Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 4.
Abstract
The Mpk1 MAPK of the yeast cell wall integrity pathway uses a non-catalytic mechanism to activate transcription of stress-induced genes by recruitment of initiation factors to target promoters. We show here that Mpk1 additionally serves a function in transcription elongation that is also independent of its catalytic activity. This function is mediated by an interaction between the docking site of Mpk1 and a D motif in the Paf1 subunit of the Paf1C elongation complex. A mutation in Paf1 that blocks this interaction causes a specific defect in transcription elongation of an Mpk1-induced gene, which results from Sen1-dependent premature termination through a Nab3-binding site within the promoter-proximal region of the gene. Our findings reveal a regulatory mechanism in which Mpk1 overcomes transcriptional attenuation by blocking recruitment of the Sen1-Nrd1-Nab3 termination complex to the elongating polymerase. Finally, we demonstrate that this mechanism is conserved in an interaction between the human ERK5 MAPK and human Paf1.
Keywords: transcription attenuation, Mpk1, Paf1, Sen1, ChIP
INTRODUCTION
The cell wall of the budding yeast S. cerevisiae is required to maintain cell shape and integrity (Klis, 1994). The cell must remodel this rigid structure during vegetative growth, and during pheromone-induced morphogenesis. Wall remodeling is monitored and regulated by the cell wall integrity (CWI) signaling pathway, which activates a MAP kinase (MAPK) cascade (reviewed in Levin, 2005). The MAPK cascade is a linear pathway comprised of Pkc1, a MEKK (Bck1), a pair of redundant MEKs (Mkk1 and Mkk2) and a MAPK (Mpk1/Slt2). Mpk1 is a functional homolog of human ERK5 (Truman et al., 2006), a MAPK activated in response to growth factors as well as physical and chemical stresses (Abe et al., 1996; Yan et al., 2001).
CWI signaling is induced in response to cell wall stressors, including elevated growth temperature, pheromone-induced morphogenesis, hypo-osmotic shock, and chemical cell wall antagonists, (Levin, 2005). This pathway activates two known transcription factors. One of these is Rlm1 (Dodou and Treisman, 1997; Watanabe et al., 1997), which is activated through phosphorylation by Mpk1 (Jung et al., 2002). The second is SBF (Madden et al., 1997; Baetz et al., 2001), a dimeric transcriptional regulator comprised of Swi4 and Swi6, which is essential for normal regulation of G1-specific transcription (reviewed in Breeden, 2003). Swi4 is the sequence-specific DNA-binding subunit, but Swi6 is required for binding to cell cycle-regulated promoters (Andrews and Moore, 1992; Sidorova and Breeden, 1993). Swi6 allows Swi4 to bind DNA at cell cycle-regulated promoters by relieving an auto-inhibitory intramolecular association of the Swi4 C-terminus with its DNA-binding domain. Additionally, Swi6 is the transcriptional activation component of SBF (Sedgwick et al., 1998).
SBF drives gene expression in response to cell wall stress in a manner that is independent of its role in G1-specific transcription (Kim et al., 2008; Truman et al., 2009). Mpk1 and its pseudokinase paralog, Mlp1, which is also activated by the MEKs of the CWI signaling pathway, use a non-catalytic mechanism to activate SBF. Activated (phosphorylated) Mpk1, or Mlp1, form a complex with Swi4 that associates with SBF-binding sites in the promoters of cell wall stress target genes independent of Swi6 (Kim et al., 2008; Kim and Levin, 2010). In this context, Mpk1 (or Mlp1) relieves the auto-inhibitory Swi4 interaction by binding to a MAPK docking site on Swi4 (a D motif) adjacent to the C-terminal Swi6-binding site (Truman et al., 2009). Although Swi6 is not required for Swi4 to bind to the promoters of cell wall stress genes, it must be recruited to the Mpk1-Swi4 complex to activate transcription.
Here, we show that Mpk1 also serves a non-catalytic function in transcription elongation through an association with the RNA polymerase II (Pol II)-associated complex (Paf1C). This complex, originally identified in yeast, is comprised of five subunits (Paf1, Cdc73, Rtf1, Ctr9, and Leo1; Mueller et al., 2004). It has been implicated in transcription start site selection (Stolinski et al., 1997), elongation (Costa and Arndt, 2000; Betz et al., 2002; Kim et al., 2010b), and as a platform for the recruitment of histone methyltransferases (Krogan et al., 2003; Wood et al., 2003) and 3′ end processing factors to the elongation complex (Mueller et al., 2004; Penheiter et al., 2005; Sheldon et al., 2005). The importance of this complex is underscored by the identification of various subunits of the human Paf1C as candidate oncogenes and tumor suppressor genes (Chaudhary et al., 2007). Despite this, little is known about the regulation of the Paf1C, or of mechanisms governing gene expression through this complex.
The Paf1C associates with the open reading frames of all Pol II-transcribed genes examined, consistent with its binding to Pol II (Pokholok et al., 2002; Krogan et al., 2002; Jaehning, 2010). However, global gene expression analyses suggested that the Paf1C is important for the expression of fewer than 5% of yeast genes (Penheiter et al., 2005), including many involved in progression of the cell cycle (Koch et al., 1999; Porter et al., 2002), and some for cell wall biosynthesis (Chang et al., 1999). Consistent with the idea that the Paf1C is important for the expression of a subset of Pol II-transcribed genes, the yeast Paf1C components are not essential, but their loss results in hypersensitivity to a variety of stresses (Betz et al., 2002), implicating this complex in the expression of stress-responsive genes. Among the conditions to which mutants in the Paf1C are hyper-sensitive is cell wall stress, prompting the suggestion that the CWI pathway plays a regulatory role in Paf1C-mediated transcription (Chang et al., 1999).
We demonstrate that the Mpk1-Paf1C association allows transcription elongation of the FKS2 gene by preventing recruitment of and premature termination by the Sen1-Nrd1-Nab3 complex. This complex is known to function specifically in the termination of short, non-polyadenylated Pol II transcripts, including small nucleolar RNAs (snoRNAs), cryptic unstable transcripts (CUTs), and at a few known mRNA attenuation sites (Ursic et al., 1997; Rasmussen and Culbertson, 1998; Steinmetz et al., 2001; 2006; Wyers et al., 2005; Arigo et al., 2006). Thus, our findings reveal a MAPK-driven mechanism for suppressing transcriptional attenuation of a stress-induced gene.
RESULTS
We showed previously by chromatin immunoprecipitation (ChIP) analysis that activated Mpk1 MAPK binds to the FKS2 promoter independent of its protein kinase activity through association with the Swi4 transcription factor (Kim et al., 2008). Because other MAPKs, most notably Hog1, have been shown to associate not only with the promoters of target genes, but with the coding regions as well (Pokholok et al., 2006; Proft et al., 2006), we extended our analysis of Mpk1 to ask if this MAPK binds to the FKS2 coding region. We found that Mpk1 associates with genomic regions along the length of the FKS2 gene in response to activation by cell wall stress induced by elevated growth temperature (Figure 1A). Moreover, the catalytically inactive Mpk1-K54R mutant form of Mpk1 behaved like wild-type in this assay. However, a mutant form of Mpk1 that cannot be phosphorylated by its upstream activating kinases (Mpk1-TAYF) failed to associate with any part of the FKS2 gene, revealing the requirement for activating signal. We demonstrated additionally that the pseudokinase paralog of Mpk1, Mlp1, also associates with the FKS2 promoter (Kim et al., 2008). In fact, it was this finding that led us to the understanding that Mpk1-driven FKS2 transcription does not require its protein kinase activity. Like Mpk1, activated Mlp1 also bound to the FKS2 coding region (Figure 1B). In contrast to the presence of Mpk1 and Mlp1 across the FKS2 gene, we found that the Swi4 and Swi6 transcription factors were present only at the promoter (Figure 1C and D), consistent with their established role in transcription initiation. This suggested a model in which Mpk1 and Mlp1 move from the transcription initiation complex to the transcription elongation complex, leaving Swi4-Swi6 behind.
