An Experimentally Validated Genome-Scale Metabolic Reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228 (original) (raw)
Abstract
Klebsiella pneumoniae is a Gram-negative bacterium of the family Enterobacteriaceae that possesses diverse metabolic capabilities: many strains are leading causes of hospital-acquired infections that are often refractory to multiple antibiotics, yet other strains are metabolically engineered and used for production of commercially valuable chemicals. To study its metabolism, we constructed a genome-scale metabolic model (_i_YL1228) for strain MGH 78578, experimentally determined its biomass composition, experimentally determined its ability to grow on a broad range of carbon, nitrogen, phosphorus and sulfur sources, and assessed the ability of the model to accurately simulate growth versus no growth on these substrates. The model contains 1,228 genes encoding 1,188 enzymes that catalyze 1,970 reactions and accurately simulates growth on 84% of the substrates tested. Furthermore, quantitative comparison of growth rates between the model and experimental data for nine of the substrates also showed good agreement. The genome-scale metabolic reconstruction for K. pneumoniae presented here thus provides an experimentally validated in silico platform for further studies of this important industrial and biomedical organism.
Klebsiella pneumoniae, a Gram-negative bacterium of the family Enterobacteriaceae, is a microorganism with significance in both medicine and biotechnology. K. pneumoniae is a common opportunistic human pathogen, causing pneumonia, urinary tract infections, and bacteremia (25). Most clinical K. pneumoniae isolates are multidrug resistant, and up to 20% of them are extended-spectrum beta-lactamase-producing strains. The situation is worsened by the recent spreading of carbapenem-resistant NDM-1 strains, which leave very few antibiotics effective and therefore pose a serious threat to human health (23). In biotechnology, K. pneumoniae is capable of metabolizing glycerol as a sole source of carbon to produce 1,3-propanediol, a chemical that has many industrial applications (16, 51). Proper functioning of the metabolic network in K. pneumoniae underlies its ability both to cause disease and to serve as a useful platform strain for metabolic engineering.
The study of metabolism in many organisms has greatly benefitted from in silico genome-scale reconstruction of their metabolic networks combined with flux balance analysis (FBA), a computational technique that incorporates many different components of metabolism and their couplings to provide insight into steady-state flux distributions among the different pathways. Such system level analyses are critical since they can reveal emergent, network level properties not readily apparent from more-focused investigation of individual genes or pathways.
The metabolic network reconstruction of Escherichia coli K-12 represents the best-developed genome-scale network to date because E. coli is arguably the most studied and best characterized microorganism in terms of genome annotation, functional characterization, and knowledge of growth behavior (11). Metabolic network reconstruction is a process through which the gene-protein reaction associations that participate in the metabolic activity of a biological system are identified to form a network. Imposition of constraints on the genome-scale network and then conversion to a mathematical representation lead to a genome-scale model that can be analyzed by FBA. FBA is based on linear programming and is used to determine the steady-state reaction flux distribution under governing constraints by maximizing an objective function, such as growth rate. Therefore, constraint-based models can help predict cellular phenotypes for particular environmental conditions. Genome-scale models have been useful in understanding the metabolism of a variety of organisms, e.g., Salmonella spp. (1, 36) and Staphylococcus aureus (24).
Small network-scale metabolic flux analysis of glycerol metabolism in K. pneumoniae, with application to production of 1,3-propandiol, has been performed (49, 50). Because genome-scale models have proven useful not only for metabolic engineering applications (6, 31) but also for predicting the outcomes of gene deletions (9), identifying potential drug targets (24), and improving gene annotation (32), a genome-scale metabolic reconstruction of K. pneumoniae is needed to fully assess the metabolic functions of this important organism. The aim of this work is to build an experimentally verified genome-scale metabolic network reconstruction of K. pneumoniae MGH 78578.
MATERIALS AND METHODS
Bacterial strains and media.
Klebsiella pneumoniae MGH 78578 (ATCC 700721) was purchased from ATCC. It was routinely cultured at 37°C in M9 minimal medium consisting of Na2HPO4 (6.9 g/liter), KH2PO4 (3 g/liter), NaCl (0.5 g/liter), NH4Cl (1 g/liter), CaCl2 (0.1 mM), MgSO4 (2 mM), and a carbon source. For growth rate measurements, the carbon source concentration was 0.2% (wt/vol). For validation of Biolog results, the carbon source concentration was 5 mM. Frozen stocks were maintained at −80°C in M9 minimal medium with 25% glycerol.
Macromolecular composition analysis.
The frozen stock of K. pneumoniae MGH 78578 was inoculated into 50 ml M9 minimal medium with 0.2% glucose and was shaken at 37°C overnight in a 250-ml Erlenmeyer flask. The culture was diluted into three batch cultures, which contained 250 ml prewarmed M9 glucose medium, to an initial optical density at 600 nm (OD600) of approximately 0.02. Cells were harvested when the OD600 reached 0.6 to measure DNA, RNA, protein, lipid, and carbohydrate content and dry cell weight. For the dry cell weight, a cell pellet from a 50-ml culture sample was resuspended in water and carefully transferred into a preweighed Eppendorf microcentrifuge tube, then dried at 85°C. The dry weight was measured on a balance with 0.1-mg accuracy (Mettler Toledo; AG285). For the macromolecular composition analysis, the amounts of DNA, protein, and carbohydrate were determined by the Burton method, Robinson-Hogden biuret method, and phenol method, respectively (14). The amount of RNA was determined by the KOH-UV method (3). The lipid content was determined by the sulfo-phospho-vanillin method (19). The amount of each macromolecule was converted to grams per liter for calculating the percentage of dry cell weight for each macromolecule.
Phenotypic microarray.
