Mitophagy Selectively Degrades Individual Damaged Mitochondria After Photoirradiation (original) (raw)
Abstract
Damaged and dysfunctional mitochondria are proposed to be removed by autophagy. However, selective degradation of damaged mitochondria by autophagy (mitophagy) has yet to be experimentally verified. In this study, we investigated the cellular fate of individual mitochondria damaged by photoirradiation in hepatocytes isolated from transgenic mice expressing green fluorescent protein fused to microtubule-associated protein 1 light chain 3, a marker of forming and newly formed autophagosomes. Photoirradiation with 488-nm light induced mitochondrial depolarization (release of tetramethylrhodamine methylester [TMRM]) in a dose-dependent fashion. At lower doses of light, mitochondria depolarized transiently with re-polarization within 3 min. With greater light, mitochondrial depolarization became irreversible. Irreversible, but not reversible, photodamage induced autophagosome formation after 32±5 min. Photodamage-induced mitophagy was independent of TMRM, as photodamage also induced mitophagy in the absence of TMRM. Photoirradiation with 543-nm light did not induce mitophagy. As revealed by uptake of LysoTracker Red, mitochondria weakly acidified after photodamage before a much stronger acidification after autophagosome formation. Photodamage-induced mitophagy was not blocked by phosphatidylinositol 3-kinase inhibition with 3-methyladenine (10 m_M_) or wortmannin (100 n_M_). In conclusion, individual damaged mitochondria become selectively degraded by mitophagy, but photodamage-induced mitophagic sequestration occurs independently of the phosphatidylinositol 3-kinase signaling pathway, the classical upstream signaling pathway of nutrient deprivation-induced autophagy. Antioxid. Redox Signal. 14, 1919–1928.
Introduction
Cellular homeostasis is maintained by a fine balance between anabolic and catabolic pathways. Together, proteasomes and the autophagic apparatus are responsible for removal of damaged and superfluous cellular constituents. Proteasomal degradation typically eliminates short-lived proteins that are damaged or misfolded; whereas autophagy removes long-lived proteins, protein aggregates, and whole organelles, including endoplasmic reticulum, peroxisomes, and mitochondria (9, 32). Autophagy is now categorized into chaperone-mediated autophagy, microautophagy, and macroautophagy (9). Autophagy as originally defined corresponds to macroautophagy whereby whole organelles and pieces of cytoplasm are first sequestered into vesicles called autophagosomes that then fuse with lysosomes for digestion. Thus, the terms autophagy and macroautophagy are often used interchangeably, a practice followed here unless otherwise indicated.
In general, insulin and other growth factors inhibit autophagy, whereas nutrient deprivation and glucagon promote autophagy (4, 41). In the normal life of cells, autophagy occurs more or less continuously to remove damaged and superfluous organelles, including dysfunctional mitochondria that could be detrimental to cells. Several studies suggest that both inadequate and excess autophagy promote cell injury and death (25, 28). Therefore, proper regulation of autophagy is fundamental to cellular well being.
During autophagy, a novel membranous structure called a phagophore elongates and encloses cellular components to form a double membrane vesicle called an autophagosome (42). Lysosomes then fuse with autophagosomes to form autolysosomes in which lysosomal hydrolases degrade the sequestered contents. Autophagosomes can contain virtually any cytoplasmic element, including cytosolic proteins and various membranous organelles, including endoplasmic reticulum, peroxisomes, and mitochondria (3, 22).
In yeast, genetic screens identified a series of evolutionarily conserved autophagy (ATG) genes that are required for autophagy. The protein products of ATG genes are called Atg proteins and are categorized according to function. For example, a mammalian homologue of yeast Atg6, Beclin 1 interacts with phosphatidylinositol 3-kinase (PI3K) to form a complex that is involved in an initial step in autophagosome formation (11). Microtubule-associated protein 1 light chain 3 (LC3) was then identified as a mammalian ortholog of yeast Atg8 (15). In mammalian cells, cleavage of a 22 amino-acid fragment from newly synthesized pro-LC3 produces LC3-I. The concerted actions of Atg7 (an E1-like activity), Atg3 (an E2-like conjugating activity), and Atg12–Atg5-Atg16L complexes (E3-like ligase activity) conjugate LC3-I with phosphatidylethanolamine to produce LC3-II (5, 46). So activated, LC3-II localizes selectively to forming and newly formed autophagosomes even after other Atg proteins dissociate. Thus, LC3-II is a marker of ongoing autophagy. After sequestration, some LC3-II become entrapped on the inner surfaces of the double-membrane autophagosomes. After fusion with lysosomes, this LC3-II is degraded. LC3-II on the outer surface also disappears, most likely by cleavage of the phospholipid conjugate, and the LC3 is reutilized. A transgenic mouse strain was created that expresses a green fluorescent protein (GFP)-LC3 fusion protein. In cells and tissues of GFP-LC3 transgenic mice, GFP fluorescence selectively identifies the membranes of forming and newly formed autophagosomes (30).
Whether or not autophagy selectively targets specific organelles has been controversial. After withdrawal of peroxisome proliferators in the presence of protease inhibitors, peroxisomes selectively accumulate in autophagosomes (51). The removal of peroxisomes by autophagy is referred to as pexophagy and occurs in yeast when methanol-containing medium is switched to glucose or ethanol-containing medium (17, 49). Similarly, selective autophagic degradation of hepatic glycogen, but not mitochondria and other organelles, occurs in the early postnatal period (23). Mitochondria of nonproliferating tissues such as heart, brain, liver, and kidney constantly turn over with a half-life of 10–25 days (29, 36), and recent evidence supports selective autophagic removal of mitochondria, a process of mitophagy (16, 26, 35, 45). In staurosporine-treated cells where apoptosis is inhibited by caspase inhibitors, mitochondria are eliminated by mitophagy in a specific and regulated manner (50). In yeast, the mitochondrial outer membrane protein Uth1 is required for efficient mitophagy in a nutrient-poor medium, but a corresponding mammalian protein has yet to be identified (21). Also, a yeast protein phosphatase homologue, Aup1, in the mitochondrial intermembrane space is required for mitophagy (45). In a recent study, parkin was recruited to mitochondria depolarized by treatment with carbonyl cyanide m-chlorophenyl hydrazone, an uncoupler, to promote autophagy (33). Although these data support mitophagy as a distinctive pathway from the autophagy of other cytoplasmic components, selectivity of mitophagy for individual damaged mitochondria in normal cellular physiology has not been directly documented.