Figure 1. Mpk1 and Mlp1, associate with the FKS2 promoter and coding region, but Swi4 and Swi6 bind only to the promoter.
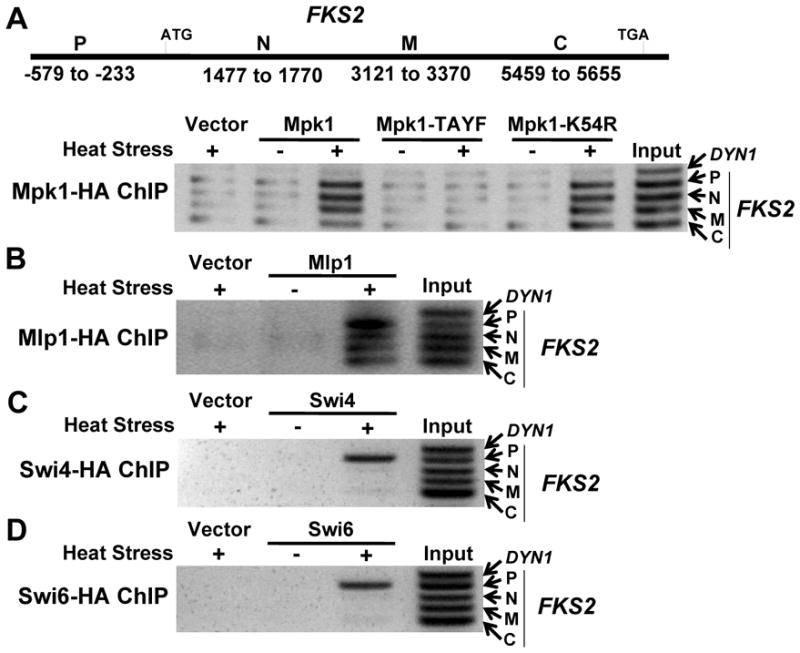
(A) Top; schematic of the FKS2 gene showing regions amplified for ChIP analyses along the length of the gene. P, promoter; N, N-terminal coding region; M, middle coding region; C, C-terminal coding region. Bottom; Mpk1 ChIP on FKS2. HA-tagged forms of Mpk1, an activation defective phosphorylation site mutant (Mpk1-TAYF), or a catalytically inactive form (Mpk1-K54R) were expressed from plasmids in an _mpk1_Δ _mlp1_Δ strain (DL3327). Cells were exposed to heat stress at 39°C for 2h, or unstressed at RT. A region from the middle of the DYN1 coding region was used as a negative control. (B) Mlp1 ChIP on FKS2. HA-tagged Mlp1 was expressed from a plasmid in an _mpk1_Δ _mlp1_Δ strain (DL3327). (C) Swi4 ChIP on FKS2. HA-tagged Swi4 was expressed from a plasmid in a _swi4_Δ strain (DL3145). (D) Swi6 ChIP on FKS2. HA-tagged Swi6 was expressed from a plasmid in a _swi6_Δ strain (DL3148).
Mpk1 associates with the Paf1C at the FKS2 promoter
Our starting assumption was that Mpk1 association with the elongation complex is integral to the mechanism by which this MAPK controls cell wall stress-induced transcription. Therefore, we wished to identify the factors required for Mpk1 to bind the FKS2 coding region. As a first step, we tested individual deletion mutants in non-essential genes that encode established or putative transcription elongation factors for their effects on FKS2-lacZ reporter expression. From among 71 deletion mutants we identified 25 that displayed impaired induction of FKS2-lacZ in response to heat stress (Table S1). Tests of two-hybrid interactions between Mpk1 and this subset of factors led us to focus on the Paf1C. All five components of this complex (Paf1, Cdc73, Rtf1, Ctr9 and Leo1; Mueller et al., 2004) interacted with Mpk1 in a cell wall stress-dependent manner (Figure S1). In addition to the Paf1C subunits, we also detected stress-induced association of Mpk1 with the Gcn5 and Ada2 subunits of the SAGA and ADA histone acetyltransferase (HAT) complexes (see below).
Because we were specifically interested in the mechanism by which Mpk1 associates with the transcription elongation complex, we next examined by co-immunoprecipitation the requirements for association of Mpk1 with Paf1. A variety of cell wall stress treatments stimulated this association (Figure 2A), indicating a requirement for activating signal to Mpk1. In support of this conclusion, we found that formation of the Mpk1-Paf1 complex was blocked in a non-phosphorylatable form of Mpk1 (Mpk1-TAYF; Figure 2B). However, Mpk1 catalytic activity was dispensable for Paf1-binding, as judged by the behavior of the catalytically inactive Mpk1-K54R protein.
Figure 2. Requirements for association of Mpk1 with Paf1.
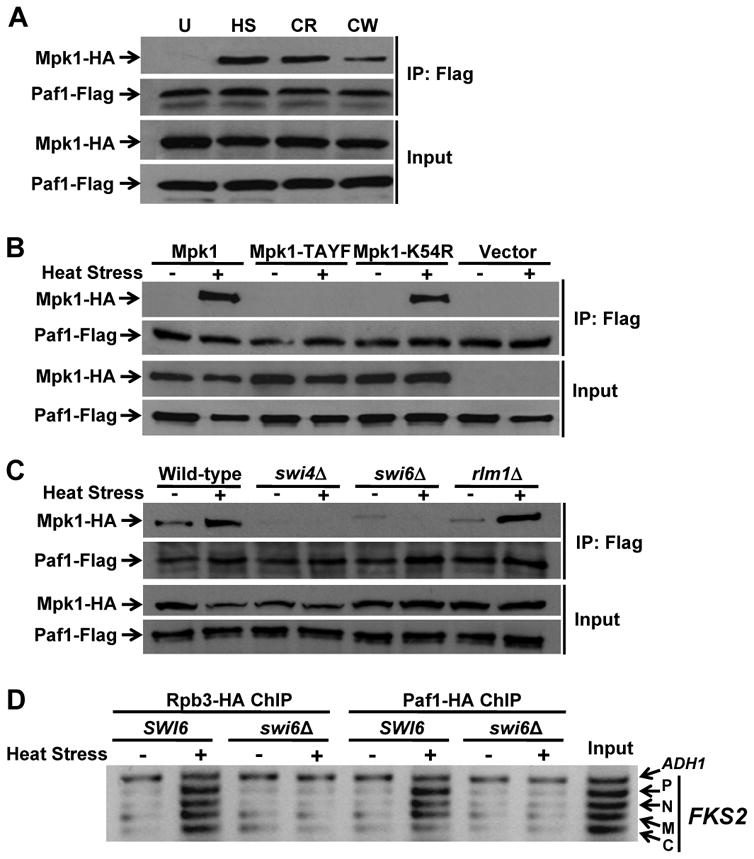
(A) The Mpk1-Paf1 interaction is stimulated by cell wall stress. Paf1-Flag was tested for co-immunoprecipitation (IP) of Mpk1-HA activated by various cell wall stresses for 2h – U, unstressed (RT); HS, heat stress at 39°C; CR, 50μg/ml Congo red; CW, 40μg/ml calcofluor white – in an _mpk1_Δ _mlp1_Δ (DL3183) strain expressing differentially tagged Mpk1 and Paf1. (B) The Paf1-Mpk1 interaction requires activating signal to Mpk1, but not catalytic activity. Mpk1-HA, an activation defective phosphorylation site mutant (Mpk1-TAYF-HA), or its catalytically inactive form (Mpk1-K54R-HA), was co-expressed with Paf1-Flag in an _mpk1_Δ _mlp1_Δ strain (DL3183). (C) The Paf1-Mpk1 association requires Swi4 and Swi6, but not Rlm1. Yeast strains were: wild-type (DL3187), _swi4_Δ (DL3405), _swi6_Δ (DL3233), and _rlm1_Δ (DL3586). (D) Neither Pol II, nor Paf1 are recruited to the FKS2 gene in the absence of Swi6. Rpb3-HA and Paf1-HA ChIP on FKS2 in wild-type cells (DL3187) and a swi6_Δ strain (DL3233). The A_DH1 promoter was used as a reference control.