Phenotypic microarray analysis of K. pneumoniae MGH 78578 was performed as a service by Biolog (Hayward, CA) (4). In total, 190 carbon, 95 nitrogen, 35 sulfur, and 59 phosphor sources were tested. To validate the Biolog results, overnight K. pneumoniae MGH 78578 precultures grown in LB medium at 37°C were harvested by centrifugation and resuspended to an OD600 of 0.2 in M9 minimal medium lacking any carbohydrates. Ten-microliter suspensions were added to 200 ml M9 minimal medium containing a 5 mM concentration of the carbohydrate to be tested in a 96-well plate. The plate was incubated at 37°C in a VERSAmax microplate reader (Molecular Devices) with shaking for 24 h, after which the OD600 was measured for each well. Each carbohydrate was tested in duplicate.
Adaptive evolution.
The frozen stock of K. pneumoniae MGH 78578 was streaked onto M9 minimal agar with 0.2% _myo_-inositol and cultured overnight at 37°C. A single colony was suspended in 100 μl sterile water. Three aliquots from this suspension were transferred into three separate 500-ml Erlenmeyer flasks containing 200 ml prewarmed M9 _myo_-inositol medium. The batch cultures were incubated at 37°C with shaking until the cell growth reached exponential phase. An aliquot from each flask was passed to three new 500-ml Erlenmeyer flasks containing 200 ml fresh prewarmed M9 _myo_-inositol medium twice a day. The volume transferred at each passage decreased as the growth rate increased in order to maintain the cells in exponential phase. We serially passaged the three cultures in this manner for 15 days, at which time their growth rates stabilized. An aliquot from each of the three endpoint evolved cultures was streaked out on M9 minimal agar with 0.2% _myo_-inositol and cultured overnight at 37°C. Final growth rate and substrate uptake measurements were made on a single colony from each evolved culture.
Growth rate and substrate uptake rate.
A K. pneumoniae MGH 78578 preculture was made from a frozen stock by inoculating it into 250-ml Erlenmeyer flasks containing 50 ml M9 minimal medium with 0.2% glucose. Each flask was incubated overnight at 37°C with stirring. The preculture was diluted into three batch cultures which contained 250 ml prewarmed M9 glucose medium in a 500-ml Erlenmeyer flask to an initial OD600 of around 0.02. The OD600 was measured periodically over the next several hours to calculate the growth rate. At the same time, the supernatant from each flask was collected by sterile filtration of it through 0.22-μm membrane filters (Millipore). The supernatant was then analyzed by high-performance liquid chromatography (HPLC) using an Aminex HPX-87H ion exchange carbohydrate-organic acid column (Bio-Rad Laboratories) operating at 65°C with refractive index (RI) detection. The mobile phase was degassed with 5 mM sulfuric acid flowing at a rate of 0.5 ml/min. Compounds were quantified by comparison to standard curves.
Metabolic network reconstruction.
Because E. coli and K. pneumoniae are closely related members of the family Enterobacteriaceae, the genome-scale metabolic model for E. coli K-12 MG1655, _i_AF1260, was used as a template to expedite a draft reconstruction for K. pneumoniae MGH 78578. The first step in our metabolic network reconstruction involved comparing protein sequence similarities. After a genomic homology search using BLASTP, the orthologs of MGH 78578 and E. coli K-12 MG1655 were obtained based on reciprocal best matches. The criteria for the bidirectional matches were an E value of ≤1 × 10−5, ≥35% amino acid sequence identity, and match lengths of at least 70% of the length of both query and subject. The initial list of metabolic reactions whose associated genes satisfied the Boolean statements (e.g., “and” logic for complexes and “or” logic for isozymes) in gene-protein reaction associations was therefore constructed. We subsequently compared the genome sequence of K. pneumoniae MGH 78578 to those of K. pneumoniae KP342 and Salmonella enterica serovar Typhimurium LT2 using BASys (48) and KAAS (29) to obtain additional genome annotation for MGH 78578 open reading frames (ORFs), especially those that did not have homologs in E. coli. Data from NCBI, KEGG (22), and Transport DB (38) were used to further refine the ORFs of MGH 78578 with metabolic or transport functions. This list of MGH 78578 ORFs constituted the initial draft reconstruction for MGH 78578 and was used as a starting point for manual curation (see Table S1 in the supplemental material). Organism-specific features gleaned from literature surveys were manually added into the draft reconstruction. On the other hand, in order to improve the reliability of the metabolic reconstruction, the genes and reactions that satisfied the following criteria were removed from the reconstruction: (i) low sequence similarity (identity < 80%), (ii) blockage of the reactions in the model, and (iii) experimental verification that the reactions were absent.
Generation of BOF.
A key element of a metabolic reconstruction is an accurate biomass objective function (BOF). The BOF directly influences growth rate calculations in the model simulation. The BOF, which is a mathematical representation of the biomass composition of a cell, was formulated from both a bibliome survey and experimental macromolecular composition data acquired in this study. Each cellular biomass macromolecule was divided into its corresponding building blocks, such as amino acids, fatty acids, and nucleotides (11). The energetic requirements for growth-associated maintenance (GAM) and non-growth-associated maintenance (NGAM) of K. pneumoniae were set as 71.7 mmol ATP/g dry weight (gDW) and 6.8 mmol ATP/gDW/h, respectively (42). A more detailed description of the biomass objective function for MGH 78578 can be found in Table S2 in the supplemental material.
Modeling simulation.
After the metabolic network was reconstructed, it was implemented in Matlab using the stoichiometric matrix (S matrix) formalism. FBA was performed using the COBRA Toolbox with the glpk solver (2). Gene essentiality analysis was performed using glucose minimal medium conditions with a glucose uptake rate of 10 mmol/gDW/h and an oxygen uptake rate of 20 mmol/gDW/h. A gene was considered computationally essential when removal of the gene resulted in no flux through the BOF.
