Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations (original) (raw)
Abstract
Many pathogens colonize different anatomical sites, but the selective pressures contributing to survival in the diverse niches are poorly understood. Group A Streptococcus (GAS) is a human-adapted bacterium that causes a range of infections. Much effort has been expended to dissect the molecular basis of invasive (sterile-site) infections, but little is known about the genomes of strains causing pharyngitis (streptococcal “sore throat”). Additionally, there is essentially nothing known about the genetic relationships between populations of invasive and pharyngitis strains. In particular, it is unclear if invasive strains represent a distinct genetic subpopulation of strains that cause pharyngitis. We compared the genomes of 86 serotype M3 GAS pharyngitis strains with those of 215 invasive M3 strains from the same geographical location. The pharyngitis and invasive groups were highly related to each other and had virtually identical phylogenetic structures, indicating they belong to the same genetic pool. Despite the overall high degree of genetic similarity, we discovered that strains from different host environments (i.e., throat, normally sterile sites) have distinct patterns of diversifying selection at the nucleotide level. In particular, the pattern of polymorphisms in the hyaluronic acid capsule synthesis operon was especially different between the two strain populations. This finding was mirrored by data obtained from full-genome analysis of strains sequentially cultured from nonhuman primates. Our results answer the long-standing question of the genetic relationship between GAS pharyngitis and invasive strains. The data provide previously undescribed information about the evolutionary history of pathogenic microbes that cause disease in different anatomical sites.
Keywords: genomics, infectious disease, microevolution, pathogenomics, population genetics
For reasons that remain poorly understood, many species of pathogenic organisms have the ability to cause a diverse range of human illnesses. For example, bacteria such as Staphylococcus aureus, Haemophilus influenzae, and Streptococcus pyogenes [group A Streptococcus (GAS)] cause many distinct diseases that vary in severity. With the exception of production of certain toxins, how these and other organisms accomplish this remains largely unknown. Importantly, the genetic relationship among strains causing different disease manifestations is also poorly understood. High-throughput DNA sequencing technologies now allow us to address these issues at the full-genome level in large populations.
GAS is a Gram-positive human bacterial pathogen that causes several diseases, ranging from relatively mild superficial infections, such as pharyngitis and impetigo, to severe soft tissue infections, necrotizing fasciitis, bacteremia, acute rheumatic fever, and streptococcal toxic shock. Study of the epidemiology of GAS infections has led to a widely accepted model that most strains causing invasive infections arise from pharyngeal or other benign infections. Very little is known about the precise genetic relationship between strains isolated from pharyngitis and severe invasive infections, however. Several studies have suggested that pharyngitis and invasive strains originate from the same genetic pool (1, 2), but no full-genome analysis of pharyngitis and invasive strain populations has been performed to resolve this important issue definitively. Unlike bacterial species that frequently exchange genetic material, a process that complicates phylogenetic inferences, GAS strains exhibit relatively limited amounts of horizontal transfer across portions of the core genome, making them useful models for studying bacterial clonal evolution (3).
In this work, we tested the hypothesis that invasive and pharyngitis strains collected in one geographical area belong to the same genetic pool. We also tested the hypothesis that the genomes of pharyngitis and invasive strains have distinct genetic signatures of diversifying selection attributable to different selective forces operating in the host oropharynx and sterile-site environments. Here, we report our findings from whole-genome sequencing of 86 pharyngitis serotype M3 GAS strains collected from Ontario, Canada between 2003 and 2009. Our study provides a comparative population genomic analysis of pharyngitis and invasive GAS strains collected in the same geographical location.
Results and Discussion
Genome Sequencing.
We sequenced the genomes of all 86 emm3 GAS strains obtained from a convenience sample of 4,635 pharyngitis strains collected at six sites in Ontario from 2002 to 2009. One hundred invasive emm3 strains collected from 2002 to 2008 as part of a prospective, comprehensive, population-based surveillance study were used as a comparison group. Whole-genome short-read length DNA sequencing was used to generate over 12 gigabases of sequence information. This number corresponds to 139.5 megabases per strain, representing an average of 71-fold genomic coverage (range: 26.4- to 189.1-fold).