In addition to an activating signal, formation of the Mpk1-Paf1 complex required the presence of Swi4 (Figure 2C). Thus, the association occurs under conditions in which activated Mpk1 forms a complex with Swi4 on the promoters of target genes. We interpret this to mean that the Mpk1-Paf1 interaction is restricted to genes regulated by Mpk1-Swi4. However, Mpk1-Swi4 promoter binding was not sufficient to allow the Mpk1-Paf1 interaction, which also required the presence of Swi6 (Figure 2C). Although Swi6 is not needed for Mpk1-Swi4 promoter occupancy, it is required for transcriptional activation of FKS2 (Kim et al., 2008). Therefore, the Mpk1-Paf1 association is dependent on the same factors required for FKS2 transcription initiation in response to cell wall stress. Consistent with this, the Mpk1-Paf1 interaction was independent of the Rlm1 transcription factor (Figure 2C), which controls cell wall stress transcription of other genes in response to Mpk1 activation (Jung and Levin, 1999). We noted additionally that, in the absence of Swi6, neither Paf1, nor the Rpb3 Pol II subunit were recruited stably to the FKS2 gene (Figure 2D), even though the Mpk1-Swi4 complex resides at the promoter under these conditions (Kim et al., 2008). These results suggest that the Pol II-Paf1C complex must also be recruited to Mpk1-Swi4-Swi6-regulated promoters for the MAPK to bind the Paf1C.
In contrast to the above results, the cell wall stress-induced association of Mpk1 with either Gcn5 or Ada2 was only modestly reduced in a _swi6_Δ mutant (Figure S2), indicating that these interactions occur largely independent of Mpk1-Swi4-Swi6-driven transcription. However, both interactions were greatly reduced in an _rlm1_Δ mutant, suggesting a more general role for HAT complexes in cell wall stress transcription. For this reason, and because the Paf1C had been implicated previously in the maintenance of cell wall integrity (Chang et al., 1999), we focused our attention on this complex.
Mpk1 associates with the Paf1C through a docking motif in the Paf1 subunit
Both Mpk1 and Paf1 are part of larger complexes on the DNA, so their interactions might be indirect. However, we speculated that Mpk1 associates directly with Paf1 for two reasons. First, Swi4 and Swi6 do not associate with the FKS2 coding region, indicating that Mpk1 does not interact with the Paf1C indirectly through either of these factors. Second, among the two-hybrid associations detected between Mpk1 and various Paf1C subunits, the strongest was with Paf1 itself, which appears to be the major scaffolding subunit of the complex (Jaehning, 2010; Kim et al., 2010b). Therefore, we first tested a set of N-terminal and C-terminal truncations of Paf1 for two-hybrid association with Mpk1. This analysis narrowed the region that is important for interaction to residues 86 – 131 (Figure 3A).
Figure 3. A Paf1 mutant that is defective for interaction with Mpk1.
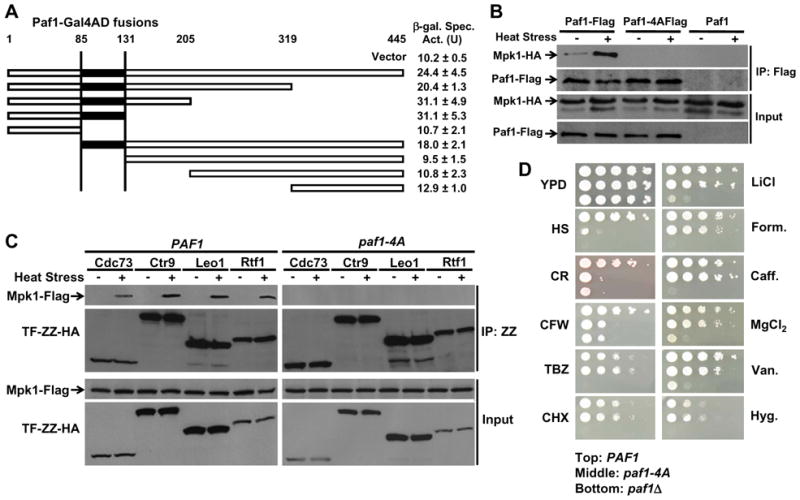
(A) Yeast 2-hybrid interaction of Mpk1 with Paf1 fragments. An Mpk1-Gal4DBD fusion was tested for interaction with the indicated Paf1-Gal4AD fusion under conditions of cell wall stress. End-points of Paf1 fusions are indicated at top. Fusions that contained residues 85–131 (black) displayed β-galactosidase activities (U, units) higher than empty vector. (B) A Paf1 mutant that fails to bind Mpk1. A _paf1_Δ strain (DL3548), expressing the indicated forms of Mpk1 and Paf1, was treated as in Figure 2. (C) Mpk1 fails to associate with any Paf1C subunit in the paf1-4A mutant. A _paf1_Δ strain (DL3548) expressing Mpk1-Flag, the indicated TAP-tagged Paf1C subunit (TF-ZZ-HA) and either Paf1, or Paf1-4A, were processed for co-immunoprecipitation of Mpk1 with the various Paf1C subunits. (D) The paf1-4A mutant is specifically hyper-sensitive to cell wall stress. A _paf1_Δ strain (DL3792), expressing PAF1 (top of each panel), paf1-4A (middle), or bearing empty vector (bottom), was subjected to the indicated stresses on YPD plates, or unstressed (YPD). Stresses were: heat stress, HS (39°C); Congo Red (CR, 50 μg/ml); Calcofluor white (CFW, 20 μg/ml); thiabendazole (TBZ, 50 μg/ml); cycloheximide (CHX, 50ng/ml); LiCl (50mM); formamide (Form., 3%); caffeine (Caff., 8mM); MgCl2 (750mM); sodium orthovanadate (Van., 3mM); and hygromycin (Hyg., 20μg/ml). Equivalent cell numbers of each strain were spotted as serial 10-fold dilutions onto plates and incubated for three days at 30°C (or 39°C).
MAPKs associate with their substrates and regulators through a canonical D motif – (Arg/Lys)1–2-(X)2–6-ΦA-x-ΦB (where Φ are hydrophobic residues Leu, Ile, or Val), recognized by a common docking site in the MAPK (Zhang et al., 2003). Because the Mpk1-Paf1 association did not require its catalytic activity, we reasoned that it might involve the Mpk1 docking site in the same manner that this MAPK associates with Swi4 (Truman et al., 2009). To explore this possibility, we next identified a single potential D motif in Paf1 within the stretch of sequence identified by the two-hybrid analysis (93-KDDRILLR-100) and constructed a mutant in which this site is ablated (residues 97–100 mutated to Ala; Paf1-4A). Consistent with the notion that this is a functional D motif, we found that the Paf1-4A protein failed to co-immunoprecipitate with Mpk1 in response to cell wall stress (Figure 3B).
To determine if the observed associations between Mpk1 and the other Paf1C subunits were indirect through Paf1, we tested by co-immunoprecipitation the effect of the paf1-4A mutation on pair-wise associations of Mpk1 with Paf1C subunits. Although Mpk1 associated in vivo with each of the Paf1C subunits, all of those associations were lost in the paf1-4A mutant (Figure 3C) suggesting that Mpk1 engages in a stable interaction with the Paf1C at a single point, which is ablated in the paf1-4A mutant.
