Enzymatic Reduction of Oxysterols Impairs LXR Signaling in Cultured Cells and the Livers of Mice (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 20.
Abstract
Liver X receptors (LXRs) are nuclear receptors that play crucial roles in lipid metabolism in vivo and are activated by oxysterol ligands in vitro. The identity of the ligand that activates LXRs in vivo is uncertain. Here we provide two lines of evidence that oxysterols are LXR ligands in vitro and in vivo. First, overexpression of an oxysterol catabolic enzyme, cholesterol sulfotransferase, inactivates LXR signaling in several cultured mammalian cell lines but does not alter receptor response to the nonsterol agonist T0901317. Adenovirus-mediated expression of the enzyme in mice prevents dietary induction of hepatic LXR target genes by cholesterol but not by T0901317. Second, triple-knockout mice deficient in the biosynthesis of three oxysterol ligands of LXRs, 24_S_-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol, respond to dietary T0901317 by inducing LXR target genes in liver but show impaired responses to dietary cholesterol. We conclude that oxysterols are in vivo ligands for LXR.
INTRODUCTION
The liver X receptors α and β (LXRα and LXRβ) are nuclear receptors and heterodimeric partners of the retinoid X receptor (Apfel et al., 1994; Song et al., 1994; Willy et al., 1995). LXRs are activated by oxysterol ligands in vitro (Forman et al., 1997; Janowski et al., 1996; Lehmann et al., 1997) and regulate the expression of genes involved in fatty acid and cholesterol metabolism in vivo (Li and Glass, 2004; Tontonoz and Mangelsdorf, 2003).
Oxysterols were initially identified as ligands for LXRs using in vitro assays in which candidate compounds were added to the medium of cultured cells transfected with LXR reporter systems. Direct binding of oxysterols to the LXR ligand binding domain was demonstrated next (Janowski et al., 1999), as was the ability of bound ligand to recruit coactivator proteins to the receptor (Schultz et al., 2000). Nonsterol agonists of LXR were identified subsequently by the pharmaceutical industry.
It has proven more difficult to identify LXR ligands responsible for activation in vivo. Controversy exists as to whether naturally occurring oxysterols such as 24_S_hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol subserve the same LXR activating function in the whole animal as they do in vitro (Bjorkhem, 2002). The strongest lines of evidence supporting an in vivo role for oxysterols are the observations that inhibitors of cholesterol biosynthesis, which presumably reduce intracellular levels of oxysterols, attenuate LXR responses in cultured cells (DeBose-Boyd et al., 2001; Wong et al., 2004) and, conversely, that cholesterol feeding, which presumably increases oxysterol levels, induces LXR target genes in mice (Peet et al., 1998). Similarly, inhibition of the enzyme 2,3-oxidosqualene:lanosterol cyclase, which elevates intracellular levels of 24,25-epoxycholesterol, activates LXR signaling in cultured macrophages (Rowe et al., 2003).
The current studies were designed to accomplish two goals: first, to identify a catabolic enzyme that could be used to inactivate oxysterol ligands and thereby determine whether oxysterols activate LXRs in different biological systems in vitro and in vivo, and second, to determine whether mice deficient in oxysterol biosynthetic enzymes exhibit impaired LXR signaling in response to dietary cholesterol. To these ends, we show that expression of the enzyme cholesterol sulfotransferase (SULT2B1b) sulfates oxysterols and attenuates LXR signaling in cultured cells and whole animals and that knockout mice that do not synthesize 24_S_-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol fail to induce certain LXR target genes in the liver when fed cholesterol.
RESULTS
Cholesterol Sulfotransferase Metabolizes Oxysterols
Sulfotransferases are a family of cytosolic and membranebound enzymes that metabolize diverse substrates ranging from xenobiotics to steroids (Strott, 2002). Family members that act on steroids transfer an SO3−1 group from the cofactor 3′-phosphoadenosine 5′-phosphosulfate to the 3-hydroxyl position of a substrate. We introduced a full-length cDNA encoding the mouse SULT2B1b cholesterol sulfotransferase (Shimizu et al., 2003) into cultured human embryonic kidney 293 (HEK293) cells and confirmed that the expressed enzyme had the capacity to sulfate cholesterol as well as several oxysterols (Javitt et al., 2001), including the LXR ligands 22-hydroxycholesterol, 24_S_-hydroxycholesterol, 25-hydroxycholesterol, 27-hydroxycholesterol, and 24,25-epoxycholesterol (see Figure S1 in the Supplemental Data available with this article online).