To that end, we used 488-nm light to photodamage individual mitochondria of primary hepatocytes from GFP-LC3 transgenic mice. Visible light of 400- to 500-nm excites and damages flavin-containing proteins in mitochondria, resulting in mitochondrial alterations and damage by reactive oxygen species (ROS) production (1, 2). Here, such photodamage depolarized and permeabilized mitochondria, which led to PI3K-independent sequestration of individual mitochondria into GFP-LC3-labeled autophagosomes.
Materials and Methods
Materials
3-Methyladenine (3MA; cat. no. 08592) and wortmannin (cat. no. w1628) were purchased from Sigma Chemical Co. (St. Louis, MO). Calcein acetoxymethyl ester (AM) (cat. no. C3100MP), LysoTracker Red (LTR; cat. no. L7528), MitoFluorFar Red (MFFR; cat. no M22423), and tetramethylrhodamine methylester (TMRM) (cat. no. T668) were obtained from Invitrogen/Molecular Probes (Carlsbad, CA). Collagenase A (cat. no. 11088793001) was obtained from Roche (Penzberg, Germany).
Hepatocyte isolation and culture
Hepatocytes from GFP-LC3 transgenic C57BL/6 mice or C57/BL6 wild-type mice were isolated by collagenase perfusion and cultured overnight in 5% CO2/95% air at 37°C on type 1 collagen-coated 35-mm glass bottom dishes at a density of 300,000 cells per plate in Waymouth's MB-752/1 growth medium supplemented with 27 m_M_ NaHCO3, 10% fetal bovine serum, 100 n_M_ insulin, and 100 n_M_ dexamethasone (WM) (13). For nutrient deprivation-induced autophagy, hepatocytes were placed in Krebs-Ringer-HEPES buffer (in m_M_: 115 NaCl, 5 KCl, 1 CaCl2, 1 KH2PO4, 1.2 MgSO4, and 25 Na-HEPES buffer, pH 7.4) plus 1 (M glucagon (KRH/G).
Loading of fluorophores
Hepatocytes isolated from GFP-LC3 transgenic mice were loaded with red-fluorescing TMRM (200 n_M_) or far red-fluorescing MFFR (200 n_M_) for 30 min in the absence or presence of 10 m_M_ 3MA or 100 n_M_ wortmannin at 37°C in WM supplemented with 25 m_M_ Na-HEPES buffer, pH 7.4. TMRM and MFFR are membrane-permeable monovalent cations that accumulate electrogenically into mitochondria (52). In other experiments, acidic compartments were labeled with LTR (500 n_M_) under identical conditions. After TMRM, MFFR, and LTR loading, one-third of the initial loading concentration was maintained in the medium to maintain a steady-state. Hepatocytes isolated from wild-type mice were loaded with LTR as just described. Subsequently, the cells were photoirradiated and loaded with 1 μ_M_ calcein AM for 10 min at 37°C to assess mitochondrial inner membrane permeabilization. After LTR and calcein loading, one-third of the initial LTR loading concentration was maintained in the medium.
Photodamage and confocal microscopy
Laser-induced photodamage and confocal microscopy were performed with a Zeiss LSM 510 Meta NLO laser scanning confocal microscope (Carl Zeiss, Thornwood, NY) using a 63×N.A. 1.4 oil-immersion planapochromat objective lens. To induce photodamage, selected areas of individual cells containing 5–10 mitochondria were exposed to 488-nm argon laser light at 100% power for times of 80, 160, 320, 640, and 1280 μs per pixel. Green (GFP-LC3), red (TMRM and LTR), and far red (MFFR) fluorescence was excited by the 488, 543, and 633-nm laser lines, respectively, of argon and helium-neon lasers in the multitracking mode. To image green and red fluorescence simultaneously, emitted light was separated by a 545-nm dichroic mirror and directed to different photomultipliers through 500- to 530-nm (green) band pass and 560-nm (red) long-pass filters. To image green, red, and far-red fluorescence simultaneously, emitted red fluorescence was additionally separated by a 635-nm dichroic mirror and directed to different photomultipliers through 565- to 615-nm (red) and 650- to 710-nm (far red) band-pass filters. For serial imaging at about one frame a minute, laser illumination was attenuated to less than 0.1% transmission power for pixel dwell times of 3.2 μs. Temperature on the microscope stage was maintained at 37°C.