Comparison of Genome-Wide Polymorphism Levels.
The DNA sequence data were mapped to the genome sequence of emm3 reference strain MGAS315 (NC_004070), and polymorphisms were identified using a variant ascertainment algorithm (VAAL) (4). We found a cumulative total of 17,648 polymorphisms in the 86 pharyngitis strains. As observed in a recent study (3), the majority (61%) of these polymorphisms mapped erroneously to repetitive regions located in prophages and were excluded from further analyses. After removal of prophage regions, 6,825 cumulative core polymorphisms remained, corresponding to an average of 79 core polymorphisms per strain (range: 55–95) relative to the reference emm3 genome. This amount of genetic variation was very similar to that observed in the 100 invasive emm3 strains obtained during the same time period (77 polymorphisms per strain). Thus, invasive and pharyngitis GAS strains have virtually identical amounts of average genetic diversity. If invasive GAS isolates represent a restricted genetic subset of the emm3 strains causing pharyngitis, a much more limited amount of diversity would have been expected.
The 6,825 cumulative core polymorphisms were distributed across 558 unique polymorphic sites. SNPs comprised 81.2% (453 of 558) of the core polymorphic sites in the 86 pharyngitis strains, of which 81.2% (368 of 453) were located in predicted coding sequence regions. Approximately half of the SNPs (43.7%) were unique to a single strain (i.e., were strain-specific), closely similar to the level (44.7%) observed in invasive strains isolated from the same time period in Ontario. The percentage of SNPs resulting in amino acid replacements (i.e., nonsynonymous nucleotide substitutions) was virtually identical in pharyngitis and invasive strains (69% and 71%, respectively) and similar to the level expected by chance. Thus, we found no significant bias toward nonsynonymous nucleotide substitutions in either strain population.
The other 18.8% of the core polymorphisms were insertions and deletions (indels). This may be an underestimation of the total number of indels, however, because of the inherent limitations of short-read-length sequencing technologies. Regardless of strain source (pharyngitis or invasive infection), 51% of all indels were located in core coding sequences. This finding was unexpected in light of our recent analysis of 95 invasive emm3 strains in which coding indels comprised only 39% of all indels (3). Detailed analysis of the coding indels in the pharyngitis strains discovered a striking overrepresentation of indels (12 of 54 coding indels) in the hasA and hasB genes. These two genes encode proteins essential for the synthesis of hyaluronic acid (5–9), a key component of the antiphagocytic GAS capsule.
Gene Regions in the GAS Genome Influenced by Diversifying Selection.
Our recent genome sequence analysis of 95 invasive emm3 strains identified several genes with a significant excess of allelic variation (i.e., greater than expected by chance alone), including ropB, covR/S, emm3, and hasB (3). Virtually all (35 of 37) SNPs in these genes resulted in amino acid replacements (3). This finding indicated that certain genes in invasive GAS strains had evidence of strong diversifying selection, presumably exerted by the host environment.
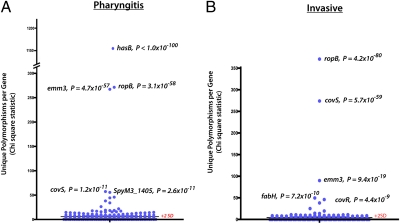
We hypothesized that selective pressures at distinct anatomical sites (e.g., upper respiratory tract, normally sterile site such as blood) influence the genomes of pharyngitis and invasive strains differently. To test this hypothesis, we first determined if, as a population, genes in pharyngitis strains had evidence of nonrandom distribution of SNPs. We discovered that genes in the has operon were under strong diversifying selection (Fig. 1). In particular, the hasB gene had an extremely high level of genetic variation, with 22 unique polymorphisms (less than 1 polymorphism was expected by chance alone). All polymorphisms in hasB were either nonsynonymous nucleotide substitutions or indels that shifted the reading frame (Fig. 2_B_). Moreover, all 3 polymorphisms in the hasA gene were single-nucleotide insertions or deletions (frameshift mutations), resulting in truncation of the HasA protein in the aminoterminal region. Because hasA and hasB are required for capsule production (5–9), we conclude that the environment GAS occupies in the host upper respiratory tract exerts considerable selective pressure against a functional hyaluronic acid biosynthesis pathway and subsequent capsule production.