Sulfotransferase Inactivates LXR Signaling in HEK293 Cells
To determine whether expression of sulfotransferase inactivates the transcriptional activity of LXR, a receptor-reporter system consisting of plasmids encoding GAL4-LXR, RXRα, and a luciferase gene linked to a GAL4-responsive promoter was introduced into HEK293 cells in the presence or absence of an expressible sulfotransferase cDNA. The addition of different oxysterol ligands stimulated luciferase expression 3- to 12-fold, and with each sterol, coexpression of sulfotransferase decreased the stimulatory response (Figure 1A). In contrast, activation of LXR-mediated luciferase gene expression mediated by a nonsterol agonist, T0901317, was unaffected by the presence of sulfotransferase at the concentration used (0.1 µM) and at lower concentrations (data not shown). To examine the receptor specificity of the inhibitory response, the ability of the sulfotransferase to inactivate bile acid ligands of the farnesoid X receptor (FXR) was determined. Sulfotransferase expression resulted in a modest (~2-fold) but reproducible stimulation of FXR transcriptional activity in the absence of ligand (Figure 1B). The addition of chenodeoxycholate led to a 32-fold stimulation of luciferase gene expression that was unaffected by coexpression of sulfotransferase (Figure 1B). Similarly, activation of FXR by the nonsterol agonist GW4064 was not influenced by sulfotransferase.
Figure 1. Inactivation of Oxysterol, but Not Nonsterol, Ligands by Sulfotransferase.

(A) HEK293 cells were transfected with plasmids of the GAL4-LXRα/GAL4-luciferase receptor-reporter system for 4 hr. Thereafter, the indicated ligands were added to the medium at either 1.25 µM (sterols) or 0.1 µM (T0901317) concentration, and the incubation was continued for 16 hr. Cells were harvested and luciferase activity was determined.
(B) HEK293 cells were transfected with plasmids of the full-length FXR/FXRE-luciferase receptor-reporter system as above. The ligands chenodeoxycholic acid (CDCA) and GW4064 were added at concentrations of 25 and 0.5 µM, respectively.
(C) HEK293 cells were transfected with plasmids of the full-length LXRα/RXRα/LXRE-luciferase receptor-reporter system. Ligands added were 24,25-epoxycholesterol (24,25-EC, 0.625 µM), T1091317 (0.1 µM), and 9-_cis_-retinoic acid at the indicated concentrations.
The ability of sulfotransferase to affect activation of the LXRα/RXRα heterodimer mediated by 9-_cis_-retinoic acid was tested next (Figure 1C). A receptor-reporter system consisting of plasmids encoding full-length LXR and RXRα and an LXR-responsive luciferase gene was introduced into HEK293 cells in the presence or absence of the sulfotransferase expression vector. When different amounts of 9-_cis_-retinoic acid were added alone to the medium of the transfected cells, a hyperbolic dose-response curve was generated. Coexpression of the sulfotransferase did not change this response. Addition of 24,25-epoxycholesterol plus 9-_cis_-retinoic acid increased luciferase gene expression in an additive fashion, and the presence of the sulfotransferase reduced this response to that observed in the presence of 9-_cis_-retinoic acid alone. The combination of the retinoid and T0901317 also activated transcription of the reporter gene, and this response was not altered by expression of sulfotransferase. We concluded that sulfation of 24,25-epoxycholesterol did not result in the production of an antagonist, as if this were the case, the response to the RXR ligand would have been diminished. Sulfation of 24,25-epoxycholesterol also did not affect the response of LXR to T0901317 (data not shown).