RESULTS
Metabolic network reconstruction.
The metabolic reconstruction of K. pneumoniae was carried out in a series of successive refinements beginning with mapping from the current E. coli reconstruction, _i_AF1260 (11), to the K. pneumoniae genome. The work flow for the reconstruction process is shown in Table 1. Since K. pneumoniae and E. coli are closely related, sharing ∼88% homology over 1,137,281 nucleotides of MGH 78578 (Fig. 1A), we can draw a draft reconstruction directly from the extensively curated _i_AF1260 metabolic reconstruction for E. coli K-12 MG1655. The genome comparison between MGH 78578 and MG1655 yielded 1,072 orthologous genes, which are responsible for 1,795 reactions in _i_AF1260. These genes and their corresponding reactions were included in the initial draft reconstruction of MGH 78578, but 10 of them were later removed during manual curation (Fig. 1B). Subsequently, genes which were annotated to have metabolic enzymatic activity (see Table S1 in the supplemental material) were added to the reconstruction based on literature surveys, even if they did not have homologs in E. coli. For example, K. pneumoniae can utilize citrate as the sole carbon source because it has citrate-utilizing enzymes encoded by the gene cluster KPN_00028 to KPN_00038 (40). E. coli K-12 MG1655 does not have this cluster and does not readily utilize citrate as a sole carbon source. For aromatic compound metabolism, MGH 78578 can grow in benzoate and 4-hydroxyphenylacetate; growth in these has been experimentally validated in this study. The gene clusters responsible for these metabolic pathways are KPN_01869 to KPN_01875 and KPN_04779 to KPN_04789, respectively (8, 34). Moreover, K. pneumoniae can anaerobically biosynthesize vitamin B12. Comparative genomic analysis revealed that the genes involved are KPN_03184 to KPN_03200 (10, 39). Meanwhile, reactions belonging to incomplete pathways were removed from the reconstruction. For example, MGH 78578 has an ORF that matches to b4090 of MG1655, which encodes allose 6-phosphate isomerase. However, MGH 78578 cannot utilize d-allose according to our experimental result. Therefore, the gene-protein reaction associations related to d-allose metabolism were removed from the reconstruction. Taken together, 1,228 genes, 1,188 enzymes, and 1,970 reactions were included in the MGH 78578 reconstruction, designated _i_YL1228. These genes account for 27% of the 4,476 annotated genes in the MGH 78578 genome.
TABLE 1.
Work flow of the metabolic network reconstruction process
| Step | Basis | End product |
|---|---|---|
| Draft generation | Sequence similarity analysis, _i_AF1260 | Draft reconstruction |
| Manual curation | Literature surveys and online databases (e.g., COG, KEGG) | Curated reconstruction |
| In silico modeling | Biomass objective function (biomass composition and maintenance verification) and phenotype screen (Biolog) | Genome-scale reconstruction model |
| Validation | Adaptive evolution and quantitative analysis | Validated genome-scale reconstruction model |
FIG. 1.
Whole-genome comparisons between K. pneumoniae MGH 78578 and E. coli K-12 MG1655. (A) Genome alignment was performed by MUMMER. Red and green lines represent forward and reverse matches, respectively. (B) Venn diagram of homology overlap shared between K. pneumoniae model _i_YL1228 and E. coli model _i_AF1260. The number in parentheses indicates the initial homologous genes present in _i_AF1260 that were included in the draft _i_YL1228 reconstruction. Ten of these genes were later removed after manual curation. (C) Coverage of characterized ORFs in different COG functional categories in the initial draft K. pneumoniae model (blue) and the curated _i_YL1228 (red) and _i_AF1260 (green). (D) Percentages of coverage of characterized ORFs in _i_YL1228 and _i_AF1260 in different COG functional categories.
Characteristics of _i_YL1228.
Among the 1,970 reactions in _i_YL1228, 1,883 have known gene associations. We then used the COG (clusters of orthologous groups) database to functionally classify these ORFs (44). As shown in Fig. 1C, _i_YL1228 has ∼20 more genes than the draft reconstruction in classes C (energy production and conversion), E (amino acid transport and metabolism), G (carbohydrate transport and metabolism), and H (coenzyme transport and metabolism). Note that the percent coverages of characterized ORFs in _i_AF1260 and _i_YL1228 differ considerably in functional classes E, G, and P (inorganic ion transport and metabolism) (Fig. 1D), mainly because MGH 78578 has more putative transporters (834 transport genes in MGH 78578 versus 536 in E. coli K-12 [12]). As more information on the transporters of MGH 78578 becomes available, the reconstruction can be expanded accordingly. Nevertheless, the current scale of _i_YL1228 is remarkable considering that the species knowledge index (SKI) value for K. pneumoniae (∼2.6) is much smaller than that for E. coli (∼61.2) (37).
Biomass composition and assessment of maintenance requirements.
The biomass objective function (BOF) was generated by combining the biomass composition of K. pneumoniae MGH 78578 with growth-associated maintenance (GAM). We have directly measured the biomass composition of K. pneumoniae, including carbohydrate, DNA, lipid, protein, and RNA content. We found a greater proportion of carbohydrates in K. pneumoniae biomass than in E. coli biomass (18.7% versus 8.4%) (Table 2), which can be explained by the fact that K. pneumoniae possesses a thick polysaccharide capsule (27).
TABLE 2.