Results
Photodamaged mitochondria are selectively removed by mitophagy
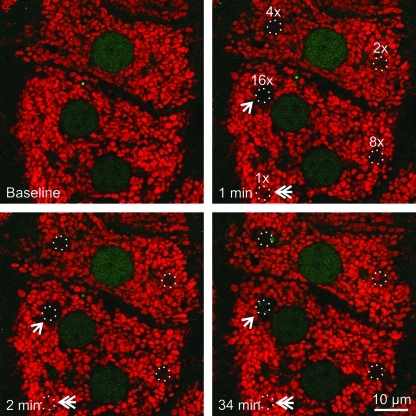
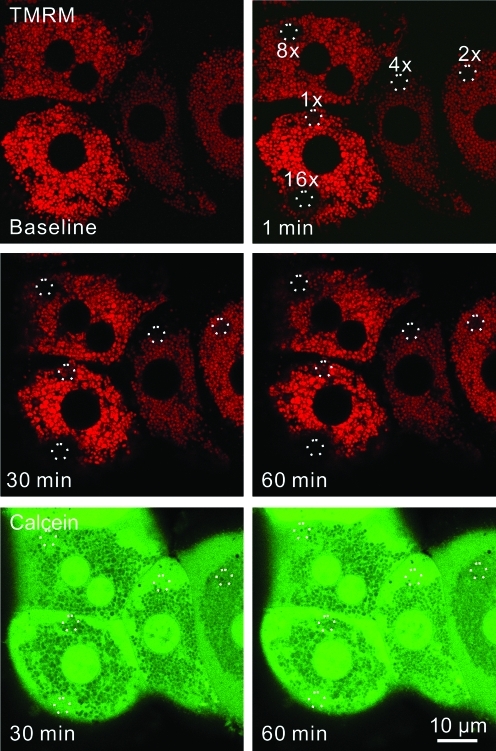
To investigate possible mitophagy after photodamage to mitochondria, cultured hepatocytes from GFP-LC3 transgenic mice were first loaded with TMRM, a red-fluorescing fluorophore that labels polarized mitochondria and is released on mitochondrial depolarization. Selected areas of hepatocytes containing 5–10 TMRM-loaded mitochondria were exposed to 488-nm argon laser light at 100% transmission for 80, 160, 320, 640, and 1280 μs. This illumination was, respectively, 2.5, 5, 10, 20, and 40×104 times greater than the power of illumination used for imaging. These pixel powers were designated in ascending order as 1×, 2×, 4×, 8×, and 16×. After selected area photoirradiation, confocal images of red TMRM and green GFP-LC3 fluorescence were collected every minute for up to 120 min. After lower laser photoirradiation (at or below 2×), mitochondria released TMRM, indicating depolarization, but subsequently recovered TMRM fluorescence (Fig. 1, double arrow). This transient depolarization signified reversible photodamage. At higher laser power (above 2×), mitochondria became irreversibly depolarized, indicating permanent mitochondrial damage (Fig. 1). At 16×power, adjacent mitochondria surrounding the illuminated mitochondria also became depolarized (Fig. 1, arrow). Presumably, this bystander injury was due to free radical generation by regions of the hepatocytes actually exposed to light (1). Overall, these data showed that photoirradiation with 488-nm laser light caused sustained mitochondrial depolarization in a dose-dependent fashion. Nonetheless, the photoirradiated hepatocytes remained viable and healthy with no morphological evidence of injury, necrosis, or apoptosis (e.g., cell surface blebbing, chromatin condensation, nuclear lobulation) over the time course of the experiments.
FIG. 1.
Mitochondrial damage occurs in a dose-dependent manner after exposure to 488-nm light. Hepatocytes isolated from GFP-LC3 transgenic mice were loaded with 200 n_M_ TMRM for 30 min. Regions of the TMRM (red)-loaded hepatocytes were exposed to different amounts of 488-nm light at 100% power at 2.7, 5.5, 11, 22, and 44×104 times greater than the pixel illumination used for imaging. These illumination powers were labeled in ascending order as 1×, 2×, 4×, 8×, and 16×. The circles represent areas of photoirradiation. Images of red TMRM and green GFP-LC3 were collected before (baseline) and every minute after photoirradiation. Loss of TMRM fluorescence indicated depolarization of mitochondria. The double arrow indicates transient TMRM loss after photodamage at 1×. The arrow illustrates TMRM loss from the mitochondria just outside the zone of illumination at 16×. GFP-LC3, green fluorescence protein fused to microtubule-associated protein1 light chain 3; TMRM, tetramethyl rhodamine methyl ester. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
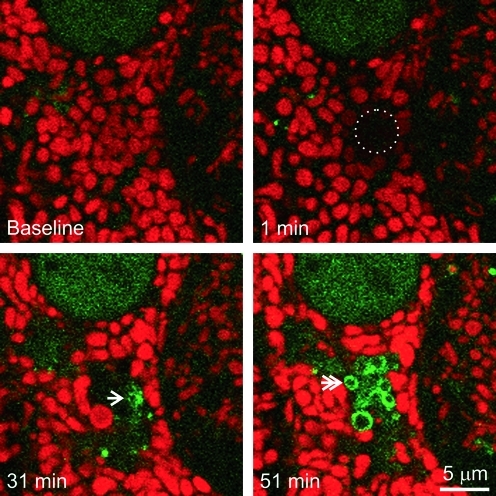
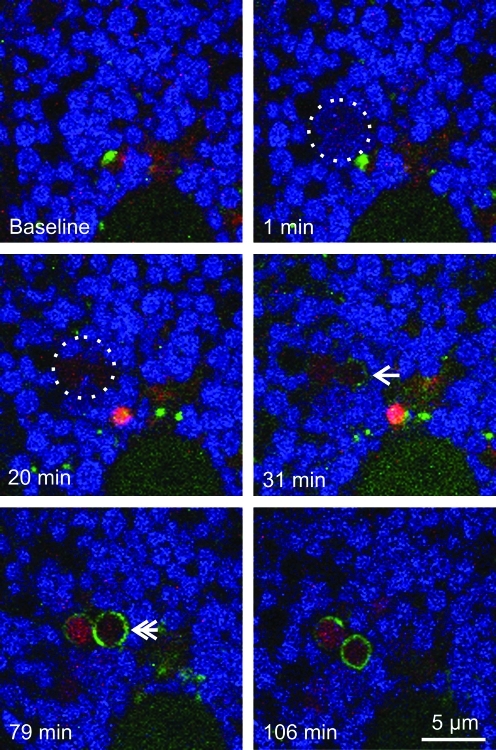
At 32±5 min after photoirradiation (4×and up), GFP-LC3 fluorescence began to associate with depolarized mitochondria as small speckled structures that eventually coalesced into ring structures (autophagosomes) (Fig. 2, bottom right panel). However, GFP-LC3 did not localize to mitochondria that transiently depolarized after low light exposure. Unlike nutrient deprivation-induced mitophagy (19), GFP-LC3-labeled cup-shaped phagophores did not grow around and sequester individual mitochondria; rather, GFP-LC3 began to decorate the surface of the photodamaged mitochondria (Fig. 2).