Sulfotransferase Inactivates Endogenous LXR Signaling in Cultured Cells
Several CHO cell lines that expressed sulfotransferase in response to ecdysone were generated, and the behavior of an endogenous LXR target gene, ABCA1 (Venkateswaran et al., 2000), was monitored. Induction of the enzyme in these cells had no effect on the ability of T0901317 to induce ABCA1; however, sulfotransferase efficiently counteracted stimulation by 24,25-epoxycholesterol (Figure S2).
RAW 264.7 cells are a transformed line of mouse macrophages sensitive to the stimulatory effects of LXR ligands. We found that infection of RAW cells with recombinant adenoviruses expressing sulfotransferase inactivated the ability of four different oxysterols to induce the LXR target genes ABCA1 and SREBP-1c but had no effect on induction mediated by T0901317 (Figure S3). RAW cells infected with a control adenovirus expressing E. coli β-galactosidase responded normally to oxysterols and the synthetic agonist.
Sulfotransferase Inactivates Insulin-Mediated Induction of SREBP-1c
Transcription of the SREBP-1c gene is activated by insulin in hepatocytes, and this stimulation is dependent on LXR (Chen et al., 2004; Repa et al., 2000). To determine whether expression of the sulfotransferase would block this activation, primary hepatocytes were prepared from rats and infected with adenoviruses encoding either sulfotransferase or β-galactosidase, and the response to insulin and T0901317 was measured. In uninfected cells, the level of SREBP-1c mRNA was increased 24-fold and 36-fold by treatment with insulin and T0901317, respectively (Figure 2A). When the sulfotransferase adenovirus was added to cells at a multiplicity of infection of 2, the response of the SREBP-1c gene to insulin was abolished, while that for T0901317 was not affected. A similar outcome was observed when a multiplicity of infection of 6 was used (Figure 2A). Neither insulin nor sulfotransferase affected the expression of other LXR target genes in these cells (data not shown).
Figure 2. Sulfotransferase Blocks the Ability of Insulin to Activate LXR in Primary Hepatocytes.
(A) Cells were prepared from rat liver and plated on collagen-coated dishes for 3–4 hr. The plating medium was replaced with fresh medium containing either no adenovirus (−) or adenovirus expressing the mouse cholesterol sulfotransferase cDNA at the designated multiplicities of infection (MOI). After an additional 14–16 hr incubation, cells were washed, and medium containing vehicle or the indicated concentrations of insulin or T0901317 was added. Nine hours later, total RNA was extracted from the cells, and levels of SREBP-1c mRNA were determined by real-time RT-PCR.
(B) Primary hepatocytes were prepared, plated, and infected with adenovirus expressing either the E. coli β-galactosidase gene (β-Gal) or the mouse cholesterol transferase gene (ST) and then treated with vehicle or insulin as described in (A). The experiments in (A) and (B) were performed on different days.
In a second experiment, uninfected cells again responded to insulin with a 27-fold induction of SREBP-1c mRNA levels (Figure 2B). This induction was not diminished by infection with the β-galactosidase adenovirus but was eliminated by the sulfotransferase adenovirus. These data allowed three conclusions to be drawn. First, sulfotransferase inactivated the response of the SREBP-1c gene to insulin in primary hepatocytes. Second, given the results shown in Figure 1C, the insulin effect on this gene was most likely mediated by LXR and not RXR. Third, given the substrate specificity of the sulfotransferase enzyme and the biology of LXR, insulin must in some manner stimulate the production of an endogenous sterol ligand of LXR that is selective for activation of the SREBP-1c gene or that affects the response of the system to an oxysterol.