Fig. 1.
Comparison of GAS genes with an excess of polymorphisms in pharyngitis and invasive strains. Shown is the distribution of χ2 statistics along with corresponding Bonferroni-adjusted P values to correct for multiple testing. Analysis for pharyngitis (A) and invasive (B) strains is shown.
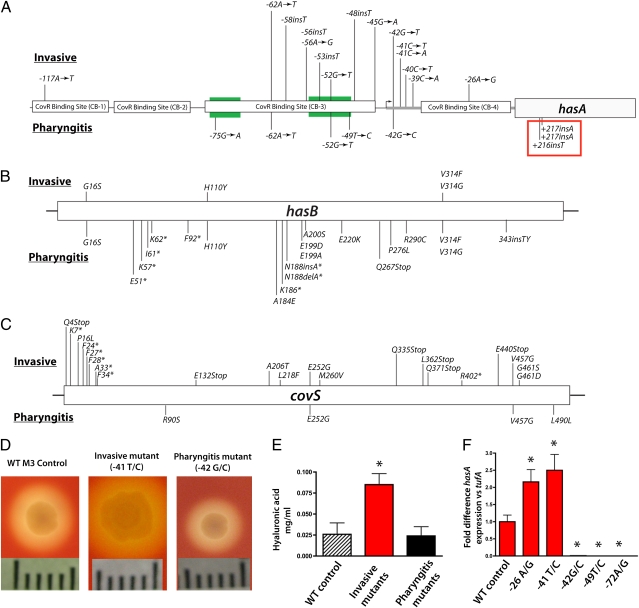
Fig. 2.
Phenotypic impact of genetic variation in the has operon. Schematic of polymorphisms within the has operon promoter (A), hasB (B), and covS (C) genes. Polymorphisms found in invasive strains are shown above the diagram, whereas polymorphisms in pharyngitis strains are indicated below. Regions involved in CovR binding are indicated in black boxes, and the −35/−10 regions are shown in green. Insertions shown in the red box were identified in strains collected from the nonhuman primate experimental pharyngitis protocol. Nucleotide positions in A are labeled according to their distance from the translation GTG start codon. In B and C, labels refer to amino acid positions and asterisks indicate insertions or deletions that result in early protein truncation. (D) Photographs showing colony morphology differences in has promoter mutants. (Scale bar: 1 cm.) (E) Hyaluronic acid quantification for invasive and pharyngitis strains. (F) Quantitative PCR measurement of hasA transcript levels for pharyngitis and invasive mutants shown in pharyngitis appear in black. Asterisks in E and F indicate statistically significant differences relative to control calculated using the Student's t test assuming unequal variances. Values for invasive strains are shown in red, and those for pharyngitis strains are shown in black.
Some GAS genes had evidence of the action of diversifying selection in both the pharyngitis and invasive strain populations. For example, the high level of allelic variation in ropB was closely similar between these two strain populations (0.114 vs. 0.102 alleles per strain). All nine ropB SNPs in the pharyngitis strains resulted in amino acid replacements, a result consistent with our recent report of ropB variation in invasive strains (3). Thus, diversifying selection appears to be acting on the ropB gene in both pharyngitis and invasive isolates.
We also observed an excess of polymorphisms in the covS gene, greater than what was expected by chance. There was a striking difference in magnitude between invasive and pharyngitis populations, with 21 unique polymorphisms in invasive strains but only 4 in pharyngitis strains (P = 0.0008, Fisher's exact test). There was also a difference in the predicted effect of these polymorphisms in invasive strains, with seven indels resulting in frameshift mutations and six producing nonsense mutations (Fig. 2_C_). In the aggregate, 14% of all polymorphisms predicted to result in premature protein truncation were found in covS.