FIG. 2.
Photodamaged mitochondria are degraded by mitophagy. A TMRM-loaded GFP-LC-3 hepatocyte was exposed to 16×illumination, as described in Figure 1. The circle indicates the area of photoirradiation. The arrow indicates first localization of GFP-LC3. The double arrow illustrates GFP-LC3 ring formation. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Photodamage-induced mitophagy is independent of TMRM loading
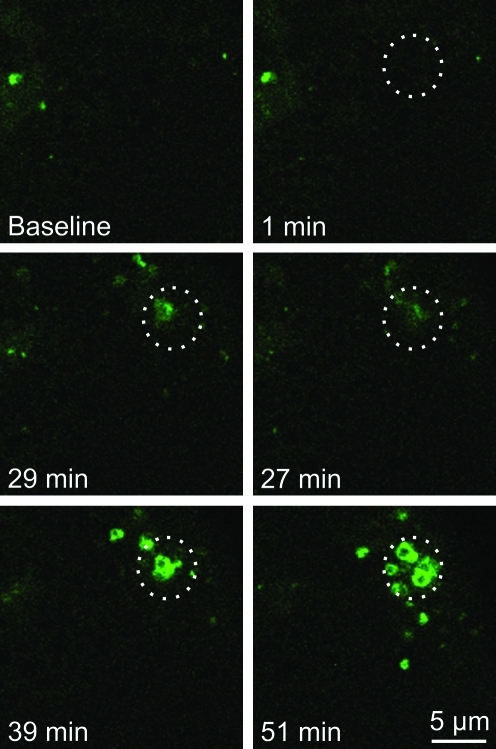
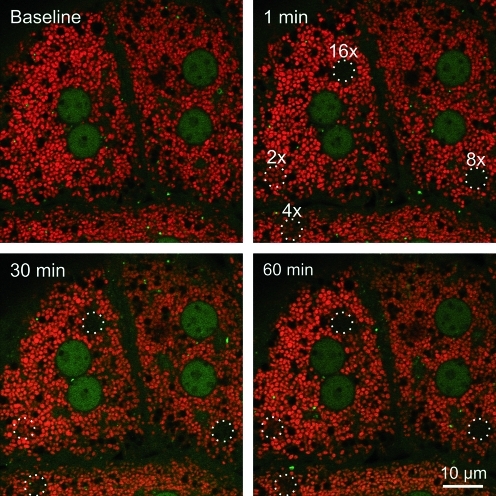
To determine whether light-induced mitophagy is dependent on the presence of TMRM as a photosensitizer, small regions of GFP-LC3 hepatocytes were exposed to 488-nm illumination at 16×laser power in the absence of TMRM. Similar to observations in the presence of TMRM, the region exposed to 16×laser power became decorated by GFP-LC3 after about 27 min (Fig. 3). The GFP-LC3 fluorescence subsequently coalesced into green rings indistinguishable from the GFP-LC3 rings that formed after photodamage in the presence of TMRM. Thus, mitophagy after photoirradiation with 488-nm light occurred independently of TMRM, which is consistent with only weak absorbance of 488-nm light by TMRM (Fig. 3). In another experiment, hepatocytes were loaded with TMRM and subjected to photodamage with different doses of 543-nm laser light, a wavelength that is absorbed by TMRM. Green excitation light at 543-nm caused photobleaching of TMRM (Fig. 4, top right panel). However, GFP-LC3 did not localize to the mitochondria that were exposed to 543-nm light (Fig. 4, bottom panels). Taken together, these observations indicate that 488-nm light induces mitochondrial photodamage that leads to mitophagy. The presence of TMRM did not sensitize the process.
FIG. 3.
Photodamage-induced mitophagy is independent of TMRM. GFP-LC3 hepatocytes were exposed to 488-nm light at 16×relative power. The circles indicate the area of photoirradiation. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 4.
Exposure to 543-nm light does not induce mitophagy. TMRM (red)-loaded GFP-LC3 hepatocytes were exposed to different amounts of 543-nm light at 100% power–5.5, 11, 22, and 44×104 times higher than the power used for imaging. These pixel powers were labeled in ascending order as 2×, 4×, 8×, and 16×. The circles represent the areas of irradiation. Images of red TMRM and green GFP-LC3 were collected every minute before (baseline) and after photodamage. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Photodamages induces the mitochondrial permeability transition
Photoirradiation generates ROS that can induce the mitochondrial permeability transition (MPT) in mitochondria (14). During the MPT, mitochondria become permeable to molecules up to 1.5 kDa, which causes mitochondrial depolarization. To investigate a role for the MPT in initiating photodamage-induced mitophagy, hepatocytes from wild-type mice were loaded with TMRM, exposed to 488-nm light, and then loaded with calcein AM. Calcein AM enters the cytosol (and nucleus) where esterases release green-fluorescing calcein free acid, which outlines mitochondria as round dark voids. When mitochondria undergo the MPT, calcein fluorescence fills the voids (27, 37, 38). After exposure to 488-nm light, mitochondria released TMRM transiently at lower doses and permanently at higher light levels (Fig. 5, compare top and middle left panels). Subsequently, after loading with calcein AM, polarized mitochondria excluded the green-fluorescing probe. By contrast, depolarized mitochondria that had been exposed to high power light (16×) filled with calcein immediately (Fig. 5, lower panels), but mitochondria exposed to intermediate power (8×) released TMRM within about a minute but only took up calcein after several minutes more (data not shown). Over time, however, all mitochondria irreversibly releasing TMRM became filled with calcein fluorescence (Fig. 5). At 1×power, mitochondria recovered TMRM fluorescence after depolarizing initially. These mitochondria did not take up calcein (Fig. 5, bottom panel). Thus, photoirradiation with higher doses of 488-nm light led to inner membrane permeabilization, which is the hallmark feature of the MPT.