Sulfotransferase Inactivates LXR Signaling In Vivo
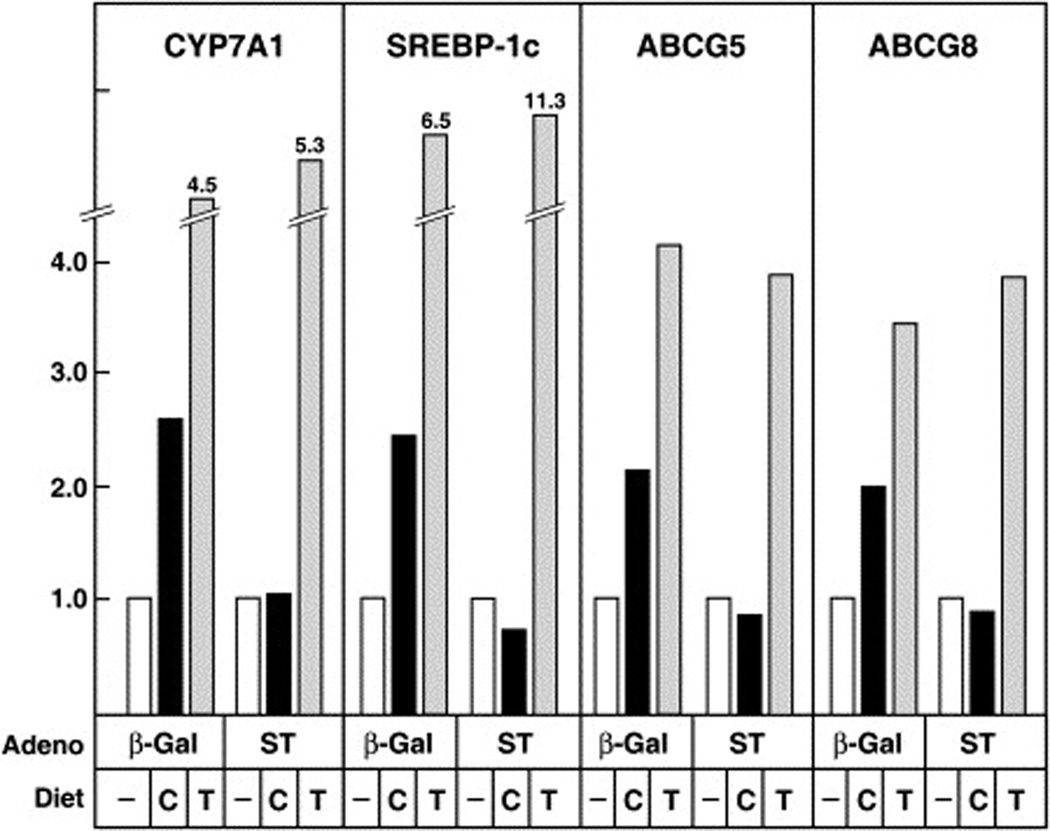
To establish whether expression of sulfotransferase affected LXR responses in vivo, wild-type C57BL/6 male mice were infected with adenovirus expressing either β-galactosidase or sulfotransferase and then fed diets consisting of normal chow or chow supplemented with 1% cholesterol or 0.025% T0901317 for a period of 4 days. The addition of cholesterol to the diet led to an induction of known LXR target genes, including cholesterol 7α-hydroxylase (CYP7A1), SREBP-1c, ABCG5, and ABCG8, in mice infected with the control β-galactosidase virus (Figure 3). Feeding T0901317 to similarly infected animals led to a more robust induction of this gene quartet. Infection with the sulfotransferase virus blocked the ability of dietary cholesterol to stimulate expression of these genes but had little effect in T0901317-fed mice.
Figure 3. Expression of Sulfotransferase in Mice Disrupts LXR-Mediated Gene Activation.
Male C57BL/6 mice (n = 3 per group) were infected with the indicated adenovirus (7.8 × 108 pfu/mouse) on day 0 of the experiment and then maintained on diets containing 0.02% cholesterol (−), 1% cholesterol (C), or 0.025% T0901317 (T). On day 4, animals were killed, and total RNA was prepared from individual livers. Equal amounts of RNA from each animal in an experimental group were pooled, and levels of four LXR target-gene mRNAs were determined by real-time RT-PCR. The results are representative of two separate infection/feeding experiments using the same number of animals of each genotype.