Biomass composition of K. pneumoniae MGH 78578
| Macromolecule | Mass (g/gDW) in: | |
|---|---|---|
| K. pneumoniae MGH 78578 | E. colib | |
| Protein | 0.521 | 0.550 |
| DNA | 0.023 | 0.031 |
| RNA | 0.131 | 0.205 |
| Lipid | 0.081 | 0.091 |
| Carbohydrate | 0.187a | 0.084 |
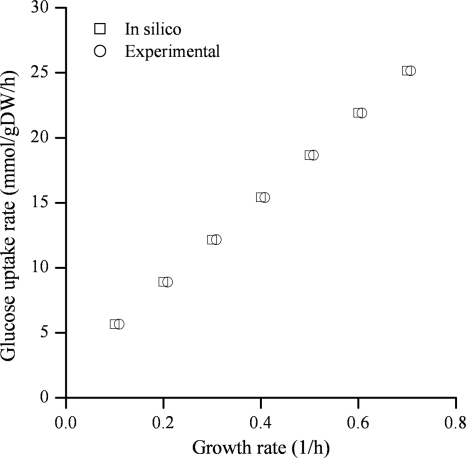
According to the sensitivity analyses performed previously (11, 35), the biomass composition plays a minor role in growth rate prediction. However, the maintenance energies, namely, GAM and non-growth-associated maintenance (NGAM), can considerably influence growth rate predictions (11, 35). Therefore, an assessment of GAM/NGAM was performed using the fitted parameters (a GAM of 71.7 mmol ATP/gDW and an NGAM of 6.8 mmol ATP/gDW/h) for anaerobic glucose-limited chemostat data for Aerobacter aerogenes (the name previously used for Klebsiella pneumoniae [42]). We thus confirmed that an NGAM value of 6.8 mmol ATP/gDW/h and a GAM value of 71.7 mmol ATP/gDW make the model fit the experimental data well, as shown in Fig. 2.
FIG. 2.
Assessment of growth- and non-growth-associated maintenance (GAM and NGAM, respectively). Taking an NGAM value of 6.8 mmol ATP/gDW/h and a GAM value of 71.7 mmol ATP/gDW enables the model to accurately predict the growth rates of K. pneumoniae in glucose.
Model refinement based on qualitative phenotypes.
In the reconstruction process summarized in Table 1, the second and third steps are iterative and recursive. With the biomass objective function, a genome-scale reconstruction model that is ready for FBA can be used to simulate the growth of K. pneumoniae in a variety of nutrition sources. We deployed the model in this way to predict growth versus no growth on different carbon, nitrogen, phosphorus, and sulfur sources and compared the simulation results with high-throughput phenotypic microarray experimental data (Biolog). Inconsistencies between simulation results and Biolog data were then used to refine the model. After reconciling the model with the Biolog data by adding missing reactions supported by literature, _i_YL1228 was able to predict metabolic phenotypes of MGH 78578 with an overall agreement rate of 84% (Table 3). The model is provided as File S1 (in Systems Biology Markup Language [SBML] format) in the supplemental material.
TABLE 3.
In silico modeling of growth conditions
| Source | No. of comparisons | No. of comparisons showinga: | Agreement rate (%) | |||
|---|---|---|---|---|---|---|
| Agreement | Disagreement | |||||
| G | NG | C, G; E, NG | C, NG; E, G | |||
| Carbon | 88 | 51 | 27 | 10 | 0 | 88.6 |
| Nitrogen | 50 | 26 | 14 | 3 | 7 | 80.0 |
| Phosphorus | 21 | 21 | 0 | 0 | 0 | 100.0 |
| Sulfur | 12 | 4 | 0 | 2 | 6 | 33.3 |
| Total | 171 | 102 | 41 | 15 | 13 | 83.6 |
Model validation and adaptive evolution.
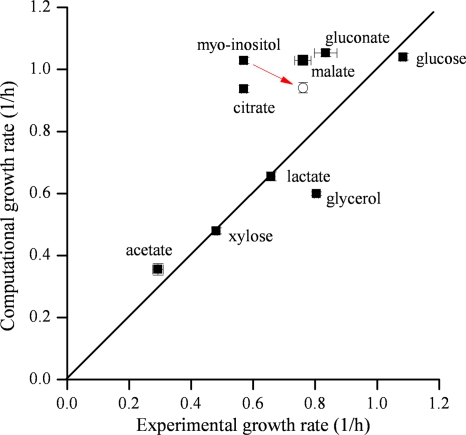
In addition to qualitatively assessing the model predictions in terms of growth versus no growth, we also quantitatively assessed the model's accuracy in predicting growth rates on nine different carbon sources under aerobic conditions. The nine carbon sources represent different entry points into the metabolic network. As shown in Table 4, the in silico predictions agreed well with the experimental results (the error rates between model prediction and experimental data were less than 31%, and most of them ranged between 0.4% and 26.3%) except for citrate and _myo_-inositol. For these two, the predicted growth rates were higher than those seen experimentally: 0.937/h versus 0.570/h and 1.029/h versus 0.570/h, respectively. We hypothesized that the overestimation arose in part because FBA-based calculations assume that an organism grows optimally in every nutrient environment it encounters, a property that is not always found. In such cases, however, it has been shown that organisms can indeed achieve faster growth rates that are consistent with model predictions if they are subjected to experimental adaptive evolution (17).
TABLE 4.
