Efficient detection of RNA–protein interactions using tethered RNAs (original) (raw)
Abstract
The diverse localization of transcripts in cells suggests that there are many specific RNA–protein interactions that have yet to be identified. Progress has been limited, however, by the lack of a robust method to detect and isolate the RNA-binding proteins. Here we describe the use of an RNA aptamer, scaffolded to a tRNA, to create an affinity matrix that efficiently pulls down transcript-specific RNA-binding proteins from cell lysates. The addition of the tRNA scaffold to a Streptavidin aptamer (tRSA) increased binding efficiency by ∼10-fold. The tRSA system with an attached G-quartet sequence also could efficiently and specifically capture endogenous Fragile X Mental Retardation Protein (FMRP), which recognizes this RNA sequence. An alternative method, using biotinylated RNA, captured FMRP less efficiently than did our tRSA method. Finally we demonstrate the identification of novel RNA-binding proteins that interact with intron2 or 3′-UTR of the polarity protein Crumbs3 transcript. Proteins captured by these RNA sequences attached to the tRNA scaffold were identified by mass spectrometry. GFP-tagged versions of these proteins also showed specific interaction with either the Crb3 intron2 or 3′-UTR. Our tRSA technique should find wide application in mapping the RNA–protein interactome.
INTRODUCTION
High throughput proteomics and protein–protein interaction screens have enabled rapid progress to be made in mapping the protein ‘interactome’. The RNA–protein interactome is likely to be much larger and more complex than this, given the huge numbers of transcripts identified by recent global analyses (1–3), and the diversity of RNA localization and function in cells (4). Yet progress in the identification of transcript-specific RNA-binding proteins (RBPs) has been surprisingly slow. In general, it has been much easier to find RNAs that bind to specific proteins, rather than vice versa. RNA is unstable and flexible, which causes problems not only during isolation of binding proteins but also for designing functional tags. Prokipcak et al. (5) were able to purify a protein binding to c-myc mRNA by classical purification and RNA affinity chromatography (5). Ross et al. (6) successfully used synthetic oligoribonuceotides with a 3′-biotinylated end spacer for affinity purification of proteins that bind to the zipcode in the 3′-UTR of actin mRNA (6). RNA affinity columns have also been used to purify a few other proteins including a complementation factor involved in Apobec-1 dependent RNA editing, in which the target RNA was transcribed in vitro then biotinylated and attached to Streptavidin beads (7). However, synthetic oligoribonucleotides are expensive, their affinity for target proteins is often low, and chemical labeling is likely to alter the secondary structure of the RNAs.
A promising alternative to chemical labeling is the use of RNA aptamers. Aptamers bind to specific molecules that can be used both to track RNA localization in living cells and in affinity chromatographic methods to isolate RBPs (8–10). Some RNA tags, including MS2, PP7 and lambda N22, are naturally occurring sequences, while others, such as Streptavidin and Sephadex aptamers, have been found by screening synthetic libraries (11). An affinity selection approach with a tobramycin aptamer was used by Hartmuth et al. (16) for the isolation of the prespliceosomal complex (12). However, these strategies have all required multiple purification steps and specialized reagents such as recombinant proteins or affinity matrices, which might explain why they have not been widely adopted.
Transfer RNA scaffolding technology was developed for the efficient expression and purification of RNAs in Escherichia coli (13). In the presence of Mg2+, tRNAs fold into stable clover-leaf structures that are resistant to unfolding and can protect RNA fusions from degradation (14). Ponchon and Dardel (13) demonstrated that intact tRNA-RNA chimeras could be produced in high yield from bacterial lysates, and that they were correctly folded. Moreover, endogenous L20 protein could be co-precipitated, though with low efficiency, from bacterial lysate using a tRNA-scaffolded sephadex aptamer-23SrRNA fusion. We reasoned, therefore, that tRNA might provide a useful scaffold for the affinity purification of transcript-specific RBPs.