The paf1-4A allele is defective for transcription elongation of FKS2
The Paf1-4A protein is generally functional, as judged by several criteria. First, the mutant subunit was detected in complex with each of the other Paf1C subunits and with the Rpb3 subunit of Pol II in vivo (Figure S3), indicating that the structural integrity of the Paf1C as well as its interaction with Pol II, which recruits Paf1C to the promoter (Jaehning, 2010), remain intact. Second, the phenotypes of the paf1-4A mutant revealed specific functional defects. Betz et al. (2002) demonstrated that a _paf1_Δ mutant is hyper-sensitive to growth inhibition by a wide variety of stress agents. Therefore, we compared the paf1-4A mutant to an isogenic _paf1_Δ mutant for growth inhibition by a panel of stressors (Figure 3D). Remarkably, the paf1-4A mutant behaved like the _paf1_Δ mutant only in response to cell wall stresses (i.e. heat stress, Congo Red, and calcofluor white). By contrast, this mutant exhibited the wild-type level of resistance to other stresses to which the _paf1_Δ mutant was sensitive. The only exception to this rule was caffeine, to which the _paf1_-_4_A mutant was resistant despite the fact that caffeine is a cell wall stress agent (Martin et al., 2000; Jung et al., 2002). However, caffeine has many cellular targets and we showed recently that although it activates Mpk1, it also provokes additional Ser phosphorylation on this MAPK through the DNA damage checkpoint pathway (Truman et al., 2009). The consequence of this novel phosphorylation of Mpk1 (on Ser423) is that it blocks its association with Swi4, thereby preventing transcription through Swi4/Swi6. Therefore, the caffeine sensitivity of the _paf1_Δ mutant cannot be the consequence of loss of the non-catalytic Mpk1 transcription pathway, consistent with the finding that the paf1-4A mutant is not sensitive to this agent.
The above results suggested that the paf1-4A mutant is specifically defective in gene expression that requires the Mpk1-Paf1 interaction. To test this directly, we compared FKS2 to CLN2 with respect to several parameters of gene expression in wild-type cells, and in the paf1-4A and _paf1_Δ mutants. We chose CLN2 because its expression, which is periodic through the cell cycle, is strongly dependent on both the Paf1C (Koch et al., 1999) and Swi4/Swi6 (Breeden, 2003), but independent of Mpk1/Mlp1 (Igual et al., 1996; Kim et al., 2008). First, transcription of CLN2 in asynchronous cultures was blocked in a _paf1_Δ mutant, but CLN2 mRNA levels were unaffected by the paf1-4A mutation, as measured by RT-PCR (Figure 4A). Second, the Paf1-4A protein retained the ability to associate with the CLN2 promoter and the CLN2 coding region (Figure 4B). Third, the Pol II subunit, Rpb3, associated with the CLN2 promoter and the CLN2 coding region in the paf1-4A mutant, but was not detected bound to the CLN2 gene at all in the _paf1_Δ mutant (Figure 4C). By all of these measures, the paf1-4A mutant is functional for CLN2 transcription.
Figure 4. The paf1-4A mutant is blocked for transcription elongation of FKS2, but not CLN2.
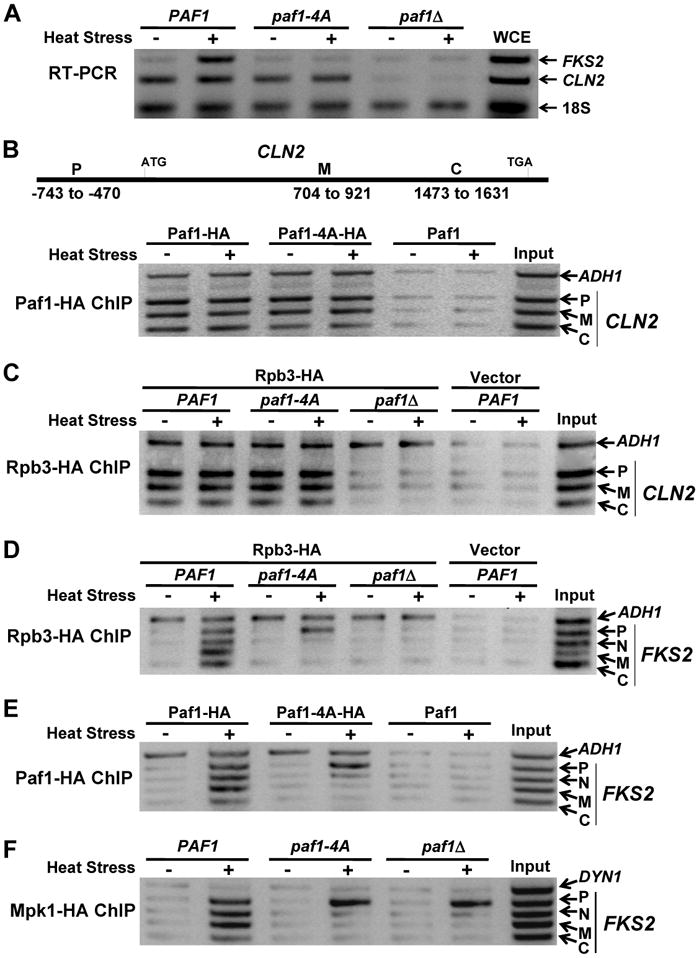
(A) RT-PCR of FKS2 and CLN2. A _paf1_Δ strain (DL3792) expressing PAF1, paf1-4A, or bearing empty vector was subjected to heat stress. Total RNA was processed for RT-PCR detection of FKS2 and CLN2 mRNA and 18S RNA. WCE; whole-cell extract. (B–F) ChIP analyses in a _paf1_Δ strain (DL3548) expressing the indicated forms of Paf1. (B) Top; schematic of the CLN2 gene showing regions amplified for ChIP along the length of the gene. P, promoter; M, middle coding region; C, C-terminal coding region. Bottom; Paf1-HA ChIP on CLN2. (C) Rpb3-HA ChIP on CLN2. (D) Rpb3-HA ChIP on FKS2. (E) Paf1-HA ChIP on FKS2. (F) Mpk1-HA ChIP on FKS2.
In contrast to the above, heat stress-induced transcription of FKS2 was blocked in both the paf1-4A and _paf1_Δ mutants (Figure 4A), consistent with the observed sensitivity of both mutants to cell wall stress agents. Although heat stress induced recruitment of Pol II (Rpb3) to the FKS2 promoter in the paf1-4A mutant, the polymerase did not progress to the coding region (Figure 4D), indicating a defect in transcription elongation of this gene. This conclusion was supported by the further finding that the Paf1-4A mutant protein was recruited to the FKS2 promoter, but did not progress to the FKS2 coding region (Figure 4E). Consistent with the absence of an elongation complex along the length of the FKS2 gene, Mpk1 that was recruited to the promoter also failed to progress to the coding region in either the _paf1_Δ or _paf1-4_A mutants (Figure 4F). Such a defect in elongation could be caused either by stalling of Pol II shortly after initiation, or by premature termination of transcription, a point to which we will return below. As was observed for the CLN2 promoter, Pol II failed even to be recruited stably to the FKS2 promoter in the _paf1_Δ mutant, supporting a previously-suggested role for the Paf1C in promoter site selection (Stolinski et al., 1997). This result contrasted with that observed for the ADH1 promoter, to which Pol II was recruited independent of Paf1 (Figure 4C and D), despite the presence of Paf1 on the ADH1 promoter in wild-type cells (Figure 4B and E; and Krogan et al., 2002).