In contrast to our previous findings in invasive emm3 strains (3), we found no evidence of diversifying selection acting on covR in pharyngitis strains. This is consistent with the hypothesis that disruption of covR and resulting derepression of virulence factor production is an important pathway by which GAS pharyngitis strains gain enhanced invasive potential (10, 11). It is important to note that our findings do not provide insight into whether acquisition of covR mutations occurs in the host upper respiratory tract and facilitates access or occurs after GAS has reached the sterile site. The importance of covR/S mutations in the pathogenesis of invasive GAS infections was illustrated by the observation that 37% of all invasive strains we studied contained at least one polymorphism in covR or covS, compared with only 4.6% of pharyngitis strains.
In summary, we discovered a complex pattern of genome-wide nucleotide variation, with some genes, such as hasA, showing evidence of diversifying selection exclusively in pharyngitis strains; some genes, such as covR, showing evidence of selection in invasive strains but not in pharyngitis strains; and some genes, such as ropB and emm3, which had similar levels of apparent diversifying selection, showing evidence of selection in pharyngitis and invasive strains.
Divergent Signals of Diversifying Selection in the has Capsule Synthesis Gene Region.
Intergenic regions frequently have DNA sequences involved in regulation of transcription of contiguous or nearby genes. Thus, we next tested the hypothesis that the invasive and pharyngitis populations differed in the abundance of intergenic polymorphisms. The intergenic region with the highest number of polymorphisms was located upstream of the has operon. This intergenic region contains binding sites for the transcription regulator CovR and sequences essential for expression of has genes (12). A high level of polymorphisms in this intergenic region also was present in both pharyngitis and invasive strain populations. The location of the genetic variants was remarkably different in the invasive and pharyngitis populations, however (Fig. 2_A_). This discovery suggests that mutations at distinct sites lead to different amounts of gene product depending on whether it is beneficial or detrimental for the organism to produce capsule. The polymorphic variants in the invasive strains were largely clustered in regions containing binding sites for CovR or were specific nucleotide changes located immediately downstream of the transcript start site that are associated with a high capsule phenotype (13). In contrast, pharyngitis strains had has promoter variants that clustered in the −35/−10 boxes (Fig. 2_A_), regions predicted to down-regulate expression of the has operon if mutated. The data provide strong population genetic evidence of differing evolutionary forces acting on the intergenic region upstream of the has operon.
Next, we tested the hypothesis that the has genetic alterations we identified altered capsule phenotype. Strains with polymorphisms in the has intergenic region had strikingly different colony morphologies compared with strains with the WT sequence (Fig. 2_D_). These differences in capsule phenotype led us to test the hypothesis that has promoter polymorphisms result in increased transcription of the has operon in invasive strains, whereas polymorphisms in pharyngitis strains would decrease has transcript levels. We measured the level of hasA transcript using TaqMan quantitative RT-PCR with specific primers and probes. Significant differences in hasA transcript levels were found between invasive and pharyngitis mutants compared with the WT strain (P < 0.001 for both; Fig. 2_F_). As predicted, we observed a 2.2- to 2.5-fold increase in hasA transcript level in invasive strains. Also, as anticipated, we found striking reductions in the hasA transcript level in pharyngitis strains with has promoter mutations, ranging from a 97- to 1,052-fold decrease relative to WT strains.
These findings indicate that differing selective pressures in the upper respiratory tract and sterile sites affect evolution of the has operon in distinctly different ways. During epidemics of invasive infection, GAS strains are often highly encapsulated and have increased virulence in animal models (14–16). This increased virulence of mucoid strains can be significantly attenuated by disruption of genes in the has operon (17, 18). In contrast, in the upper respiratory tract, selection operates against the production of hyaluronic acid. Reduced capsule production may increase the exposure of GAS surface proteins, such as M protein, and facilitate interaction with host proteins. Numerous other host and nonhost selective forces need to be considered as well, however. For example, the presence of certain cocolonizing bacteria in the upper respiratory tract, or bacterial stress induced by differences in nutrient conditions, could provide differential selective forces.