We now describe an approach that uses available materials and provides for flexible, robust and efficient purification of transcript-specific RBPs.
MATERIALS AND METHODS
Plasmid construction and antibodies
Streptavidin aptamer (SA) tags were generated by primer annealing and PCR, using the following sequences:
tRNA scaffolded SA tag: 5′-CAATTGAAAAAAAAAAAAAGCCCGGATAGCTCAGTCGGTAGAGCAGCGGCCTCGACCAGAATCATGCAAGTGCGTAAGATAGTCGCGGGTCGAGGCCGCGTCCAGGGTTCAAGTCCCTGTTCGGGCGCCACTGCAGAAAAAAAAAAAAGAATTC (Figure 1a)
1× SA tag: CAATTGGTCGACCGACCAGAATCATGCAAGTGCGTAAGATAGTCGCGGGCCGGGGGCGTATTATGTGCGTCTACATGAATTC
DNA fragments were digested with MunI and EcoRI, and ligated into the EcoRI site of pcDNA3 (Invitrogen). Since MunI and EcoR1 sites are cohesive, only the 3′-EcoR1 site is retained after ligation. The 6× stretavidin aptamer (6× SA) was generated by repeatedly cloning the 1× SA fragment into this surviving EcoRI site. These plasmids were designated pcDNA3-tRNA scaffolded SA (tRSA), pcDNA3-1× SA and pcDNA3-6× SA, respectively.
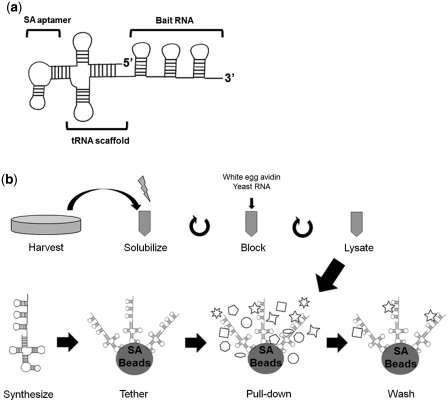
Figure 1.
tRSA construction and procedure. (a) Schematic of the tRNA-scaffolded streptavidin aptamer employed for this assay. Bait RNAs are attached to the 3′-end of a transfer RNA-SA fusion. (b) Schematic of the method. Precleared cell lysate and RNA beads are prepared in parallel.
To generate expression plasmid encoding MS2-GFP without nuclear localization signal (NLS), pcNMS2-GFP-NLS was digested with HindIII and XhoI, and the MS2-GFP fragment was cloned into corresponding sites of pcDNA3.
18× MS2 binding sequences (MBS) was amplified by PCR from β-globin-24bs/- plasmid with following primers,
| MBS forward: | 5′-GAGCTGTACAAGGGCGAATTCGCTTGGTCTAGCTC, |
|---|---|
| MBS reverse: | 5′-GCCCTCGAGCGATTCTAGACAGCAG |
The 18× MBSs PCR fragment was digested with EcoRI and XhoI, and cloned into corresponding sites of pcDNA3-tRSA, pcDNA3-1× SA and pcDNA3-6× SA, 3′ to the tags.
The G quartet fragment with EcoRI and XhoI sites was generated with the following primers;
| G quartet forward: | 5′-AATTCGGCTGCGGTGTGGAAGGAGTGGCTGGGTTGCGCAGCTC |
|---|---|
| G quartet reverse: | 5′-TCGAGAGCTGCGCAACCCAGCCACTCCTTCCACACCGCAGCCG |
The primers were annealed and cloned into corresponding sites of pcDNA3-tRSA.