Human Paf1 complements the Mpk1-dependent function of yeast Paf1
When expressed in yeast, the human ERK5 MAPK (hERK5) is activated in response to cell wall stress and suppresses the phenotypic defects of an _mpk1_Δ mutant (Truman et al., 2006). hERK5 also carries out the non-catalytic transcriptional function of Mpk1/Mlp1 (Kim et al., 2008), strongly suggesting that it interacts with Paf1 in the same manner as does Mpk1. This finding further suggests that the human Paf1C may be subject to similar regulation. Therefore, we examined the human PD2/PAF1 (hPAF1) sequence for potential MAPK-binding D motifs analogous to that present in yeast Paf1. Although yeast Paf1 shares only 24% amino acid sequence identity with the hPAF1 protein, we identified a potential D motif (93-RIDPNVLL-100; underlined residues defined by the D motif) within the same region of hPAF1 as the D motif in yeast Paf1 and constructed a mutant predicted to ablate this site (residues Val98 and Leu100 mutated to Ala; hPAF1-AA). hPAF1 and its point mutant were expressed in a yeast _paf1_Δ mutant and tested for the ability to complement its deficiencies in FKS2 and CLN2 transcription. The wild-type hPAF1 was able to restore CLN2 transcription and to restore partially cell wall stress-induced FKS2 transcription (Figure 5A), indicating functional complementation of the _paf1_Δ mutation. Significantly, the hPAF1-AA mutant was specifically blocked for FKS2 induction, suggesting that MAPK regulation of the Paf1C is conserved in humans.
Figure 5. hERK5 and hPAF1 interact to drive FKS2 expression.
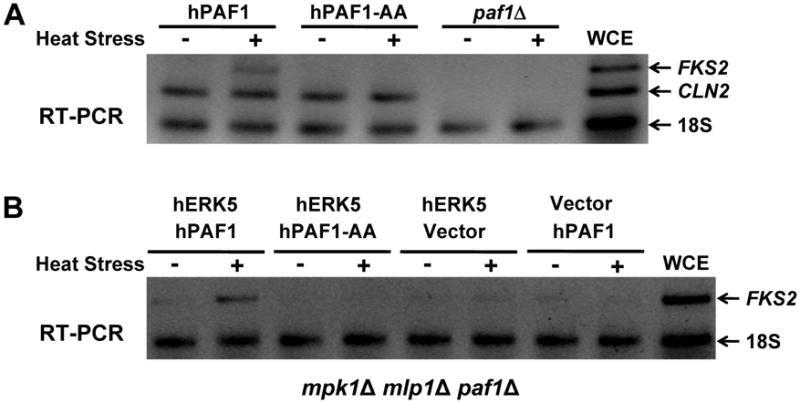
(A) and (B) RT-PCR of FKS2. (A) Expression of either hPAF1, or hPAF1-AA in a _paf1_Δ mutant (DL3792). (B) Co-expression of hERK5 and hPAF1 in a _paf1_Δ _mpk1_Δ _mlp1_Δ mutant (DL3980). WCE; whole-cell extract.
To determine if hERK5 can function in combination with hPAF1 to drive FKS2 expression in a yeast cell, we expressed both heterologous proteins in an _mpk1_Δ _mlp1_Δ _paf1_Δ mutant. FKS2 expression was induced in response to cell wall stress in this strain, but not in a strain expressing the hPAF1-AA mutant (Figure 5B), supporting the conclusion that hERK5 can interact with the D motif in hPaf1 to drive transcription elongation.
Sen1-Nrd1-Nab3-mediated transcription attenuation of FKS2
A large number of short (200 – 400nt) sense transcripts that terminate between −243 and −213 of FKS2 (relative to the ATG) have been identified through global transcription analyses conducted under non-inducing conditions (Miura et al., 2006, Neil et al., 2009). This suggests that attenuation (premature termination) shortly after initiation may be used to minimize the basal level of FKS2 transcription. Therefore, we explored the possibility that the critical function of the Mpk1-Paf1 association in transcription elongation was to prevent attenuation under inducing conditions. The paf1-4A mutant was tested for the production of prematurely terminated FKS2 transcripts using RT-PCR to detect the 5′ regions of FKS2 RNAs. Consistent with previous reports, we detected attenuated transcripts across the FKS2 promoter-proximal region under non-inducing conditions in both wild-type and paf1-4A cells (Figure 6A). Strikingly, although the paf1-4A mutant did not express extended FKS2 mRNA under stress conditions, it did induce attenuated transcripts, confirming that this mutant is defective in transcription elongation and suggesting that the Mpk1-Paf1 interaction serves an anti-termination function.
Figure 6. The Mpk1-Paf1 interaction prevents Sen1-mediated premature termination.
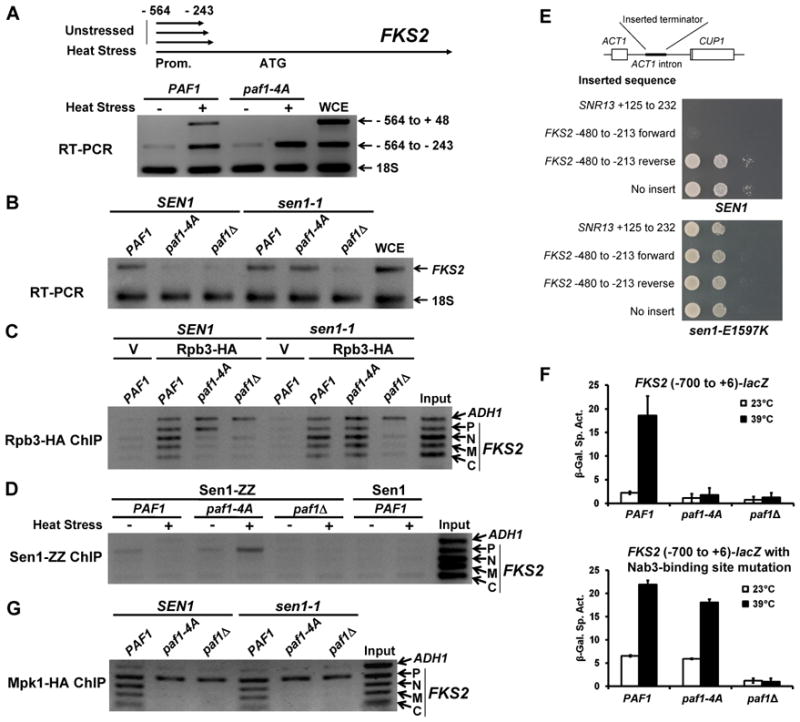
(A) RT-PCR of the FKS2 5′ region. A _paf1_Δ strain (DL3977) expressing PAF1, or paf1-4A was processed for RT-PCR detection of FKS2 transcripts. One primer pair detects only extended mRNA (−564 to +48). A second pair detects both attenuated transcripts and extended mRNA (−564 to −243). (B) RT-PCR of FKS2. A _SEN1 paf1_Δ strain (DL3977) and a _sen1-1 paf1_Δ strain (DL3978) expressing PAF1, paf1-4A, or bearing empty vector, were processed for RT-PCR detection of FKS2 expression after heat stress. (C) Rpb3-HA ChIP on FKS2. The same strains as in (B), but expressing Rpb3-ZZ-HA, were processed for ChIP after heat stress. V; vector. (D) Sen1-ZZ ChIP on FKS2 in a _paf1_Δ strain expressing Sen1-CBD-ZZ (DL3979) and the indicated forms of Paf1. (E) A region of the FKS2 promoter functions as a Sen1-dependent terminator. The indicated region of FKS2 sequence was inserted in both orientations into the ACT1 intron of an ACT1-CUP1 fusion plasmid, which was transformed into _SEN1 cup1_Δ (DL3990) and _sen1-E1597K cup1_Δ (DL3995) strains. The SNR13 terminator was used as a positive control. Serial dilutions were spotted onto plates with 0.4 mM CuSO4 and incubated for three days at the semi-permissive temperature for sen1-E1597K of 30°C. (F) The Nab3-binding site at position −476 in FKS2 is responsible for transcriptional dependence on the Mpk1-Paf1 interaction. Expression of the indicated FKS2-lacZ fusion was measured before and after heat stress in a _paf1_Δ strain (DL3548) expressing PAF1, paf1-4A, or bearing empty vector. (G) Mpk1-HA ChIP on FKS2. The same strains as in (B), but expressing Mpk1-HA, were processed for ChIP analysis on FKS2 after heat stress.