FIG. 5.
Inner membrane permeabilization occurs after photodamage. Hepatocytes from a wild-type mouse were loaded with TMRM and exposed to 488-nm laser light at relative powers of 1×, 2×, 4×, 8×, and 16×, as described in Figure 1. Images were collected before (baseline) and after photoirradiation. After exposure, hepatocytes were loaded with 1 μM calcein acetoxymethyl ester for 10 min, and images of red TMRM and green calcein fluorescence were collected. The circles indicate areas of photoirradiation. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Photodamage-induced mitophagosomes undergo acidification
In general after autophagic sequestration, autophagosomes fuse with lysosomes and acidify. To investigate acidification of mitophagosomes (autophagosomes containing mitochondria), GFP-LC3 transgenic hepatocytes were loaded with MFFR, a far red-fluorescing fluorophore that accumulates electrophoretically into polarized mitochondria like TMRM (40). MFFR-labeled mitochondria were photodamaged with 488-nm light at 16×power, and then red-fluorescing LTR, a weakly basic fluorophore that accumulates into acidic compartments, was loaded. After photodamage, mitochondria released their MFFR fluorescence signifying depolarization (Fig. 6, top panels). After 20 min, LTR started accumulating weakly in the photodamaged mitochondria (Fig. 6, middle left panel), and GFP-LC3 (green) particles began to decorate the photodamaged mitochondria (Fig. 6, middle right panel, arrow). Red LTR fluorescence progressively intensified afterward (Fig. 6, bottom panels). Thus, after photodamage, mitochondria became weakly acidic as they began to associate with GFP-LC3. Subsequently, after envelopment by GFP-LC3, the structures strongly acidified.
FIG. 6.
Mitophagosomes formed after photodamage acidify. GFP-LC3 hepatocytes were loaded with 300 n_M_ MFFR and 500 n_M_ LTR for 30 min. Circles show the area of photoirradiation with 488-nm light at a relative intensity of 16 ×. After photoirradiation, mitochondria lost MFFR fluorescence (pseudocolored blue) and took up red LTR fluorescence, which signified acidication. Arrow indicates localization of green GFP-LC3 to damaged mitochondria. Double arrow indicates GFP-LC3 rings (autophagosomes) around LTR-labeled photodamaged mitochondria. LTR, Lysotracker Red. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Photodamage-induced mitophagy activates downstream of PI3K signaling
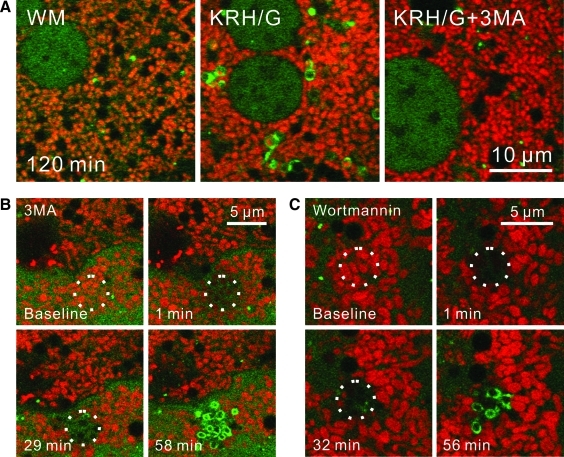
3MA, the classical inhibitor of autophagy, and wortmannin block autophagy after nutrient deprivation by inhibition of class III PI3K (8, 43). To illustrate this effect of 3MA, GFP-LC3 hepatocytes labeled with TMRM were incubated in complete growth medium (WM) or KRH/G with and without 3MA. In WM, GFP-LC3-labeled rings and disks were rarely observed (Fig. 7A, left panel). By contrast, after incubation of hepatocytes for 120 min in KRH/G, GFP-LC3-labeled structures increased markedly, including green-fluorescing rings containing TMRM-labeled red-fluorescing mitochondria (mitophagosomes) (Fig. 7A, middle panel). Formation of GFP-LC3-labeled rings and disks during incubation in KRH/G was virtually completely blocked by 3MA (Fig. 7A, right panel). A similar inhibition of nutrient deprivation-induced autophagosome proliferation in hepatocytes under identical conditions occurs after treatment with wortmannin (39). To determine the role of PI3K on mitophagy after photodamage, GFP-LC3 hepatocytes were loaded with TMRM and treated with 10 m_M_ 3MA for 30 min before photoirradiation of a small group of mitochondria with 488-nm laser light at 16×laser power. Photodamaged mitochondria again irreversibly lost TMRM fluorescence, indicating loss of membrane potential (Fig. 7B, C, top panels). In the presence of 3MA, green ring structures again started to form around photodamaged mitochondria after about 30 min (Fig. 7B). To further confirm that photodamage-induced mitophagy occurred after inhibition of PI3K, GFP-LC3 hepatocytes were treated with 100 n_M_ wortmannin for 30 min before photoirradiation. Wortmannin did not inhibit photodamage-induced mitophagy (Fig. 7C). In the presence of either 3MA or wortmannin, the number of GFP-LC3 rings and the strength of labeling were greater than in their absence (Fig. 7).
FIG. 7.