Nonhuman Primate Upper Respiratory Tract Infection.
We sequentially sampled GAS strains isolated from nonhuman primates using a previously described experimental pharyngitis protocol (19). Strains collected in the course of this study permitted us to test the hypothesis that function-altering polymorphisms in the hyaluronic acid biosynthesis operon would arise in the course of longitudinal interaction of GAS with the host. Full-genome sequencing of 11 strains obtained at 35 and 44 d after inoculation identified truncation mutations in hasA in 3 strains from two individual monkeys (Fig. 2_A_). These polymorphisms were not present in the input strain, indicating that they rose to abundance in vivo. Additional large-scale longitudinal studies of human cohorts may provide further details about the role of inactivation of the has operon during GAS infection.
Analysis of Prophage Content in Pharyngitis Genomes.
Mobile genetic elements, such as prophages, that encode virulence factors play an important role in host-pathogen interactions (20–22). Typically the sequenced emm3 GAS strains have as many as six prophages per strain, and each prophage typically encodes one or two virulence factors (3, 20). Inasmuch as these prophage-encoded genes are important for virulence, we tested the hypothesis that, as a population, the pharyngitis and invasive strains differed in prophage content. Analysis of the short-read sequence data with the MOSAIK and Velvet assemblers (23) did not reveal significant differences in the prophage content of the pharyngitis and invasive strain populations.
Genetic Population Structure Comparison.
One of the key goals of our study was to analyze the population genetic relationship between pharyngitis and invasive GAS type emm3 strains. Specifically, we sought to test the hypothesis that the invasive emm3 strains represent a genetic subset of strains causing pharyngitis. One possibility is that strains causing invasive infections represent a distinct phylogenetic lineage of emm3 strains that have enhanced invasive potential because they have special genetic attributes, such as unique virulence factors. Alternatively, it is possible that invasive strains originate from essentially any phylogenetic lineage represented among the pharyngitis population through acquisition of genetic changes that increase their ability to cause invasive infection. The full-genome strategy permitted us to differentiate precisely between these two possibilities.
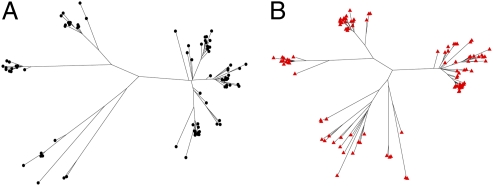
To address this issue, we generated a phylogenetic tree using the genome data for the 86 pharyngitis strains and compared its structure with one previously formulated for invasive emm3 strains from Ontario collected in the same time frame. The phylogenetic tree of pharyngitis strains was constructed based on a set of 453 concatenated biallelic core SNPs identified by the VAAL. The overall structure of the resulting tree (Fig. 3_A_) was strikingly similar to that of the tree for the invasive strains (Fig. 3_B_) and included all four primary genetic lineages previously identified among the Ontario invasive M3s (3). The similarity in tree topology suggested that pharyngitis and invasive strains had common phylogenetic structures.
Fig. 3.
Unrooted neighbor-joining phylogenetic trees assembled from the complete list of all core biallelic SNPs. The trees for the 86 pharyngitis strains (A) and for 100 temporally matched invasive strains (B) are shown. Despite being assembled completely independently from each other, both phylogenetic trees show a remarkably similar overall structure, suggesting common evolutionary histories.