The Sen1-Nrd1-Nab3 transcription termination complex associates with Pol II near promoters and appears to be most important for termination within approximately 500 bp of transcription start sites (Steinmetz et al., 2006; Kim et al., 2010a). Significantly, the Paf1C has been shown to be important for recruitment of the Sen1-Nrd1-Nab3 complex to snoRNA genes, and mutations in genes encoding Paf1C subunits result in transcriptional read-through products of these genes (Sheldon et al., 2005). To test the involvement of the Sen1-Nrd1-Nab3 termination complex in FKS2 expression, we asked if a temperature-sensitive allele of SEN1 (sen1-1; Ursic et al., 1997) would suppress the FKS2 transcription defect in the paf1-4A mutant. In this experiment, raising the growth temperature to 39°C had two effects. First, it induced cell wall stress as in other experiments. Second, it inactivated Sen1 function. Figure 6B shows that FKS2 expression was restored to the paf1-4A mutant when Sen1 function was disabled, indicating that the FKS2 elongation defect that results from ablation of the Mpk1-Paf1 interaction is the consequence of Sen1-dependent premature termination. ChIP analysis also revealed that Pol II occupancy of the entire FKS2 gene was restored to the paf1-4A mutant in the absence of Sen1 function (Figure 6C).
To determine if the Mpk1-Paf1 interaction blocks recruitment or function of the Sen1-Nrd1-Nab3 termination complex in this setting, we next conducted Sen1 ChIP on FKS2. In wild-type cells, Sen1 was detected weakly in association with the FKS2 promoter under non-inducing conditions, but not with other parts of the FKS2 gene (Figure 6D). However, this weak association was not evident under inducing conditions, lending support to the suggestion that Sen1-dependent attenuation plays a role in repressing FKS2 expression under non-inducing conditions. Sen1 was also detected on the FKS2 promoter in the paf1-4A mutant under non-inducing conditions. However, in contrast to wild-type cells, its association increased under inducing conditions (Figure 6D), suggesting that the Mpk1-Paf1 interaction normally interferes with recruitment of the termination complex.
Recruitment of the Sen1-Nrd1-Nab3 complex to Pol II is influenced by the phosphorylation status of the C-terminal heptapeptide repeat domain (CTD) of the Pol II largest subunit (Rpb1). Both Nrd1 and Sen1 possess CTD-interaction domains (Steinmetz and Brow, 1998; Finkel et al., 2010) that contribute to binding of the Sen1 complex to CTD Ser5-phosphorylated Pol II (Vasiljeva et al., 2008). In contrast to this, CTD Ser2 phosphorylation appears to inhibit Sen1 complex binding (Gudipati et al., 2008). Additionally, a positional correlation between CTD Ser7 phosphorylation and recruitment of Nrd1 was observed recently (Kim et al., 2010a). Therefore, to determine if the Mpk1-Paf1 interaction affects recruitment of the Sen1 complex indirectly by altering the phosphorylation status of Pol II during FKS2 transcription, we carried out FKS2 ChIP analyses using antibodies specific for the Ser5-, Ser2-, or Ser7-phosphorylated forms of the CTD. For this experiment, we compared PAF1 to the paf1-4A mutant whose elongation defect was suppressed by the sen1-1 mutation. The Mpk1-Paf1 interaction did not affect the CTD phosphorylation status on FKS2 (Supplemental Figure S4), thus ruling out this potential mechanism for control of Sen1 recruitment.
Sen1-mediated termination is dictated by the recognition of specific sequences in the nascent RNA by Nrd1 (GTA[A/G]) and/or Nab3 (TCTT) (Steinmetz et al., 2001; Morlando et al., 2002; Carroll et al., 2004). As noted above, a large number of short transcripts are initiated from numerous start sites across the FKS2 promoter (Miura et al., 2006, Neil et al., 2009). Therefore, we tested several regions of the FKS2 promoter for the presence of a Sen1-dependent terminator using an ACT1-CUP1 fusion described by Steinmetz et al. (2001). Briefly, a terminator sequence inserted into the ACT1 intron (Figure 6E) disrupts expression of the full-length message, resulting in copper sensitivity of a yeast strain lacking the endogenous CUP1 gene. The region of FKS2 from −480 to −213 (relative to the ATG) behaved as a Sen1-dependent terminator (Figure 6E). Within this sequence, we found a single potential Nab3-recognition site at position −476. A point mutation at this site relieved dependence on the Mpk1-Paf1 interaction for transcription in the context of an FKS2-lacZ reporter (Figure 6F), revealing that the Sen1 termination complex acts through this solitary Nab3 site. Additionally, mutation of the Nab3 site caused a 3-fold elevation of basal _FKS2_-lacZ expression, supporting the conclusion that transcriptional attenuation by the Sen1 complex is used under non-inducing conditions to minimize FKS2 expression.
Finally, returning to our initial question concerning how Mpk1 associates with the FKS2 coding region during transcription, we tested the requirement for the Mpk1-Paf1 interaction for this association. Figure 6G shows that Mpk1 failed to associate with the FKS2 coding region in the paf1-4A mutant whose elongation defect was suppressed by the sen1-1 mutation. This result indicates that Mpk1 is tethered to the elongation complex through its interaction with Paf1.
DISCUSSION
MAPKs regulate transcription not only by phosphorylation of target transcription factors, but also through the formation of stable interactions on the DNA. This has been studied most thoroughly in the yeast Hog1 MAPK, which is activated by osmotic stress and uses several distinct mechanisms for inducing gene expression including recruitment of transcription factors, chromatin remodelers, and Pol II to responsive promoters (reviewed in de Nadal and Posas, 2010). All of these mechanisms require Hog1 protein kinase activity. By contrast, the yeast Mpk1 MAPK and its pseudokinase paralog, Mlp1, drive a portion of the cell wall stress-induced transcriptional program through a pathway that does not require protein kinase catalytic activity (Kim et al., 2008; Truman et al., 2009; Kim and Levin, 2010). Mpk1 and Mlp1 recruit the Swi4/Swi6 (SBF) transcription factor to target promoters through direct binding to Swi4. In this study, we investigated the role of Mpk1 association with the transcription elongation complex on a transcriptional target whose expression is driven through this non-catalytic pathway.
Mpk1 moves from the initiation complex to the elongation complex on the Paf1C scaffold
We found that active (phosphorylated) Mpk1 and Mlp1 associate with the coding region of the FKS2 gene during transcription by binding to the Paf1C elongation complex. MAPKs possess a docking site (called the D motif-binding site) for binding targets and regulators, which resides on the opposite side of the protein from the catalytic site (Zhang et al., 2003). The docking site on active Mpk1 (Truman et al., 2009), engages the Paf1C through contact with a D motif near the N-terminus of the Paf1 subunit. It appears that this is the only stable association made between Mpk1 and the Paf1C, because all of the other in vivo associations detected between Mpk1 and Paf1C subunits were lost upon mutational ablation of the D motif in Paf1.
An interesting aspect of the Mpk1-Paf1 interaction is that it requires both activating signal to Mpk1 and the presence of both Swi4 and Swi6. Mpk1 recruits the latter two factors to the FKS2 promoter through association of its docking site with a D motif in Swi4 (Kim et al., 2008; Truman et al., 2009). Although an Mpk1-Swi4 complex binds to the FKS2 promoter in the absence of the Swi6 transcription activation subunit (Kim et al., 2008), we found that Swi6 was required for stable recruitment of Pol Il and the Paf1C to this promoter. We conclude from these results that the Mpk1-Paf1 association occurs only under conditions in which Pol II and the Paf1C have also been recruited to the promoters of Mpk1-Swi4-Swi6-regulated genes. It seems likely, therefore, that the Mpk1-Paf1 interaction occurs specifically in the local context of such genes and is the consequence of a handoff of Mpk1 from Swi4 to Paf1 on the assembled initiation complex (Figure 7).
Figure 7.