3-Methyladenine and wortmanin do not prevent mitophagy after photodamage. GFP-LC3 transgenic hepatocytes were loaded with TMRM. In (A), hepatocytes were incubated 120 min in WM (left panel), KRH/G (middle panel) or KRH/G plus 10 m_M_ 3MA (right panel). In (B) and (C), hepatocytes were pretreated with 10 m_M_ 3MA (B) or 100 n_M_ wortmannin (C) for 30 min. Photoirradiation with a 488-nm laser at a relative intensity of 16×(circles) was then performed. Images were collected every minute. WM, Waymouth's medium/10% fetal bovine serum/insulin/dexamethasone; KRH/G, Krebs-Ringer-HEPES plus glucagons; 3MA, 3-methyladenine. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Discussion
Our results provide direct evidence that mitophagy selectively removes and degrades damaged mitochondria. In hepatocytes incubated in nutritionally replete growth medium, only photoirradiated mitochondria that depolarized were sequestered into autophagosomes. When mitochondria were photoirradiated at lower laser power, mitochondria initially depolarized but then recovered polarization as indicated by reuptake of TMRM. Such mitochondria did not undergo mitophagy. Thus, photodamage leading to sustained mitochondrial depolarization was required to initiate sequestration into autophagosomes.
Mitochondrial depolarization occurred in a dose-dependent manner after photoirradiation with 488-nm light (Fig. 1). The lowest light exposure, namely that used to image hepatocytes, did not cause mitochondrial depolarization or induce mitophagy. Initially, after a light exposure about 2.5×103 times greater than that used for imaging, mitochondria depolarized, but they then recovered their membrane potential within about 3 min. At greater photoirradiation, mitochondria depolarized in a sustained fashion. At highest illumination, a bystander effect occurred in which depolarization not only occurred in mitochondria under the light beam but also in adjacent mitochondria outside the illuminated region (Fig. 2). Bystander photoirradiation suggests that a toxic agent formed in illuminated regions diffused into immediately adjacent areas. This toxic agent is most likely ROS, such as singlet oxygen, which is produced during exposure to strong light. Photoxicity-dependent mitophagy occurred in the absence of TMRM or other added fluorophore and, thus, did not require photosensitization of an exogenous absorber (Fig. 3). In addition, mitophagy did not occur with 543-nm light (Fig. 4). Previous work shows that photoirradiation of 400- to 500-nm light causes oxygen-dependent inactivation of flavoproteins and succinate dehydrogenase that is mediated by production of ROS (1). Thus, photodamage to mitochondria in our experiments is likely via photoexcitation of succinate dehydrogenase and other mitochondrial flavoproteins.
ROS induce the MPT in mitochondria, leading to depolarization, uncoupling, and more ROS formation (34). After photodamage, sustained mitochondrial depolarization appeared to be a prerequisite for subsequent mitophagy. Mitochondria that depolarized transiently after light exposure did not undergo subsequent mitophagy, whereas photodamaged mitochondria that underwent sustained depolarization were reproducibly sequestered into autophagosomes (Fig. 2). Moreover, this latter group of mitochondria became permeable to calcein, indicative of the inner membrane permeabilization of the MPT (Fig. 5). Thus, sustained mitochondrial depolarization and associated inner membrane permeabilization seemed to be required for autophagy signaling. These results are consistent with involvement of the MPT in photodamage-induced mitophagy, as previously proposed for autophagy stimulated by nutritional deprivation (10). Moreover, when cells are treated with a mitochondrial uncoupler such as carbonylcyanide m-chlorophenylhydrazone, depolarized mitochondria are removed (33).
Photodamage-induced mitophagy, however, differed from nutrient deprivation-induced mitophagy in several ways. In nutrient deprivation-induced mitophagy, small (0.2–0.3 μm) pre-autophagosomal structures associate with polarized mitochondria and grow into crescent-shaped phagophores that envelope and enclose individual polarized mitochondria into mitophagosomes (19). Mitochondrial depolarization only occurs at or after formation of mitophagosomes. Subsequently, as the mitophagosomal vesicles acidify and fuse with lysosomes, GFP-LC3 is released and/or degraded (20). However, when the MPT is inhibited, autophagy also becomes inhibited, indicating that the MPT likely plays a role in the nutrient deprivation-induced mitophagy, possibly in coordination with autophagosomal sequestration.
By contrast, in mitophagy induced by photodamage with 488-nm light, only depolarized mitochondria were targeted for autophagic degradation. Moreover, instead of being enveloped by a crescent-shaped phagophore, the periphery of photodamaged mitochondria became decorated with small speckled aggregates of GFP-LC3 that later coalesced into rings enveloping the entire mitochondrion (Figs. 2 and 6). Additionally, mild acidification of photodamaged mitochondria occurred before assembly of continuous GFP-LC3-decorated rings. Subsequently, after completion of the GFP-LC3 ring, the mitophagosomal vesicle became more intensely acidic. Future studies will be needed to determine whether depolarized mitochondria themselves undergo mild acidification or whether a sequestration membrane encloses photodamaged mitochondria before recruitment of GFP-LC3 (Fig. 6).
Mitophagy stimulated by 488-nm light also occurred in the absence of TMRM, indicating that TMRM was not acting as a photosensitizer (Fig. 3). Photoirradiation with 543-nm did not induce mitophagy (Fig. 4). Unexpectedly, photoirradiation with 543-nm light did lead to loss of mitochondrial TMRM fluorescence (Fig. 4). This may reflect direct photobleaching of TMRM rather than photodamage and depolarization of mitochondria. However, if mitochondria remained polarized, reaccumulation of TMRM from the cytosol and extracellular medium into mitochondria would be expected to occur over time, and such reuptake did not occur. Alternatively, mitophagy after photoirradiation may depend on the nature of the mitochondrial injury, and damage to flavoproteins by 488-nm irradiation may be the more potent mitophagy inducer. Additional studies will be needed to characterize what specific mitochondrial injuries induce mitophagy the most.
