –20/24 complex and components of the translational machinery required for NMD.
Materials and methods
In vitro splicing
Uniformly labeled splicing substrates with a GpppG dinucleotide cap were synthesized by standard run-off transcription. TPI pre-mRNA corresponded to a portion of exon 6 through exon 7 of the human TPI gene. DNA templates for AdML (HMS81) (Gozani et al., 1994), β-globin (Reed and Maniatis, 1988) and PIP85.B (Query et al., 1994) were described previously. DNA templates for corresponding intronless control mRNAs were constructed by PCR. AdML pre-mRNAs and control mRNA containing a single labeled phosphate were synthesized by splinted ligation (Moore and Query, 1998). In vitro splicing reactions, analysis of splicing products and glycerol gradients were performed as previously described (Le Hir et al., 2000). Glycerol gradients (10–30% glycerol) contained 8 mM Tris–HCl pH 7.6, 40 mM KCl, 20 mM potassium glutamate, 2 mM magnesium acetate.
RNase digestions
Activation of endogenous RNase H activity was accomplished by adding each cDNA oligo (1 µM) directly to splicing reactions and incubating the reaction for an additional 10 min at 30°C. When mRNAs were purified by glycerol gradient fractionation, they were supplemented with exogenous RNase H (0.1 U/µl; USB). For complete RNase digestion, splicing reactions were supplemented with 0.5 mg/ml heparin, RNase T1 (1 U/µl; Sigma) and/or RNase A (0.02 µg/µl; Sigma) and incubated for an additional 10 min at 30°C. For gel filtration analysis, a 10 µl splicing reaction containing 4 nM singly labeled AdML pre-mRNA was incubated for 90 min at 30°C, supplemented with heparin, treated with RNase A, diluted with 300 µl of IP-150 buffer [150 mM NaCl, 50 mM Tris–HCl pH 7.6, 2 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.05% NP-40] and loaded onto a Superose 6 FPLC column (Pharmacia) equilibrated with the same buffer. Molecular weight standards were analyzed in parallel under the same conditions.
Xenopus laevis oocyte microinjections
Uniformly labeled AdML and β-globin pre-mRNAs and control mRNAs were synthesized as above, except with a 7mGpppG dinucleotide cap. Oocyte injections and analysis of microinjected RNA by denaturing gel electrophoresis and autoradiography were performed as previously described (Jarmolowski et al., 1994). For RNase H cleavage, 10 nuclei were collected 90 min after injection in 35 µl of RNase H buffer (100 mM Tris–HCl pH 7.5, 50 mM MgCl2, 250 mM KCl, 0.5 mM DTT) containing the corresponding cDNA oligo at 5 µM and incubated for 15 min at 30°C. RNase H cleavage was stopped by the addition of 200 µl of SDS–proteinase K buffer (300 mM NaCl, 50 mM Tris–HCl pH 7.5, 5 mM EDTA, 1.5% SDS, 2 mg/ml proteinase K). RNAs were extracted with phenol, precipitated and separated by 15% (AdML) and 10% (β-globin) denaturing PAGE, respectively.
Antibodies and IPs
The following antibodies were kindly provided by the person indicated: anti-SRm160 [monoclonal antibody (mAb) B1C8; B.J.Blencowe]; anti-DEK (G.Grosveld); anti-RNPS1 (A.Mayeda); anti-REF and anti-TAP-C (raised again the C-terminal domain of TAP; E.Izaurralde); anti-Y14 (mAb-4C4; G.Dreyfuss); anti-hnRNP A1 (mAb-4B10), anti-hnRNP C1/C2 (mAb-4F4) and anti-Sm (Ab-Y12; S.Piñol-Roma). Anti-3mCAP antibodies were purchased from Roche. All antibodies (except anti-3mCAP) were independently confirmed by western blotting; each detected a major band with the expected mobility in HeLa cell nuclear extract (data not shown). mAbs B1C8 and 4C4 were bound to protein A–Sepharose (PAS) beads (Pharmacia) via rabbit anti-mouse IgG + IgM (Pierce). All other antibodies were bound directly to PAS beads. IPs were performed as previously described (Le Hir et al., 2000) in IP-100 buffer (the same as IP-150, except 100 mM NaCl). IP efficiencies were calculated by dividing the intensity of a given precipitated fragment (measured by PhosphorImager; Molecular Dynamics) by the intensity of the same fragment present in the input reaction, taking into account the fact that only one-fifteenth of each input reaction was loaded on every gel.
Acknowledgments
Acknowledgements
We are grateful to the following people for providing antibodies: Ben Blencowe, Gideon Dreyfuss, Gerard Grosveld, Akila Mayeda and Serafin Piñol-Roma. We thank Natasha Levin, Cecilia Lu and Maidung Nguyen for technical support; members of the Moore and Maquat laboratories for helpful discussions; Torben Heick-Jensen, Charles Query and Michael Rosbash for critical comments on the manuscript; and Ben Blencowe and Gideon Dreyfuss for communicating results before publication. M.J.M. is an HHMI assistant investigator and H.L.H. was partially supported by a fellowship from Philippe Foundation. L.E.M. was funded by NIH grant DK33938 and E.I. was supported by the European Molecular Biology Organization (EMBO).