Quantitative comparison between in silico growth rate and experimental growth rate
| Carbon source (uptake rate [mmol/gDW/h]) | Growth rate (1/h) | Oxygen uptake rate (mmol/gDW/h) | Error rate (%) | |
|---|---|---|---|---|
| Exptl | In silico | |||
| Acetate (14.291) | 0.293 | 0.355 | 14.657 | 21.13 |
| Citrate (14.017) | 0.570 | 0.937 | 21.837 | 64.41 |
| d-Xylose (6.006) | 0.481 | 0.479 | 11.229 | 0.44 |
| Gluconate (17.909) | 0.965 | 1.264 | 21.837 | 30.93 |
| Glucose (10.457) | 1.084 | 1.040 | 21.744 | 4.02 |
| Glycerol (10.609) | 0.804 | 0.599 | 13.618 | 25.50 |
| l-Lactate (22.686) | 0.658 | 0.655 | 21.837 | 0.44 |
| l-Malate (34.572) | 0.834 | 1.053 | 21.837 | 26.24 |
| _myo_-Inositol (13.802) | 0.570 | 1.029 | 21.837 | 80.38 |
| _myo_-Inositola (11.024) | 0.760 | 0.941 | 21.837 | 23.67 |
We tested this hypothesis for _myo_-inositol by serial passage of K. pneumoniae for 15 days in minimal media containing _myo_-inositol as the sole carbon source. As shown in Fig. 3, the growth rate for K. pneumoniae increased from 0.570/h to 0.760/h after 15 days adaptive evolution, which reduced the growth rate error from 80% to 24% (Table 4). Note that the computational growth rate decreased from 1.029/h to 0.941/h because the uptake rate of _myo_-inositol was reduced from 13.802 to 11.024 mmol/gDW/h after the adaptation. Taken together, the results show that the genome-scale reconstruction model _i_YL1228 is well established for representing metabolism in K. pneumoniae MGH 78578.
FIG. 3.
Quantitative comparison of in silico and experimental growth rates of K. pneumoniae MGH 78578. The open circle represents the growth rate of K. pneumoniae MGH 78578 after 15 days of adaptive evolution in 0.2% _myo_-inositol minimal media.
Comparison to other members of Enterobacteriaceae.
We evaluated the differences in metabolic phenotypes among E. coli K-12 MG1655, Salmonella enterica serovar Typhimurium LT2, and K. pneumoniae MGH 78578 by comparing the Biolog phenotype microarrays of the three species. There is a metabolic reconstruction available for a fourth enterobacterium, Yersinia pestis 91001 (30), but no Biolog data for this organism have been reported. Major differences in metabolic capability of MGH 78578 in comparison with Salmonella and E. coli are summarized in Table 5 and in more detail in Table S3 in the supplemental material. The Biolog data show that MGH 78578 can utilize 18 substrates that E. coli and Salmonella cannot, whereas MGH 78578 cannot utilize 26 substrates that the other two can. One explanation for the differences in metabolic capacity is the MGH 78578-specific presence or absence of metabolic genes or operons. For example, MGH 78578 has KPN_02823 (bglK, encoding β-glucoside kinase) and KPN_01234 (celF, encoding cellobiose-6-phosphate hydrolase), which are responsible, respectively, for phosphorylation and hydration of β-glucosides, such as cellobiose, arbutin, gentiobiose, and salicin. However, due to the lack of information about β-glucoside transporters in K. pneumoniae, we did not add transporters for β-glucosides to the model even though MGH 78578 can grow on β-glucosides in vivo. Among the MGH 78578-specific substrates, d-cellobiose, d-arabitol, and d-raffinose can be utilized by all tested K. pneumoniae strains (5). Unlike the commensal E. coli strain MG1655, K. pneumoniae MGH 78578 and Salmonella Typhimurium SLT2 are pathogens, and their unique metabolic capabilities, as shown in the last 7 rows of Table 5, might be associated with their pathogenicity.
TABLE 5.
Differences in metabolic phenotypes among E. coli, Salmonella Typhimurium LT2, and K. pneumoniae MGH 78578
| Source | Compound | Exchange reactionb | Resulta for: | Gene(s)/operon | Reference(s) | ||
|---|---|---|---|---|---|---|---|
| E. coli | Salmonella | MGH 78578 | |||||
| Carbon | Sucrose | EX_sucr(e) | NG | NG | G | scrABY | 46 |
| Carbon | d-Cellobiose | EX_cellb(e) | NG | NG | G | celF and bglK | 45,47 |
| Carbon | d-Arabinose | NG | NG | G | |||
| Carbon | d-Arabitol | EX_abt_D(e) | NG | NG | G | dalDKT | 15 |
| Carbon | Arbutin | NG | NG | G | |||
| Carbon | Gentiobiose | NG | NG | G | |||
| Carbon | Palatinose | NG | NG | G | |||
| Carbon | d-Raffinose | NG | NG | G | |||
| Carbon | Salicin | NG | NG | G | |||
| Carbon | l-Sorbose | NG | NG | G | |||
| Carbon | Stachyose | NG | NG | G | |||
| Carbon | Dihydroxyacetone | EX_dha(e) | NG | NG | G | dhaK | 43 |
| Nitrogen | Urea | EX_urea(e) | NG | NG | G | ureABC | 28 |
| Nitrogen | d-Glutamic acid | NG | NG | G | |||
| Nitrogen | Ethanolamine | EX_etha(e) | NG | NG | G | eutH | 33 |
| Nitrogen | Guanine | EX_gua(e) | NG | NG | G | hpx operon | 7 |
| Nitrogen | Parabanic acid | NG | NG | G | |||
| Sulfur | _N_-Acetyl-l-cysteine | NG | NG | G | |||
| Carbon | Dulcitol | EX_galt(e) | NG | G | G | gatD | 41 |
| Carbon | l-Glutamic acid | EX_glu_L(e) | NG | G | G | gdhA | 21 |
| Carbon | Citric acid | EX_cit(e) | NG | G | G | cit operon | 40 |
| Carbon | _p_-Hydroxyphenyl acetic acid | EX_4hphac(e) | NG | G | G | hpa operon | 13,26,34 |
| Carbon | _m_-Hydroxyphenyl acetic acid | NG | G | G | |||
| Carbon | 2-Deoxy-d-ribose | NG | G | G | |||
| Nitrogen | Tyramine | EX_tym(e) | NG | G | G |
In addition to comparing metabolic phenotypes to those of other members of the Enterobacteriaceae, we predicted gene essentiality for in silico modeling of aerobic growth in glucose minimal medium using _i_YL1228 and compared the results with the computationally essential genes predicted by the models of E. coli (_i_AF1260) and Salmonella (_i_RR1083 [36]). We found that 8 of the 118 computationally essential genes predicted by _i_YL1228 were unique to K. pneumoniae MGH 78578. These included the genes needed for lipopolysaccharide biosynthesis (KPN_02202, KPN_02492, KPN_02493, and KPN_03963); capsule synthesis (KPN_01515); and lipid biosynthesis (KPN_01093). Also included are the genes that encode ornithine carbamoyltransferase (argI [KPN_04659], whose isozyme argF is absent in MGH 78578 but present in E. coli and Salmonella) and inositol-5-monophosphate dehydrogenase (guaB [KPN_02834], whose orthologous gene b2508 in E. coli was experimentally verified as an essential gene [20]). Considering that large-scale gene essentiality screens for K. pneumoniae are not available, the essential gene prediction derived from _i_YL1228 provides a good alternative resource to facilitate drug target identification.