A particularly noteworthy difference between nutrient deprivation-induced mitophagy and photodamage-induced mitophagy is that the latter was not blocked by PI3K inhibition with 3MA (10 m_M_) or wortmannin (100 n_M_) (Fig. 7) (18). Class III PI3K/p150 interacts with Beclin 1, a mammalian homologue of Atg6 that is required for an early stage of autophagosome formation during nutrient deprivation (47). Rather, in our study, PI3K inhibition appeared to augment GFP-LC3 association with photodamaged mitochondria (Fig. 7). These findings indicate that activation of mitophagy after photodamage occurs independently of PI3K signaling, which is in striking contrast to autophagy after other stimuli, such as nutritent deprivation, that is blocked by PI3K inhibitors (see Fig. 7A). Indeed, photodamage-induced GFP-LC3 ring formation was more robust after PI3K inhibition, which suggests that subsequent processing of mitophagosomes may require PI3K, as shown for the processing of mitophagosomes in nutrient deprivation-induced autophagy (19, 31, 47).
Mitochondria of nonproliferating tissues such as heart, brain, liver, and kidney have a half-life of 10 to 25 days (29, 36). In this normal turnover, old and presumably dysfunctional mitochondria are removed by mitophagy and replaced by biogenesis of new mitochondria. Such mitophagy serves the physiological function of segregating and degrading dysfunctional mitochondria that might otherwise release ROS, pro-apoptotic proteins, and other toxic mediators. Since mitochondria are a primary site of ROS generation, mitochondrial DNA (mtDNA) is prone to oxidative damage. Due to limited mtDNA repair mechanisms, damaged mtDNA likely accumulates with time. Since mtDNA is nearly 100% active in transcription (compared to 2% or 3% for nuclear DNA), damage to mtDNA will lead quickly to mitochondrial dysfunction. Decreased mitophagy may promote accumulation of mtDNA mutations in aging rodents (6, 26, 48), whereas caloric restriction and rapamycin, inducers of autophagy, increase longevity in rodents (7, 12). Moreover, mitochondria with mutant mtDNA are selectively removed in heteroplasmic cells (44).
In conclusion, photoirradiation by 488-nm light caused mitochondrial depolarization, inner membrane permeabilization, and subsequent selective mitophagy, consistent with previous reports of photodynamic induction of the MPT and involvement of the MPT in mitophagy (10, 24, 53). However, upstream signaling for photodamage-induced mitophagy bypassed the classical PI3K signaling pathway of nutrient deprivation-induced autophagy, although PI3K still appeared to be needed for downstream processing of newly formed mitophagosomes. Thus, mitophagy is an important mechanism to sequester and degrade damaged mitochondria in otherwise viable and healthy cells.
Abbreviations Used
3MA
3-methyladenine
AM
acetoxymethyl ester
ATG
autophagic related-genes
GFP
green fluorescence protein
KRH/G
Krebs-Ringer-HEPES plus glucagon
LC3
microtubule-associated protein1 light chain 3
LTR
LysoTracker Red
MFFR
MitoFluor Far Red
MPT
mitochondrial permeability transition
mtDNA
mitochondrial DNA
PI3K
phosphatidylinositol 3-kinase
ROS
reactive oxygen species
TMRM
tetramethyl rhodamine methyl ester
WM
Waymouth's medium/10% fetal bovine serum/insulin/dexamethasone
Footnotes
This work is in partial fulfillment of the requirements for a Ph.D. to I. Kim from the University of North Carolina at Chapel Hill.
Acknowledgments
The authors thank Dr. Noboru Mizushima at Tokyo Medical and Dental University for GFP-LC3 mice. This work was supported, in part, by Grants 5 R01 CA119079, 2-R01 DK37034, 1 P01 DK59340, 1 R01 DK073336, and C06 RR015455 from the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aggarwal BB. Quintanilha AT. Cammack R. Packer L. Damage to mitochondrial electron transport and energy coupling by visible light. Biochim Biophys Acta. 1978;502:367–382. doi: 10.1016/0005-2728(78)90057-9. [DOI] [PubMed] [Google Scholar]
- 2.Alexandratou E. Yova D. Handris P. Kletsas D. Loukas S. Human fibroblast alterations induced by low power laser irradiation at the single cell level using confocal microscopy. Photochem Photobiol Sci. 2002;1:547–552. doi: 10.1039/b110213n. [DOI] [PubMed] [Google Scholar]
- 3.Arstila AU. Shelburne JD. Trump BF. Studies on cellular autophagocytosis. A histochemical study on sequential alterations of mitochondria in the glucagon-induced autophagic vacuoles of rat liver. Lab Invest. 1972;27:317–323. [PubMed] [Google Scholar]
- 4.Arstila AU. Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733. [PMC free article] [PubMed] [Google Scholar]
- 5.Barth S. Glick D. Macleod KF. Autophagy: assays and artifacts. J Pathol. 2010;221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergamini E. Autophagy: a cell repair mechanism that retards ageing and age-associated diseases and can be intensified pharmacologically. Mol Aspects Med. 2006;27:403–410. doi: 10.1016/j.mam.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Bergamini E. Cavallini G. Donati A. Gori Z. The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomed Pharmacother. 2003;57:203–208. doi: 10.1016/s0753-3322(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 8.Blommaart EF. Krause U. Schellens JP. Vreeling-Sindelarova H. Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 9.Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 10.Elmore SP. Qian T. Grissom SF. Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 11.Furuya N. Yu J. Byfield M. Pattingre S. Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 12.Harrison DE. Strong R. Sharp ZD. Nelson JF. Astle CM. Flurkey K. Nadon NL. Wilkinson JE. Frenkel K. Carter CS. Pahor M. Javors MA. Fernandez E. Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatano E. Bradham CA. Stark A. Iimuro Y. Lemasters JJ. Brenner DA. The mitochondrial permeability transition augments Fas-induced apoptosis in mouse hepatocytes. J Biol Chem. 2000;275:11814–11823. doi: 10.1074/jbc.275.16.11814. [DOI] [PubMed] [Google Scholar]