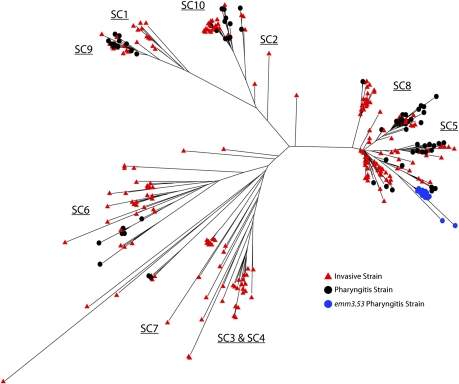
We next sought to determine if pharyngitis and invasive strains clustered in identical or distinct branches of the phylogenetic tree. We constructed a combined tree based on a set of 1,434 biallelic core SNPs identified by the VAAL in the 86 pharyngitis and 215 invasive emm3 strains from Ontario. The analysis revealed that overall, pharyngitis and invasive strains are interspersed and clustered tightly with one another in the tree (Fig. 4). This finding effectively ruled out the idea that invasive GAS isolates represent only one or a limited subset of GAS lineages; that is, as a population, strains causing invasive infections and pharyngitis have had closely similar evolutionary histories. These data strongly suggest that individual invasive isolates are derived repeatedly from the population of pharyngitis strains. Further, these recently derived invasive strains likely do not survive long in the population and are seldom transmitted among patients with invasive infections. Thus, the sterile-site niche undergoes frequent repopulation. Because pharyngitis and invasive strains do not differ as a population in virulence gene content, the most likely explanation for the virulence differences is acquisition of relatively minor genetic alterations (e.g., SNPs, short indels) that enhance virulence, such as function-altering mutations that dysregulate CovR/S signaling. Future analyses of other GAS populations, such as asymptomatic skin and upper respiratory tract carrier strains, will add needed information about their phylogenetic relationships and provide a more complete picture of GAS population genetics.
Fig. 4.
Combined phylogenetic tree of invasive and pharyngitis strains. Unrooted neighbor-joining phylogenetic trees were assembled from the complete list of all core biallelic SNPs.
Recently Emerged Phylogenetic Lineages.
Based on targeted gene sequencing of serotype M1 organisms, Hoe et al. (2) reported that, as a population, strains causing invasive episodes emerge as distinct subclones that cause pharyngitis. These investigators reported that distinct subclones emerged in pharyngitis cases approximately 9 mo before their recovery from invasive episodes. It was hypothesized that this lag was related to the time required for a subclone to become abundant in the pharyngitis population (2). Thus, we tested the hypothesis that there existed recently emerged subclone lineages in the pharyngitis strains that were not present in the invasive strain population. Consistent with this hypothesis, we identified 16 strains, all from Ottawa, with a rare emm3 allele, the emm3.53 allele. This allele differs from the most common emm3 allele present in Ontario (emm3.2) by a single nonsynonymous nucleotide substitution. The full-genome analysis demonstrated that all strains with the emm3.53 allele are very closely related (Fig. 4), differing, on average, by only eight core SNPs from each other. The greatly restricted genomic variation among emm3.53 strains strongly indicates identity by recent descent. Further, the topology of the tree supports the idea that emm3.53 strains emerged from a precursor strain with the emm3.2 allele (Fig. 4). Despite the high frequency of pharyngitis strains in Ottawa with the emm3.53 allele, none of the invasive strains in all of Ontario had the emm3.53 allele. Further analysis will be required to determine if this clone continues to disseminate throughout Ontario and whether this lineage specifically causes pharyngitis and not invasive episodes.
Concluding Comment.
In summary, our study describes results of the largest whole-genome comparative analysis of a bacterial pathogen to date and is a genome-wide investigation of GAS strains causing upper respiratory tract infection. The data provide information about how differing host selective forces operate to shape genome evolution of a bacterial pathogen occupying different anatomical sites within the host. Our findings clearly illustrate a model of GAS evolution in which invasive strains originate from all major lineages of strains causing pharyngitis. Invasive strains are genetically more similar to the population of pharyngitis strains from which they evolved rather than other invasive strains as a whole, confirming, on a genome level, observations that during epidemics of invasive disease, circulating pharyngitis strains frequently share morphological characteristics found in invasive strains (16, 24). Further, we did not identify any single highly prevalent genetic variant that would explain the differences in disease phenotype between pharyngitis and invasive strains. Rather, we found that diversifying selection in the invasive population drives the accumulation of rare variants that alter function, such as the CovR/S global gene regulatory system. This finding indicates an important role for rare genetic variants in bacterial pathogenesis, an observation strikingly similar to the recent attention devoted to the role of rare human genetic variants in common diseases in man (25).