DISCUSSION
This study describes an experimentally validated genome-scale metabolic model, _i_YL1228, of the Gram-negative pathogen K. pneumoniae MGH 78578. The model has been reconstructed by carrying out a series of refinements on the basis of mapping from the E. coli reconstruction (_i_AF1260) to the MGH 78578 genome and reconciling with experimental data. Macromolecular composition data for MGH 78578 determined in this study combined with energetic requirements for maintenance were verified to fit experimental data well. The model agreed with phenotypic microarray data (84%) and predicted accurate growth rates (less than 30% error). Our model _i_YL1228 can serve as a platform for understanding K. pneumoniae MGH 78578 metabolism and also as a predictive model for gene essentiality analysis.
Comparative genomic analysis between K. pneumoniae MGH 78578 and E. coli K-12 MG1655 shows a high degree of similarity between the two genomes. Based on the sequence similarity analysis, we found 2,938 putative orthologs between MGH 78578 and MG1655, with an average protein percent identity of 83%. Of these orthologs, 1,308 have metabolic/transport function. An additional 302 genes in MG1655 are involved in metabolism but not present in MGH 78578. On the other hand, 478 metabolic genes in MGH 78578 are not present in MG1655. These differences imply that the metabolic capabilities might have been transferred, lost, and/or gained after the divergence of the two organisms. Notably, many of the MGH 78578-specific metabolic ORFs are annotated as putative genes. Nevertheless, the known genes or operons that are present only in MGH 78578 might have contributed to its species/strain-specific metabolic capabilities. Examples of genes/operons that are present in K. pneumoniae MGH 78578 but absent from E. coli K-12 MG1655 include the cbi, pdu, hpa, and rml operons, which confer on MGH 78578 the abilities to synthesize cofactor B12, utilize propanediol, metabolize 4-hydroxyphenylacetate, and synthesize dTDP-l-rhamnose, respectively.
Although K. pneumoniae MGH 78578, Salmonella, and E. coli K-12 MG1655 are closely related microorganisms, their metabolic phenotypes are known to be distinct. Here we used a phenotypic microarray (Biolog) to characterize the metabolic differences between MGH 78578 and MG1655 and Salmonella. A detailed summary of the result can be found in Table S3 in the supplemental material. It is worth noting that the Biolog data may have a technical error rate of approximately 15%, which was estimated by two data sets performed for E. coli MG1655 (1, 11). Therefore, caution must be taken when using Biolog data for model refinements. In this study, we manually verified 37 carbon source conditions to confirm the true metabolic capabilities of MGH 78578 for the refinement of the reconstruction. Unlike the small metabolic difference (36 out of 379 conditions, 9.5%) between E. coli and Salmonella (1), a large difference (94 out of 379, 24.8%) between E. coli MG1655 and K. pneumoniae MGH 78578 was found when comparing the Biolog result for E. coli MG1655 (11) to that for K. pneumoniae MGH 78578. Although the difference was reduced to 20.1% (65 out of 323) after validation of the consensus Biolog result obtained from the two data sets with our experimental result for MGH 78578, it highlights the different metabolic capabilities of these enterobacteria. Among the 26 MGH 78578-specific nonutilized substrates, some carboxylic acids that MGH 78578 cannot utilize as a sole carbon source are intermediates in the tricarboxylic acid (TCA) cycle, so they should be assimilated by MGH 78578, which suggests that MGH 78578 lacks the corresponding transporters to transport them into cells, especially during dicarboxylic acid transport.
Furthermore, we assessed in silico whether K. pneumoniae model _i_YL1228 and E. coli model _i_AF1260 can both utilize 171 different substrates and found an agreement rate of 78.5% for these 171 substrates, which are shared between the two models. This agreement rate of metabolic capability between _i_AF1260 and _i_YL1228 accords well with that derived from Biolog results (75.2 to 79.9%). Since 1,072 ORFs in _i_YL1228 were directly mapped from _i_AF1260, the two models are supposed to share approximately 87% (1,072/1,228) of metabolic capacity. However, the lower-than-expected metabolic capacity in common suggests that small genetic difference might lead to large metabolic difference.