Mice with Oxysterol Biosynthetic Deficiencies Have Altered LXR Responses
Because sulfotransferase utilizes both cholesterol and oxygenated sterols as substrates (Figure S1), it was not clear from the above data which sterol class was acting as a ligand for LXR. To clarify this issue, dietary studies were carried out in mice deficient in three known oxysterol biosynthetic enzymes, cholesterol 24-hydroxylase, cholesterol 25-hydroxylase, and sterol 27-hydroxylase. The introduced mutations eliminated enzyme activity in each case and reduced endogenous levels of side-chain oxysterols to below the levels of detection by mass spectrometry (unpublished data). Male triple-knockout mice together with wild-type controls were fed unsupplemented chow or chow supplemented with 1% cholesterol or 0.025% T0901317. A small amount of cholic acid (0.025%) was included in all diets. This supplementation restored bile acid pool size and composition in the mutant mice but did not affect these parameters in wild-type mice.
As expected, feeding cholesterol and T0901317 to wild-type mice for 7 days induced the expression of known LXR target genes, including CYP7A1, lipoprotein lipase (LPL), SREBP-1c, ABCG5, and ABCG8 (Figure 4). In contrast, while the responses of the mutant mice to dietary T0901317 were similar to those of wild-type controls, LXR target-gene induction by cholesterol feeding was eliminated (LPL, ABCG5, and ABCG8), impaired (SREBP-1c), or unaffected (CYP7A1). These data suggested that the side-chain oxysterols 24_S_-, 25-, and 27-hydroxycholesterol activate LXR to induce LPL, ABCG5, and ABCG8 in mouse liver and that other ligands, e.g., 24,25-epoxycholesterol, produced by different biosynthetic enzymes, served this purpose for the CYP7A1 and SREBP-1c genes. In support of this interpretation, infection of cholesterol-fed triple-knockout mice with the sulfotransferase adenovirus prevented the induction of all LXR target genes by dietary cholesterol (Figure S4).
Figure 4. Knockout of Oxysterol Biosynthetic Genes Attenuates LXR-Mediated Gene Transcription in Liver.
Male wild-type (+/+) or _Cyp46a1_−/− _Ch25h_−/− _Cyp27a1_−/− (3KO) C57BL/6;129S6/SvEv mice (n = 6 per group) were fed chow with 0.025% cholic acid and 0.02% cholesterol (−), 1% cholesterol (C), or 0.025% T0901317 (T) for 7 days. Animals were killed, total RNA was prepared from individual livers, and equal amounts of RNA from each animal were pooled. The levels of five LXR target-gene mRNAs were determined by real-time RT-PCR. The results are representative of two separate feeding experiments using the same number of animals for each genotype.
DISCUSSION
The current data provide two lines of evidence that support a role for oxysterols as endogenous ligands for LXR. First, forced expression of cholesterol sulfotransferase, an enzyme that metabolizes oxysterol ligands, led to inactivation of LXR signaling in HEK293 cells, CHO cells, RAW mouse macrophages, primary hepatocytes, and cholesterol-fed mice. Second, genetic elimination of three oxysterol biosynthetic enzymes attenuated the response of some, but not all, LXR target genes in mouse liver.
The levels of ligands for nuclear receptors are regulated by catabolic enzymes to ensure that excess signaling does not take place. The enzymes involved in these degradative pathways are often redundant and expressed in both target and nontarget tissues (Penning, 2003). In some cases, the expression of catabolic enzymes, including sulfotransferases, protects a tissue from the actions of a circulating ligand (Tong et al., 2005); in others, the ligand induces the expression of genes encoding catabolic enzymes in a feed-forward regulatory loop (Chawla et al., 2001). Here, we exploited this knowledge to develop a generally applicable method in which ectopic expression of a catabolic enzyme is used to gain insight into nuclear receptor signaling.
Circulating oxysterols are normally metabolized to bile acids in the liver through a multienzyme pathway in which 7α-hydroxylation is a key step (Russell, 2003). That extra-hepatic pathways for inactivation must also exist is suggested by the observation that elimination of the mouse CYP7B1 oxysterol 7α-hydroxylase, which blocks the synthesis of bile acids from oxysterols and causes their accumulation in the plasma, does not hyperactivate LXR signaling (Li-Hawkins et al., 2000). Cholesterol sulfotransferase is not expressed in the liver but is expressed in the skin and epididymis (Shimizu et al., 2003) and thus may represent an example of an extrahepatic oxysterol-metabolizing enzyme. In agreement with this notion, LXR ligands activate several biological processes in the dermis (Man et al., 2006) and increase sulfotransferase expression in human keratinocytes (Jiang et al., 2005).