Methods
Strain Collection.
Eighty-six serotype emm3 pharyngitis strains were collected by convenience sampling from six regional laboratories across Ontario from 2002 to 2010. Two hundred fifteen invasive M3 isolates were collected as part of a prospective population-based surveillance study of invasive GAS infections from 1992 to 2009. Invasive strains (organisms from normally sterile sites) were cultured from patients with soft tissue infections (n = 45), bacteremia (n = 36), lower respiratory infections (n = 30), unknown invasive infections (n = 26), septic arthritis (n = 24), necrotizing fasciitis (n = 18), or other invasive infections (e.g., meningitis, toxic shock syndrome, peritonitis; n = 36).
Genome Sequencing and Data Processing.
Genome sequencing was performed using an Illumina Genome Analyzer II instrument according to the manufacturer's instructions. Initial preprocessing of sequencing reads was performed using Tagdust. The FastQC and FastX toolkits were then used to parse the multiplexed sequencing reads, remove barcode information, and perform run quality control analyses. Polymorphism discovery was performed using the VAAL (4). Sequencing reads were then aligned to the MGAS315 reference genome using the MOSAIK assembler. Unaligned reads were placed into contigs using the Velvet de novo assembler. Contigs greater than 100 nucleotides in length were then used to search the National Center for Biotechnology Information nonredundant database using BLAST.
Phylogenetic Analysis.
A matrix file containing the genotype of all strains at each polymorphic locus was created from the VAAL polymorphism output data using a custom Perl script. Polymorphisms located within the six known integrated prophage elements in the M3 genome were removed, as were all indel polymorphisms. SNPs were concatenated and converted to FASTA sequences corresponding to each individual strain. ClustalX was then used to align and generate a guide tree for the FASTA sequences. A neighbor-joining phylogenetic tree was created using SplitsTree (26), and bootstrap analysis of 1,000 replicates was performed using MEGA4 (27). Tree annotation and final image generation were done using Dendroscope (28).
Targeted Gene Sequencing.
The has promoter, hasA/B, and ropB coding regions were sequenced by Sanger sequencing in all pharyngitis strains and invasive strains (PCR primer sequences are available in SI Methods). DNA sequencing was performed on an Applied Biosystems 3730 capillary sequencer using a BigDye v3.1 reagent kit.
Hyaluronic Acid Capsule Assay.
GAS strains grown overnight in Todd Hewitt broth containing 0.2% yeast extract (THY) were used to inoculate THY broth preheated to 37 °C. Strains were grown to midexponential phase, pelleted, washed, and suspended in 0.5 mL of nuclease-free water. Capsule was released using 1 mL of chloroform, and phase separation was performed using Phase-Lock tubes (5 PRIME). Quantification of hyaluronic acid was performed by colorimetric assay as previously described (29).
hasA Transcript Analysis.
GAS strains were grown overnight in THY, diluted 1:100, and incubated until midexponential phase (OD600 = 0.5) in THY. RNA was isolated using the RNeasy Mini kit (Qiagen), and cDNA was created using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). TaqMan quantitative real-time PCR was performed with an ABI 7500 Fast Real-Time system (Applied Biosystems) using primers and probes specific for hasA (sequences and detailed methods are available in SI Methods).
Experimental Pharyngitis in Nonhuman Primates.
The protocol used for experimental pharyngitis was similar to that described previously (19). Briefly, we used four healthy cynomolgus macaques matched for age and gender. Monkeys were infected with strain MGAS5005 (NCBI accession no. CP000017) and sampled sequentially for GAS as previously described (19, 30). The study protocol was approved by the Institutional Animal Care and Use Committee of the University of Houston.
Supplementary Material
Supporting Information
Acknowledgments
We thank K. Stockbauer for suggestions to improve the manuscript. N. Green, T. Humbird, J. Greaver, L. Jenkins, and T. Blasdel assisted with the nonhuman primate pharyngitis study.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Illumina sequence reported in this paper has been deposited in the NCBI Sequence Read Archive database (accession nos. SRP000775 and SRA030436.1).