References
- Alexiadis V., Waldmann,T., Andersen,J., Mann,M., Knippers,R. and Gruss,C. (2000) The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev., 14, 1308–1312. [PMC free article] [PubMed] [Google Scholar]
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J., Tan,W., Zolotukhin,A.S., Tabernero,C., Hudson,E.A. and Felber,B.K. (1999) Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol., 19, 6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Nickerson,J.A., Issner,R., Penman,S. and Sharp,P.A. (1994) Association of nuclear matrix antigens with exon-containing splicing complexes. J. Cell Biol., 127, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Issner,R., Nickerson,J.A. and Sharp,P.A. (1998) A coactivator of pre-mRNA splicing. Genes Dev., 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Bauren,G., Eldridge,A.G., Issner,R., Nickerson,J.A., Rosonina,E. and Sharp,P.A. (2000) The SRm160/300 splicing coactivator subunits. RNA, 6, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach,E., Schmitt,C. and Izaurralde,E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J., 13, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M.D., Gozani,O., Bennett,M., Champion-Arnaud,P., Palandjian,L. and Reed,R. (1996) Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol. Cell. Biol., 16, 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A. and Guthrie,C. (1999) Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev., 13, 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis,M.J., Piñol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Duchene M., Low,A., Schweizer,A. and Domdey,H. (1988) Molecular consequences of truncations of the first exon for in vitro splicing of yeast actin pre-mRNA. Nucleic Acids Res., 16, 7233–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge A.G., Li,Y., Sharp,P.A. and Blencowe,B.J. (1999) The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl Acad. Sci. USA, 96, 6125–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P., Inada,T., Preiss,T., Hentze,M.W., Mattaj,I.W. and Sachs,A.B. (2000) The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell, 6, 191–196. [PubMed] [Google Scholar]
- Gorlach M., Burd,C.G. and Dreyfuss,G. (1994) The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res., 211, 400–407. [DOI] [PubMed] [Google Scholar]
- Gozani O., Patton,J.G. and Reed,R. (1994) A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J., 13, 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Feld,R. and Reed,R. (1996) Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Gruter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Gunzl A. and Bindereif,A. (1999) Oligonucleotide-targeted RNase H protection analysis of RNA–protein complexes. Methods Mol. Biol., 118, 93–103. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis,J., Gamberi,C., Jarmolowski,A., McGuigan,C. and Mattaj,I.W. (1995) A cap-binding protein complex mediating U snRNA export. Nature, 376, 709–712. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens,W.C., Izaurralde,E. and Mattaj,I.W. (1994) Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol., 124, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser,K., Podtelejnikov,A., Mann,M., Jung,J.U. and Hurt,E. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Yong,J., Kim,V.N., Velazkuez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- Krause S., Fakan,S., Weis,K. and Wahle,E. (1994) Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res., 214, 75–82. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.J. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Luo H.R., Moreau,G.A., Levin,N. and Moore,M.J. (1999) The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA, 5, 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. (2000) Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 827–868. [Google Scholar]
- Mayeda A., Badolato,J., Kobayashi,R., Zhang,M.Q., Gardiner,E.M. and Krainer,A.R. (1999) Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J., 18, 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T. et al. (2000) The acute myeloid leukemia-associated protein DEK forms a splicing-dependent interaction with exon–product complexes. J. Cell Biol., 150, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J. and Query,C.C. (1998) Uses of site-specifically modified RNAs constructed by RNA ligation. In Smith,C.W.J. (ed.), RNA–Protein Interactions: A Practical Approach. IRL Press, Oxford, UK, pp. 75–108. [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1996) The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol., 134, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Query C.C., Moore,M.J. and Sharp,P.A. (1994) Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev., 8, 587–597. [DOI] [PubMed] [Google Scholar]
- Reed R. and Maniatis,T. (1988) The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev., 2, 1268–1276. [DOI] [PubMed] [Google Scholar]
- Sachs A.B. and Varani,G. (2000) Eukaryotic translation initiation: there are (at least) two sides to every story. Nature Struct. Biol., 7, 356–361. [DOI] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Siatecka M., Reyes,J.L. and Konarska,M.M. (1999) Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev., 13, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K. and Hurt,E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Moriarty,P.M. and Maquat,L.E. (2000) Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J., 19, 4734–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., Newman,A.J. and Beggs,J.D. (1995) Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J., 14, 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Izaurralde,E., Ferreira,J., Daneholt,B. and Mattaj,I.W. (1996) A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol., 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell, 66, 759–768. [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y., LaDuca,J.P. and Maquat,L.E. (1998a) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998b) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]