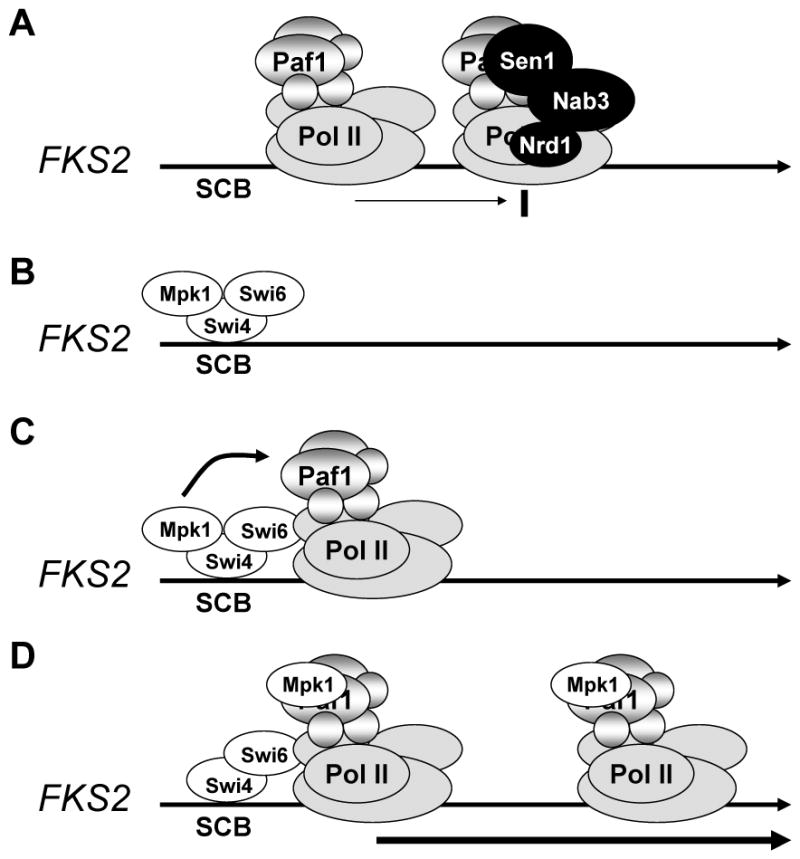
Model for Mpk1-driven FKS2 transcription. (A) Under non-inducing conditions, weak transcription initiation is attenuated by association of the Sen1-Nrd1-Nab3 complex to the elongation complex. (B) Under inducing conditions, activated Mpk1 recruits Swi4-Swi6 to the FKS2 promoter; (C) Pol II and the Paf1C are recruited to the promoter in a Swi6-dependent manner; (D) Mpk1 associates with Paf1, possibly by hand off from Swi4. Mpk1 functions as an anti-termination factor by blocking recruitment of the termination complex.
The Mpk1-Paf1 interaction is required for transcription elongation of FKS2
A mutation that ablated the D motif in Paf1 (paf1-4A) had the specific effect of blocking Mpk1-dependent functions of the Paf1C. Strikingly, the paf1-4A mutation blocked FKS2 transcription, but not the transcription of another gene (CLN2) whose expression is dependent on both SBF and the Paf1C, but not dependent on Mpk1. Consequently, this mutation caused hyper-sensitivity to cell wall stress agents, but not to other agents to which a _paf1_Δ mutant is hypersensitive. However, the most interesting behavior of the paf1-4A mutation is that it blocked transcription elongation of FKS2. Although in this mutant, Mpk1, Pol II, and Paf1-4A were all recruited to the FKS2 promoter in response to cell wall stress, none of these proteins was detected along the FKS2 coding region.
Mpk1 drives FKS2 transcription elongation by blocking recruitment of the Sen1-Nrd1-Nab3 termination complex
In principle, the observed failure of the elongation complex to progress beyond the FKS2 promoter in the paf1-4A mutant could result either from stalling of Pol II early in the elongation cycle, or from premature termination of transcription at a point close to the promoter. We found that, under inducing conditions, the paf1-4A mutant accumulated short attenuated transcripts across the FKS2 promoter-proximal region, indicating premature termination. Moreover, the paf1-4A transcription defect was suppressed by disabling the Sen1-Nrd1-Nab3 termination complex. This led to our identification of a Sen1-dependent terminator within the FKS2 promoter-proximal region, and the conclusion that expression of this gene is regulated in part by transcriptional attenuation. Under inducing conditions, we detected Sen1 on the FKS2 promoter in the paf1-4A mutant, but not in wild-type cells, suggesting that the Mpk1-Paf1 interaction interferes with recruitment of the Sen1 complex to Pol II, rather than preventing its activity after recruitment. Therefore, in this setting, active Mpk1 functions as an anti-termination factor that allows transcription of FKS2 by blocking recruitment of the Sen1 complex to the elongation complex early in the transcription cycle (Figure 7). It was reported previously that the Paf1C facilitates recruitment of the Sen1 complex to snoRNA genes (Sheldon et al., 2005), but the mechanism remains obscure. Our data suggest that, whatever the nature of the interaction between the Paf1C elongation complex and the Sen1 termination complex, it extends beyond snoRNA genes.
It is noteworthy that active Mpk1 is required for two steps in the FKS2 transcription cycle – initiation and anti-termination. We propose that, because FKS2 is expressed only under emergent conditions, Sen1-mediated termination is used to minimize FKS2 basal transcription during periods of unstressed growth (Figure 7). Attenuation would, therefore, have to be suppressed under inducing conditions. This hypothesis is supported by three findings. First, in the absence of cell wall stress, a large number of attenuated sense transcripts are produced across the FKS2 promoter-proximal region (Miura et al., 2006; Neil et al., 2009; and our Fig. 6A). Second, under non-inducing conditions we detected weak recruitment of Sen1 specifically to the FKS2 promoter (Fig. 6D). Third, mutation of the Sen1-dependent terminator in the FKS2 promoter-proximal region caused an elevation of basal FKS2 expression (Fig. 6F). Considering that the Mpk1-Paf1 interaction serves its anti-termination function shortly after initiation, it is not clear why Mpk1 remains associated with the elongation complex across most of the FKS2 gene. This MAPK may serve additional, as yet undiscovered, functions later in the FKS2 transcription cycle.
The Sen1 complex is required for termination and 3′ end formation of Pol II transcripts that are not polyadenylated (Ursic et al., 1997; Rasmussen and Culbertson, 1998). However, it has also been implicated in the transcriptional attenuation of several protein coding genes, including NRD1, HRP1, and IMD2 (Steinmetz et al., 2001, 2006; Arigo et al., 2006). Both NRD1 and HRP1 encode proteins involved in Sen1-mediated termination. Thus, their auto-regulation represents a specialized feedback mechanism. The IMD2 gene represents a similarly special case (Steinmetz et al., 2006). Our finding of the requirement for Mpk1-Paf1 association in blocking premature termination of FKS2 transcription reveals both an expanded repertoire for the Sen1-Nrd1-Nab3 termination complex on protein coding genes and a regulatory mechanism for its action. Indeed, two recent findings suggest a wider role for Sen1-dependent termination in transcription regulation than previously thought. First, Nrd1 is recruited widely to Pol II transcription units of all types (Kim et al., 2010a). Second, the genome-wide distribution of Pol II across both non-coding and protein coding genes is strongly influenced by disabling the Sen1 helicase (Steinmetz et al., 2006). We note additionally that short, sense transcripts are produced across the promoter regions of a significant fraction of protein coding genes in yeast (approximately 10%; Neil et al., 2009), suggesting that transcriptional attenuation may be a widespread phenomenon.
Paf1 is required for stable recruitment of Pol II to some promoters
We found that Paf1 is required for stable recruitment of Pol II to the CLN2 and FKS2 promoters, both genes whose expression requires Paf1. However, Paf1 is not universally required for Pol II recruitment. Indeed, that a _paf1_Δ mutant is not lethal indicates that a requirement for the Paf1C in Pol II recruitment is restricted to a small number of genes, despite its ubiquitous association with Pol II transcription units (Jaehning, 2010).