To validate our reconstruction model, we quantitatively predicted the growth rates of MGH 78578 in M9 minimal medium plus different carbon sources. In the beginning, we did not constrain oxygen uptake rates. After simulations to optimize biomass formation, oxygen uptake rates under each condition were obtained. Then, we constrained the lower bound of oxygen uptake rate in each condition to 21.837 mmol/gDW/h, which is the in silico oxygen uptake rate under the glucose condition. An oxygen uptake rate of 21.837 mmol/gDW/h is higher than the one used in _i_AF1260, but it seems reasonable because the higher glucose uptake rate was measured for K. pneumoniae. One notable observation from our result is that the in silico growth rates on some carbon sources are higher than the experimental results (Fig. 3). There are two possible explanations for these differences: (i) the reconstructed model is unrealistically efficient and (ii) the strain used here has not adapted well to the examined conditions. The first explanation is likely true because the mechanism responsible for gene regulation is not considered in the current model. In this case, addition of regulatory mechanisms to the model may reduce the discrepancy. Meanwhile, the second possibility has been experimentally supported in the case of growth on _myo_-inositol. Therefore, the overall agreement rate between in silico and experimental growth rates is expected to increase if MGH 78578 is allowed adequate time to adapt to the examined media. On the other hand, our model underestimates the growth rate with glycerol as the sole carbon source. This observation indicates that the simulation parameters such as GAM, NGAM, and BOF, which were determined with glucose as the carbon source, are not suitable for the growth of MGH 78578 in glycerol.
We present here an extensively curated and validated genome-scale metabolic model for the biomedically and biotechnologically important organism K. pneumoniae. Given the importance of this organism, this model is likely to find wide use, and we also note that it represents the third curated genome-scale model (in addition to _i_AF1260 and _i_RR1083) for Enterobacteriaceae. With these three curated genome-scale models we can anticipate that new vistas in comparative genomics will open up and will allow scientists to study actual organism capabilities and phenotypic functions that arise from highly homologous genomes.
Supplementary Material
[Supplemental material]
Acknowledgments
We give special thanks to Ines Thiele for her assistance in metabolic network reconstruction. We thank Jessica De Ingeniis from Andrei Osterman's lab at the Burnham Institute for kindly sharing the protocol of macromolecular composition measurement.
This work was supported by National Health Research Institutes intramural funding (PH-099-SP-10). The computational facilities for this work were partly supported by the National Research Program for Genomic Medicine, National Science Council, Taiwan (NSC99-3112-B-400-012). Yu-Chieh Liao and Tzu-Wen Huang are postdoctoral fellows at NHRI and visiting scholars at UCSD.
Footnotes
▿
Published ahead of print on 4 February 2011.
REFERENCES
- 1.AbuOun, M., et al. 2009. Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J. Biol. Chem. 284**:**29480-29488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, S. A., et al. 2007. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2**:**727-738. [DOI] [PubMed] [Google Scholar]
- 3.Benthin, S., J. Nielsen, and J. Villadsen. 1991. A simple and reliable method for the determination of cellular RNA content. Biotechnol. Tech. 5**:**39-42. doi: 10.1007/BF00152753. [DOI] [Google Scholar]
- 4.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11**:**1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisse, S., et al. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4**:**e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgard, A. P., P. Pharkya, and C. D. Maranas. 2003. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84**:**647-657. [DOI] [PubMed] [Google Scholar]
- 7.de la Riva, L., J. Badia, J. Aguilar, R. A. Bender, and L. Baldoma. 2008. The hpx genetic system for hypoxanthine assimilation as a nitrogen source in Klebsiella pneumoniae: gene organization and transcriptional regulation. J. Bacteriol. 190**:**7892-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doten, R. C., and L. N. Ornston. 1987. Protocatechuate is not metabolized via catechol in Enterobacter aerogenes. J. Bacteriol. 169**:**5827-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte, N. C., M. J. Herrgard, and B. O. Palsson. 2004. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 14**:**1298-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazzio, T. G., and J. R. Roth. 1996. Evidence that the CysG protein catalyzes the first reaction specific to B12 synthesis in Salmonella typhimurium, insertion of cobalt. J. Bacteriol. 178**:**6952-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feist, A. M., et al. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3**:**121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouts, D. E., et al. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4**:**e1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibello, A., M. Suarez, J. L. Allende, and M. Martin. 1997. Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch. Microbiol. 167**:**160-166. [PubMed] [Google Scholar]
- 14.Herbert, D., P. J. Phipps, and R. E. Strange. 1971. Chemical analysis of microbial cells. Methods Microbiol. 5b**:**210-344. [Google Scholar]
- 15.Heuel, H., S. Turgut, K. Schmid, and J. W. Lengeler. 1997. Substrate recognition domains as revealed by active hybrids between the d-arabinitol and ribitol transporters from Klebsiella pneumoniae. J. Bacteriol. 179**:**6014-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horng, Y. T., et al. 2010. Inactivation of dhaD and dhaK abolishes by-product accumulation during 1,3-propanediol production in Klebsiella pneumoniae. J. Ind. Microbiol. Biotechnol. 37**:**707-716. [DOI] [PubMed] [Google Scholar]
- 17.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420**:**186-189. [DOI] [PubMed] [Google Scholar]
- 18.Ingraham, J. L., O. Maaloe, and F. C. Neidhardt. 1983. Growth of the bacterial cell, p. 3. Sinauer Associates, Inc., Sunderland, MA.