Overexpression of sulfotransferase inactivated the response of LXR to multiple oxysterol ligands when the latter were added exogenously to cultured cells. Dose-response experiments with 9-_cis_-retinoic acid (Figure 1C) and T0901317 (data not shown) suggested that the mechanism of attenuation involved inactivation of the oxysterol rather than conversion to an antagonist. In two experimental situations, reduction of the LXR response was less robust. In the first, incubation of RAW cells with acetylated LDL led to an approximately 70-fold increase in the level of ABCA1 mRNA, and this increase was reduced only 50% by expression of sulfotransferase (Figure S3). This attenuated response may be due to the large amounts of cholesterol delivered to cells upon endocytosis of acetylated LDL, which would be predicted to compete with oxysterol substrates for sulfotransferase access. Alternatively, the intracellular itinerary taken by endocytosed sterols may be different from that of sterols added exogenously in an organic solvent. It also is possible that acetylated LDL contains LXR agonists that are not substrates for sulfotransferase.
The second situation in which the LXR response was varied occurred in mice in which three oxysterol biosynthetic genes were knocked out (Figure 4). Cholesterol feeding induced five established LXR target genes in wild-type mice, and three of these, lipoprotein lipase, ABCG5, and ABCG8, were not induced in the triple-knockout mice. This is an expected result if the side-chain oxysterols synthesized by the three biosynthetic enzymes are responsible for activating LXR upon consumption of dietary cholesterol. One target gene, SREBP-1c, responded partially to cholesterol feeding in the mutant mice, and another, cholesterol 7α-hydroxylase, responded normally. The latter responses may reflect prior observations showing that nuclear receptors activate different classes of target genes depending on the ligand present (Downes et al., 2003; Dussault et al., 2003; Li-Hawkins et al., 2002; Miao et al., 2004; Quinet et al., 2004). Here, endogenous ligands that are not synthesized by the deleted biosynthetic enzymes, such as 24,25-epoxycholesterol (Zhang et al., 2001), sterol intermediates in the cholesterol biosynthetic pathway (Yang et al., 2006), or nonsterol ligands, may regulate the SREBP-1c and cholesterol 7α-hydroxylase genes. The observation (Figure S4) that adenoviral infection prevented the induction of these genes by cholesterol feeding in the mutant mice supports this notion.
EXPERIMENTAL PROCEDURES
HEK293 Cell Transfections
Cells (CRL 1573, American Type Culture Collection) were grown, transfected, and treated with ligands as described in detail in Supplemental Experimental Procedures. Cells were harvested 16 hr posttreatment and disrupted by addition of 0.5 ml of detergent buffer, and the resulting lysates were assayed for luciferase and β-galactosidase enzyme activities as described (Cheng et al., 2003).
Primary Hepatocytes
Hepatocytes were prepared from male Sprague-Dawley rats as described (Chen et al., 2004). Adherent cells were washed once with 4 ml of PBS and then incubated for 14–16 hr in medium B (medium 199 supplemented with 100 nM dexamethasone, 100 nM 3,3′,5-triiodo-L-thyronine, 100 units/ml penicillin, and 100 µg/ml streptomycin sulfate). Insulin (1 nM) and purified recombinant adenovirus (Ad-β-gal or Ad-mouseST) were then added to the medium at the indicated multiplicities of infection, and the incubation was continued an additional 14–16 hr. Cells were then washed once with 4 ml PBS and incubated with 3 ml of medium B supplemented with the indicated ligands and/or hormone for 9 hr prior to harvest and isolation of total RNA.
Adenovirus Infection of Mice
Recombinant adenoviruses expressing E. coli β-galactosidase (lacZ) or mouse cholesterol sulfotransferase were constructed using a kit from Qbiogene. On day 0 of each experiment, 3- to 4-month-old C57BL/6 male mice were injected via the tail vein with 7 × 108 pfu per mouse of recombinant adenovirus and then placed on powdered chow diets (Harlan Teklad #7001) supplemented with 0.02% cholesterol (ICN, #101380, lot 2530F), 1% cholesterol, or 0.025% T0901317 (all w/w). Animals were killed on day 4 at the middle of the light cycle. Total hepatic RNA was isolated using RNA STAT-60 (Tel-Test “B” Inc.), and individual mRNA levels were quantitated by real-time RT-PCR.