Two lines of evidence previously implicated the Paf1C in transcription initiation. First, the gene encoding the Rtf1 (Restores TBP Function) subunit of the Paf1C was isolated initially through the ability of a mutant allele to alter transcription start site selection (Stolinski et al., 1997). Second, the gene encoding the Ctr9 (CLN Three Requiring) subunit was isolated in two independent genetic screens through the defect of ctr9 mutants in Swi4/Swi6 (SBF)-driven gene expression (Di Como et al., 1995; Koch et al., 1999). Notably, the results of Koch et al. (1999) implicate the Paf1C in transcriptional activation of SBF-dependent promoters. Our results reveal that the Paf1C is required for stable recruitment of Pol II to at least a subset of SBF-dependent promoters. Because SBF is a key cell cycle-regulated transcription factor and cell cycle-regulated genes represent a large fraction of those whose expression is at least partially dependent on Paf1 (Porter et al., 2002), this may reflect a general role for the Paf1C in recruitment of Pol II to SBF-dependent promoters.
MAPK regulation of Paf1C function is conserved in human PAF1
We found that hPAF1 complemented the transcription defects of a _paf1_Δ mutant, at least with regard to FKS2 and CLN2 expression. Remarkably, mutation of a predicted D motif in hPAF1 within the same region as that found in yeast Paf1 specifically blocked FKS2 induction by cell wall stress, strongly suggesting that MAPK regulation of the Paf1C is conserved in humans. Moreover, because human ERK5 complements the defects associated with loss of yeast MPK1 and MLP1 function, including FKS2 induction (Truman et al., 2006; Kim et al., 2008), it seems likely that there is a role for an ERK5-PAF1 pairing in the regulation of human gene expression. Our finding that an _mpk1_Δ, _mlp1_Δ, _paf1_Δ yeast strain was complemented for FKS2 induction with hERK5 and hPAF1 supports this conclusion.
It is important to note that our findings are necessarily restricted to a small subset of stress-induced genes. Even among those activated in response to cell wall stress, most are not under the control of the non-catalytic Mpk1/Mlp1 pathway described here. It remains to be seen if the D motif in Paf1 is recognized by additional yeast MAPKs in response to other signals. In any case, that a _paf1_Δ mutant displays hyper-sensitivity to such a wide array of stress agents (Betz et al., 2002) suggests the Paf1C serves a poorly understood function in stress-induced gene expression. We were able to separate its function in cell wall stress transcription from its other functions by a point mutation in Paf1. Perhaps expression of other Paf1C-dependent genes is subject to a similar mode of regulation by factors yet to be identified.
EXPERIMENTAL PROCEDURES
Plasmids and strains
The S. cerevisiae strains and plasmids used in this study are listed in Tables S2 and S3, respectively. Their construction and growth conditions are described in Extended Experimental Procedures.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were conducted as described by Kim et al., (2008). Primers used to amplify specific regions of FKS2, CLN2, DYN1, and ADH1 are listed in Table S4. For each experiment, representative gels were chosen from at least three independent sets of extracts. See Extended Experimental Procedures for additional details.
Co-immunoprecipitation Assays
Protein extractions and co-immunoprecipitations were conducted as described previously (Kamada et al., 1995) with modification as described in Extended Experimental Procedures.
RT-PCR Assays
Total RNA was prepared as described previously (Schmitt et al., 1990) and cDNAs were made using Superscript III reverse transcriptase (Invitrogen), as per the manufacture’s instructions, and amplified by PCR with Taq polymerase. Primers used to amplify FKS2, CLN2, 18S cDNA sequences are listed in Table S4. See Extended Experimental Procedures for additional details.
Supplementary Material
01
02
Acknowledgments
We thank Judith Jaehning for comments on the manuscript; Steve Buratowski for helpful discussions; David Brow for the ACT1-CUP1 reporter system; Michael Culbertson for the sen1-1 mutant; Peter Piper for 2-hybrid fusions; Elizabeth Yranski for assistance with strain construction, and two anonymous reviewers for insightful comments. This work was supported by a grant from the NIH (GM48533) to D.E.L.
Footnotes
Supplemental Information including Extended Experimental Procedures, three figures, and four tables and can be found online at: XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe J, Kusuhara M, Ulevitch RJ, Berk BC, Lee JD. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J Biol Chem. 1996;271:16586–16590. doi: 10.1074/jbc.271.28.16586. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Moore LA. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc Natl Acad Sci USA. 1992;89:11852–11856. doi: 10.1073/pnas.89.24.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Baetz K, Moffat J, Haynes J, Chang M, Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol Cell Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Breeden LL. Periodic transcription: a cycle within a cycle. Curr Biol. 2003;13:R31–38. doi: 10.1016/s0960-9822(02)01386-6. [DOI] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the S. cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010;29:4–13. doi: 10.1038/emboj.2009.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Chang H, Arndt KT. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Treisman R. The S. cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Chinchilla K, Ursic D, Culbertson MR. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in S. cerevisiae. Genetics. 2010;184:107–188. doi: 10.1534/genetics.109.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Igual JC, Johnson AL, Johnston LH. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung US, Sobering AK, Romeo MJ, Levin DE. Regulation of the yest Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol. 2002;46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE. The protein kinase C-activated MAP kinase pathway of S. cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- Kim H, Erickson B, Lou W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010a;17:1279–1287. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010b;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-Y, Levin DE. Transcriptional reporters for genes activated by cell wall stress through a non-catalytic mechanism involving Mpk1 and SBF. Yeast. 2010;27:541–548. doi: 10.1002/yea.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-Y, Truman AW, Levin DE. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol. 2008;28:2579–2589. doi: 10.1128/MCB.01795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis FM. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- Koch C, Wollmann P, Dahl M, Lottspeich F. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res. 1999;27:2126–2134. doi: 10.1093/nar/27.10.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of S. cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;20:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in S. cerevisiae. Micro Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Sheu YJ, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in S. cerevisiae. J Biol Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511. [DOI] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Greco P, Dichtl B, Fatica A, Keller W, Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol Cell Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu X, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;4:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Porter SE, Washburn TM, Chang M, Jaehning JA. The yeast Pafl-RNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot Cell. 2002;1:830–842. doi: 10.1128/EC.1.5.830-842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Rasmussen TP, Culbertson MR. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in S. cerevisiae. Mol Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from S. cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick SG, Taylor IA, Adam AC, Spanos A, Howell S, Morgan BA, Treiber MK, Kanuga N, Banks GR, Foord R, et al. Structural and functional architecture of the yeast cell-cycle transcription factor Swi6. J Mol Biol. 1998;281:763–75. doi: 10.1006/jmbi.1998.1996. [DOI] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. A Requirement for the S. cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Breeden LL. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in S. cerevisiae. Mol Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in S. cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW, Kim K-Y, Levin DE. Mechanism of Mpk1 MAPK binding to the Swi4 transcription factor and its regulation by a novel caffeine-induced phosphorylation. Mol Cell Biol. 2009;29:2649–2661. doi: 10.1128/MCB.00794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW, Millson SH, Nuttall JM, King V, Mollapour M, Prodromou C, Pearl LH, Piper PW. Expressed in the yeast S. cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2 (Mpk1) cell integrity stress-activated protein kinase. Euk. Cell. 2006;5:1914–1924. doi: 10.1128/EC.00263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Luo H, Lee JD, Abe J, Berk BC. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J Biol Chem. 2001;276:10870–10878. doi: 10.1074/jbc.M009286200. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in S. cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhou B, Zheng C-F, Zhang Z-Y. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J Biol Chem. 2003;278:29901–29912. doi: 10.1074/jbc.M303909200. [DOI] [PubMed] [Google Scholar]
- Zhao C, Jung US, Garrett-Engele P, Roe T, Cyert MS, Levin DE. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol Cell Biol. 1998;18:1013–1022. doi: 10.1128/mcb.18.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02