- 19.Izard, J., and R. J. Limberger. 2003. Rapid screening method for quantitation of bacterial cell lipids from whole cells. J. Microbiol. Methods 55**:**411-418. [DOI] [PubMed] [Google Scholar]
- 20.Joyce, A. R., et al. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 188**:**8259-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanamori, K., R. L. Weiss, and J. D. Roberts. 1987. Role of glutamate dehydrogenase in ammonia assimilation in nitrogen-fixing Bacillus macerans. J. Bacteriol. 169**:**4692-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M., et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Research 36**:**D480-D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumarasamy, K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10**:**597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, D. S., et al. 2009. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J. Bacteriol. 191**:**4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, Y. T., T. L. Chen, L. K. Siu, S. F. Hsu, and C. P. Fung. 2010. Clinical and microbiological characteristics of community-acquired thoracic empyema or complicated parapneumonic effusion caused by Klebsiella pneumoniae in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 29**:**1003-1010. [DOI] [PubMed] [Google Scholar]
- 26.Martin, M., A. Gibello, J. Fernandez, E. Ferrer, and A. Garrido-Pertierra. 1991. Catabolism of 3- and 4-hydroxyphenylacetic acid by Klebsiella pneumoniae. J. Gen. Microbiol. 137**:**621-628. [DOI] [PubMed] [Google Scholar]
- 27.Mengistu, Y., C. Edwards, and J. R. Saunders. 1994. Continuous culture studies on the synthesis of capsular polysaccharide by Klebsiella pneumoniae K1. J. Appl. Bacteriol. 76**:**424-430. [DOI] [PubMed] [Google Scholar]
- 28.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59**:**451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriya, Y., M. Itoh, S. Okuda, A. C. Yoshizawa, and M. Kanehisa. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research 35**:**W182-W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navid, A., and E. Almaas. 2009. Genome-scale reconstruction of the metabolic network in Yersinia pestis, strain 91001. Mol. Biosyst. 5**:**368-375. [DOI] [PubMed] [Google Scholar]
- 31.Patil, K. R., I. Rocha, J. Forster, and J. Nielsen. 2005. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics 6**:**308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pel, H. J., et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25**:**221-231. [DOI] [PubMed] [Google Scholar]
- 33.Penrod, J. T., C. C. Mace, and J. R. Roth. 2004. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J. Bacteriol. 186**:**6885-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prieto, M. A., E. Diaz, and J. L. Garcia. 1996. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J. Bacteriol. 178**:**111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puchalka, J., et al. 2008. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput. Biol. 4**:**e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raghunathan, A., J. Reed, S. Shin, B. Palsson, and S. Daefler. 2009. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst. Biol. 3**:**38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, J. L., I. Famili, I. Thiele, and B. O. Palsson. 2006. Towards multidimensional genome annotation. Nat. Rev. 7**:**130-141. [DOI] [PubMed] [Google Scholar]
- 38.Ren, Q., and I. T. Paulsen. 2005. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1**:**e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278**:**41148-41159. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, K., et al. 2002. Identification of a gene cluster in Klebsiella pneumoniae which includes citX, a gene required for biosynthesis of the citrate lyase prosthetic group. J. Bacteriol. 184**:**2439-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakeri-Garakani, A., A. Brinkkotter, K. Schmid, S. Turgut, and J. W. Lengeler. 2004. The genes and enzymes for the catabolism of galactitol, D-tagatose, and related carbohydrates in Klebsiella oxytoca M5a1 and other enteric bacteria display convergent evolution. Mol. Genet. Genomics 271**:**717-728. [DOI] [PubMed] [Google Scholar]
- 42.Stouthamer, A. H., and C. W. Bettenhaussen. 1975. Determination of the efficiency of oxidative phosphorylation in continuous cultures of Aerobacter aerogenes. Arch. Microbiol. 102**:**187-192. [DOI] [PubMed] [Google Scholar]
- 43.Sun, J., J. van den Heuvel, P. Soucaille, Y. Qu, and A. P. Zeng. 2003. Comparative genomic analysis of dha regulon and related genes for anaerobic glycerol metabolism in bacteria. Biotechnol. Prog. 19**:**263-272. [DOI] [PubMed] [Google Scholar]
- 44.Tatusov, R. L., et al. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29**:**22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J., F. W. Lichtenthaler, S. Peters, and A. Pikis. 2002. Beta-glucoside kinase (BglK) from Klebsiella pneumoniae. Purification, properties, and preparative synthesis of 6-phospho-beta-d-glucosides. J. Biol. Chem. 277**:**34310-34321. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J., et al. 2001. Metabolism of sucrose and its five linkage-isomeric alpha-d-glucosyl-d-fructoses by Klebsiella pneumoniae. Participation and properties of sucrose-6-phosphate hydrolase and phospho-alpha-glucosidase. J. Biol. Chem. 276**:**37415-37425. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J., S. B. Ruvinov, D. I. Freedberg, and B. G. Hall. 1999. Cellobiose-6-phosphate hydrolase (CelF) of Escherichia coli: characterization and assignment to the unusual family 4 of glycosylhydrolases. J. Bacteriol. 181**:**7339-7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Domselaar, G. H., et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Research 33**:**W455-W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Q., H. Teng, Y. Sun, Z. Xiu, and A. Zeng. 2008. Metabolic flux and robustness analysis of glycerol metabolism in Klebsiella pneumoniae. Bioprocess Biosyst. Eng. 31**:**127-135. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Q., and Z. Xiu. 2009. Metabolic pathway analysis of glycerol metabolism in Klebsiella pneumoniae incorporating oxygen regulatory system. Biotechnol. Prog. 25**:**103-115. [DOI] [PubMed] [Google Scholar]
- 51.Zhuge, B., C. Zhang, H. Fang, J. Zhuge, and K. Permaul. 2010. Expression of 1,3-propanediol oxidoreductase and its isoenzyme in Klebsiella pneumoniae for bioconversion of glycerol into 1,3-propanediol. Appl. Microbiol. Biotechnol. 87**:**2177-2184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]