Diet Studies in Oxysterol-Deficient Mice
The triple-knockout mice used in these studies were generated by crossing cholesterol 24-hydroxylase-deficient (Lund et al., 2003), cholesterol 25-hydroxylase-deficient (unpublished data), and sterol 27-hydroxylase-deficient (Rosen et al., 1998) mice. The introduced mutation in each case produced a null allele. The genetic background of the animals was mixed (C57BL/6;129S6/SvEv). On day 0 of each experiment, male 3- to 4-month-old animals were placed on powdered chow diets (Harlan Teklad #7001) supplemented with 0.025% cholic acid (Sigma) and 0.02% cholesterol, 1% cholesterol, or 0.025% T0901317 (all w/w). Animals were killed on day 7 at the middle of the light cycle. Gene expression was monitored by real-time RT-PCR.
Supplementary Material
Acknowledgment
We thank Lisa Beatty, Jill Fairless, Shomanike Head, and Bonne Thompson for excellent technical assistance and Joe Goldstein, Helen Hobbs, and Steve Kliewer for critical review of the manuscript. This work was supported by NIH grant AR51943 (D.W.R.), the Howard Hughes Medical Institute (D.J.M.), The Robert A. Welch Foundation grants I-0971 (D.W.R.) and I-1275 (D.J.M.), and the Perot Family Foundation (D.W.R.). D.J.M. is a consultant to Exelixis. D.W.R. is a member of the scientific advisory boards of Alcon Laboratories, Inc. and Sirna Therapeutics, Inc.
REFERENCES
- Apfel R, Benbrook D, Lernhardt E, Ortiz MA, Salbert G, Pfahl M. A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor family. Mol. Cell. Biol. 1994;14:7025–7035. doi: 10.1128/mcb.14.10.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I. Do oxysterols control cholesterol homeostasis? J. Clin. Invest. 2002;110:725–730. doi: 10.1172/JCI16388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. USA. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxylase. J. Biol. Chem. 2003;278:38084–38093. doi: 10.1074/jbc.M307028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. USA. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, et al. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Beard R, Lin M, Hollister K, Chen J, Xiao JH, Chandraratna R, Forman BF. Identification of gene-selective modulators of the bile acid receptor FXR. J. Biol. Chem. 2003;278:7027–7033. doi: 10.1074/jbc.M209863200. [DOI] [PubMed] [Google Scholar]
- Forman BF, Ruan B, Chen J, Schroepfer GJ, Jr, Evans RM. The orphan nuclear receptor LXRalpha is positively and negatively regulated by distinct products of mevalonate metabolism. Proc. Natl. Acad. Sci. USA. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc. Natl. Acad. Sci. USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitt NB, Lee YC, Shimizu C, Fuda H, Strott CA. Cholesterol and hydroxycholesterolsulfotransferases: identification, distinction from dehydroepiandrosterone sulfotransferase, and differential tissue expression. Endocrinology. 2001;142:2978–2984. doi: 10.1210/endo.142.7.8244. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Kim P, Elias PM, Feingold KR. LXR and PPAR activators stimulate cholesterol sulfotransferase type 2 isoform 1b in human keratinocytes. J. Lipid Res. 2005;46:2657–2666. doi: 10.1194/jlr.M500235-JLR200. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Kliewer ST, Moore LB, Smith-Oliver TA, Oliver BB, Su J, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- Li AC, Glass CK. PPAR- and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J. Lipid Res. 2004;45:2161–2173. doi: 10.1194/jlr.R400010-JLR200. [DOI] [PubMed] [Google Scholar]
- Li-Hawkins J, Lund EG, Turley SD, Russell DW. Disruption of the oxysterol 7α-hydroxylase gene in mice. J. Biol. Chem. 2000;275:16536–16542. doi: 10.1074/jbc.M001811200. [DOI] [PubMed] [Google Scholar]
- Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, Schuster G, Bjorkhem I, Russell DW, Eggertsen G. Cholic acid mediates negative feedback regulation of bile acid synthesis. J. Clin. Invest. 2002;110:1191–1200. doi: 10.1172/JCI16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund EG, Xie C, Kotti T, Turley SD, Dietschy JM, Russell DW. Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J. Biol. Chem. 2003;278:22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- Man MQ, Choi EH, Schmuth M, Crumrine D, Uchida Y, Elias PM, Holleran WM, Feingold KR. Basis for improved permeability barrier homeostasis induced by PPAR and LXR activators: liposensors stimulate lipid synthesis, lamellar body secretion, and post-secretory lipid processing. J. Invest. Dermatol. 2006;126:386–392. doi: 10.1038/sj.jid.5700046. [DOI] [PubMed] [Google Scholar]
- Miao B, Zondlo S, Gibbs S, Cromley D, Hosagrahara VP, Kirchgessner TG, Billheimer JT, Mukherjee R. Raising HDL cholesterol without inducing hepatic steatosis and hypertriglyceridemia by a selective LXR modulator. J. Lipid Res. 2004;45:1410–1417. doi: 10.1194/jlr.M300450-JLR200. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro J-MA, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Penning TM. Hydroxysteroid dehydrogenases and pre-receptor regulation of steroid hormone action. Hum. Reprod. Update. 2003;9:193–205. doi: 10.1093/humupd/dmg022. [DOI] [PubMed] [Google Scholar]
- Quinet EM, Savio DA, Halpern AR, Chen L, Miller CP, Nambi P. Gene-selective modulation by a synthetic oxysterol ligand of the liver X receptor. J. Lipid Res. 2004;45:1929–1942. doi: 10.1194/jlr.M400257-JLR200. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro J-MA, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H, Reshef A, Maeda N, Lippoldt A, Shpizen S, Triger L, Eggertsen G, Bjorkhem I, Leitersdorf E. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J. Biol. Chem. 1998;273:14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
- Rowe AH, Argmann CA, Edwards JY, Sawyez CG, Morand OH, Hegele RA, Huff MW. Enhanced synthesis of the oxysterol 24(S),25-epoxycholesterol in macrophages by inhibitors of 2,3-oxidosqualene:lanosterol cyclase: A novel mechanism for the attenuation of foam cell formation. Circ. Res. 2003;93:717–725. doi: 10.1161/01.RES.0000097606.43659.F4. [DOI] [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C, Fuda H, Yanai H, Strott CA. Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal expression, kinetic analysis of isoforms, and comparison with prototypical SULT2A1. Endocrinology. 2003;144:1186–1193. doi: 10.1210/en.2002-221011. [DOI] [PubMed] [Google Scholar]
- Song C, Kokontis JM, Hiipakka RA, Liao S. Ubiquitous receptor: A receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA. 1994;91:10809–10813. doi: 10.1073/pnas.91.23.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strott CA. Sulfonation and molecular action. Endocr. Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- Tong MH, Jiang H, Liu P, Lawson JA, Brass LF, Song WC. Spontaneous fetal loss caused by placental thrombosis in estrogen sulfotransferase-deficient mice. Nat. Med. 2005;11:153–159. doi: 10.1038/nm1184. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. USA. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Wong J, Quinn CM, Brown AJ. Statins inhibit synthesis of an oxysterol ligand for the liver x receptor in human macrophages with consequences for cholesterol flux. Arterioscler. Thromb. Vasc. Biol. 2004;24:2365–2371. doi: 10.1161/01.ATV.0000148707.93054.7d. [DOI] [PubMed] [Google Scholar]
- Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, Hobbs HH. Sterol intermediates from cholesterol biosynthetic pathway as liver x receptor ligands. J. Biol. Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li D, Blanchard DE, Lear SR, Erickson SK, Spencer TA. Key regulatory oxysterols in liver: analysis as 4-3-ketone derivatives by HPLC and response to physiological perturbations. J. Lipid Res. 2001;42:649–658. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.