- 14.Jou MJ. Jou SB. Chen HM. Lin CH. Peng TI. Critical role of mitochondrial reactive oxygen species formation in visible laser irradiation-induced apoptosis in rat brain astrocytes (RBA-1) J Biomed Sci. 2002;9:507–516. doi: 10.1159/000064723. [DOI] [PubMed] [Google Scholar]
- 15.Kabeya Y. Mizushima N. Ueno T. Yamamoto A. Kirisako T. Noda T. Kominami E. Ohsumi Y. Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanki T. Wang K. Cao Y. Baba M. Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiel JA. Komduur JA. van der Klei IJ. Veenhuis M. Macropexophagy in Hansenula polymorpha: facts and views. FEBS Lett. 2003;549:1–6. doi: 10.1016/s0014-5793(03)00794-4. [DOI] [PubMed] [Google Scholar]
- 18.Kim I. Mizushima N. Lemasters JJ. Selective removal of damaged mitochondria by autophagy (mitophagy) Hepatology. 2006;44(Suppl. 1):241A. (abstract) [Google Scholar]
- 19.Kim I. Rodriguez-Enriquez S. Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim I. Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am J Physiol. 2011;300:c308–c317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kissova I. Deffieu M. Manon S. Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 22.Kopitz J. Kisen GO. Gordon PB. Bohley P. Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotoulas OB. Kalamidas SA. Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Lam M. Oleinick NL. Nieminen AL. Photodynamic therapy-induced apoptosis in epidermoid carcinoma cells. Reactive oxygen species and mitochondrial inner membrane permeabilization. J Biol Chem. 2001;276:47379–47386. doi: 10.1074/jbc.M107678200. [DOI] [PubMed] [Google Scholar]
- 25.Lemasters JJ. V. Necrapoptosis and the mitochondrial permeability transition: shared pathways to necrosis and apoptosis. Am J Physiol. 1999;276:G1–G6. doi: 10.1152/ajpgi.1999.276.1.G1. [DOI] [PubMed] [Google Scholar]
- 26.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 27.Lemasters JJ. Nieminen AL. Qian T. Trost LC. Elmore SP. Nishimura Y. Crowe RA. Cascio WE. Bradham CA. Brenner DA. Herman B. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 28.Levine B. Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 29.Menzies RA. Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J Biol Chem. 1971;246:2425–2429. [PubMed] [Google Scholar]
- 30.Mizushima N. Yamamoto A. Matsui M. Yoshimori T. Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mousavi SA. Brech A. Berg T. Kjeken R. Phosphoinositide 3-kinase regulates maturation of lysosomes in rat hepatocytes. Biochem J. 2003;372:861–869. doi: 10.1042/BJ20021136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myung J. Kim KB. Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev. 2001;21:245–273. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narendra D. Tanaka A. Suen DF. Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieminen AL. Byrne AM. Herman B. Lemasters JJ. Mitochondrial permeability transition in hepatocytes induced by t-BuOOH: NAD(P)H and reactive oxygen species. Am J Physiol. 1997;272:C1286–C1294. doi: 10.1152/ajpcell.1997.272.4.C1286. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto K. Kondo-Okamoto N. Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 36.Pfeifer U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver. A morphometric approach to the kinetics of intracellular degradation by autophagy. J Cell Biol. 1978;78:152–167. doi: 10.1083/jcb.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian T. Herman B. Lemasters JJ. The mitochondrial permeability transition mediates both necrotic and apoptotic death of hepatocytes exposed to Br-A23187. Toxicol Appl Pharmacol. 1999;154:117–125. doi: 10.1006/taap.1998.8580. [DOI] [PubMed] [Google Scholar]
- 38.Qian T. Nieminen AL. Herman B. Lemasters JJ. Mitochondrial permeability transition in pH-dependent reperfusion injury to rat hepatocytes. Am J Physiol. 1997;273:C1783–C1792. doi: 10.1152/ajpcell.1997.273.6.C1783. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Enriquez S. Kim I. Currin RT. Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakanoue J. Ichikawa K. Nomura Y. Tamura M. Rhodamine 800 as a probe of energization of cells and tissues in the near-infrared region: a study with isolated rat liver mitochondria and hepatocytes. J Biochem. 1997;121:29–37. doi: 10.1093/oxfordjournals.jbchem.a021565. [DOI] [PubMed] [Google Scholar]
- 41.Schworer CM. Mortimore GE. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc Natl Acad Sci U S A. 1979;76:3169–3173. doi: 10.1073/pnas.76.7.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seglen PO. Berg TO. Blankson H. Fengsrud M. Holen I. Stromhaug PE. Structural aspects of autophagy. Adv Exp Med Biol. 1996;389:103–111. doi: 10.1007/978-1-4613-0335-0_12. [DOI] [PubMed] [Google Scholar]
- 43.Seglen PO. Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suen DF. Narendra DP. Tanaka A. Manfredi G. Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tal R. Winter G. Ecker N. Klionsky DJ. Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- 46.Tanida I. Ueno T. Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tassa A. Roux MP. Attaix D. Bechet DM. Class III phosphoinositide 3-kinase—Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–586. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terman A. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology. 1995;41(Suppl 2):319–326. doi: 10.1159/000213753. [DOI] [PubMed] [Google Scholar]
- 49.Tuttle DL. Lewin AS. Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- 50.Xue L. Fletcher GC. Tolkovsky AM. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 51.Yokota S. Degradation of normal and proliferated peroxisomes in rat hepatocytes: regulation of peroxisomes quantity in cells. Microsc Res Tech. 2003;61:151–160. doi: 10.1002/jemt.10324. [DOI] [PubMed] [Google Scholar]
- 52.Zahrebelski G. Nieminen AL. al-Ghoul K. Qian T. Herman B. Lemasters JJ. Progression of subcellular changes during chemical hypoxia to cultured rat hepatocytes: a laser scanning confocal microscopic study. Hepatology. 1995;21:1361–1372. [PubMed] [Google Scholar]
- 53.Zorov DB. Juhaszova M. Sollott SJ. Mitochondrial ROS-induced ROS release: an update and review. Biochim Biophys Acta. 2006;1757:509–517. doi: 10.1016/j.bbabio.2006.04.029. [DOI] [PubMed] [Google Scholar]