*This Direct Submission article had a prearranged editor.
References
- 1.Cockerill FR, 3rd, et al. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA. 1997;277:38–43. [PubMed] [Google Scholar]
- 2.Hoe NP, et al. Distribution of streptococcal inhibitor of complement variants in pharyngitis and invasive isolates in an epidemic of serotype M1 group A Streptococcus infection. J Infect Dis. 2001;183:633–639. doi: 10.1086/318543. [DOI] [PubMed] [Google Scholar]
- 3.Beres SB, et al. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA. 2010;107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nusbaum C, et al. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat Methods. 2009;6:67–69. doi: 10.1038/nmeth.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeAngelis PL, Papaconstantinou J, Weigel PH. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 1993;268:19181–19184. [PubMed] [Google Scholar]
- 6.Dougherty BA, van de Rijn I. Molecular characterization of hasA from an operon required for hyaluronic acid synthesis in group A streptococci. J Biol Chem. 1994;269:169–175. [PubMed] [Google Scholar]
- 7.Dougherty BA, van de Rijn I. Molecular characterization of hasB from an operon required for hyaluronic acid synthesis in group A streptococci. Demonstration of UDP-glucose dehydrogenase activity. J Biol Chem. 1993;268:7118–7124. [PubMed] [Google Scholar]
- 8.DeAngelis PL, Papaconstantinou J, Weigel PH. Isolation of a Streptococcus pyogenes gene locus that directs hyaluronan biosynthesis in acapsular mutants and in heterologous bacteria. J Biol Chem. 1993;268:14568–14571. [PubMed] [Google Scholar]
- 9.Crater DL, van de Rijn I. Hyaluronic acid synthesis operon (has) expression in group A streptococci. J Biol Chem. 1995;270:18452–18458. doi: 10.1074/jbc.270.31.18452. [DOI] [PubMed] [Google Scholar]
- 10.Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federle MJ, Scott JR. Identification of binding sites for the group A streptococcal global regulator CovR. Mol Microbiol. 2002;43:1161–1172. doi: 10.1046/j.1365-2958.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- 13.Albertí S, Ashbaugh CD, Wessels MR. Structure of the has operon promoter and regulation of hyaluronic acid capsule expression in group A Streptococcus. Mol Microbiol. 1998;28:343–353. doi: 10.1046/j.1365-2958.1998.00800.x. [DOI] [PubMed] [Google Scholar]
- 14.Seastone CV. The occurrence of mucoid polysaccharide in hemolytic streptococci of human origin. J Exp Med. 1943;77:21–28. doi: 10.1084/jem.77.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todd EW, Lancefield RC. Variants of hemolytic streptococci; their relation to type-specific substance, virulence, and toxin. J Exp Med. 1928;48:751–767. doi: 10.1084/jem.48.6.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stollerman GH, Dale JB. The importance of the group a streptococcus capsule in the pathogenesis of human infections: A historical perspective. Clin Infect Dis. 2008;46:1038–1045. doi: 10.1086/529194. [DOI] [PubMed] [Google Scholar]
- 17.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessels MR, Goldberg JB, Moses AE, DiCesare TJ. Effects on virulence of mutations in a locus essential for hyaluronic acid capsule expression in group A streptococci. Infect Immun. 1994;62:433–441. doi: 10.1128/iai.62.2.433-441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virtaneva K, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beres SB, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: Phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 22.O'Brien AD, et al. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 23.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamayo E, Montes M, García-Medina G, García-Arenzana JM, Pérez-Trallero E. Spread of a highly mucoid Streptococcus pyogenes emm3/ST15 clone. BMC Infect Dis. 2010;10:233. doi: 10.1186/1471-2334-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrager HM, Rheinwald JG, Wessels MR. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J Clin Invest. 1996;98:1954–1958. doi: 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virtaneva K, et al. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71:2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information