Human Crumbs3 intron2 and 3′-UTR fragments were amplified using the CalTech human BAC clone (Clone#: CTD-2396E7-BHS1214, Open Biosystems) as PCR template. Primers were as follows,
| Intron2 forward: | 5′-GCCGAATTCGTAGGTACCAGCTGAGAGCGC, |
|---|---|
| Intron2 reverse: | 5′-GCCCTCGAGTGGAGGGTGAAGGCAGAGAATAAC, |
| 3′-UTR forward: | 5′-GCCGAATTCTAGGTCCCCTCTCCTGCATCT, |
| 3′-UTR reverse: | 5′-ATACTCGAGACATCTCACTACTAATTTTATATAAATATA. |
PCR products were all ligated into pcDNA3-tRSA cut with EcoRI and XhoI. For the GFP-tagged RBP assay, EGFP lacking a stop codon was amplified from pEGFP-C1 (Clontech), and cloned between the BamHI and EcoRI sites of pcDNA3, designated pcDNA-nEGFP. ADAR1, GRSF1, hnRNP M and hnRNP F genes were amplified from Caco-2 cell cDNA, and Nucleolin was amplified from pRSET-Nucleolin (Plasmid 13037, addgene) using the following primers.
| hADAR1 forward Mun: | 5′-GGCCAATTGACCATGGCCGAGATCAAGGAGAAAATCT |
|---|---|
| hADAR1 reverse Xho: | 5′-GCCCTCGAGCTATACTGGGCAGAGATAAAAGTTC |
| Nucleolin forward Eco: | 5′-GCCGAATTCACCATGGTGAAGCTCGCGAAGGCAGGTA |
| Nucleolin reverse Xho: | 5′-GCCCTCGAGCTATTCAAACTTCGTCTTCTTTCC |
| GRSF1 Mun: | 5′-GGCCAATTGACCATGGCCGGCACGCGCTGGGTACTCG |
| GRSF1 Xho: | 5′-GGCCTCGAGTTATTTTCCTTTTGGACATGAATTC |
| hnRNPF forward Mun: | 5′-GCCCAATTGACCATGATGCTGGGCCCTGAGGGAGGT |
| hnRNPF reverse Xho: | 5′-GCCCTCGAGCTAGTCATAGCCACCCATGCTGTT |
| hnRNPMb. forward Mun: | 5′-GCCCAATTGACCATGGCGGCAGGGGTCGAAGCGGC |
| hnRNPMb reverse Sal: | 5′-GCCGTCGACTTAAGCGTTTCTATCAATTCGAAC |
PCR products of all these genes were digested with EcoRI or MunI and XhoI or SalI, and ligated into cut pcDNA3-nEGFP cut with EcoRI and XhoI. Plasmids were confirmed by sequencing.
Anti-GFP (Ab13970, Abcam), anti-TLS/FUS (ab23439, Abcam) or anti-Fragile X Mentor Retardation Protein **(**FMRP) (MAB2160, Chemicon) were used for western blot detection.
In vitro RNA synthesis
In vitro RNA synthesis using AmpliScribe™ T7-_Flash_™ Transcription Kit or AmpliScribe™ T7-_Flash_™ Biotin-RNA Transcription Kit (Epicentre Biotechnologies) were performed according to the manufacturers’ instructions. Template DNAs were prepared by PCR using T7 primer and specific primers, and purified with QIAquick PCR Purification Kit (Qiagen). Synthesized RNAs were analyzed qualitatively and quantitatively by electrophoresis and spectrometry, then stored at −80°C.
RNA pull-down assay
A schematic of the method for affinity purification of RBPs is shown in Figure 1b. For the experiments reported in Figures 2a and 3b, HEK293 cells were plated on 10 cm tissue culture dishes. About 20 µg MS2-GFP plasmid was transfected using Lipofectamine 2000, using the manufacturer’s protocol. Cells were maintained in DMEM with 10% FBS and penicillin–streptomycin at 37°C in 5% CO2. Two days post-transfection, cells were harvested by adding 5 ml lysis buffer (10 mM HEPES pH 7.0, 200 mM NaCl, 1% Triton ×-100, 10 mM MgCl2, 1 mM DTT and protease inhibitors). Lysis buffer was prepared with 0.1% DEP-treated water.
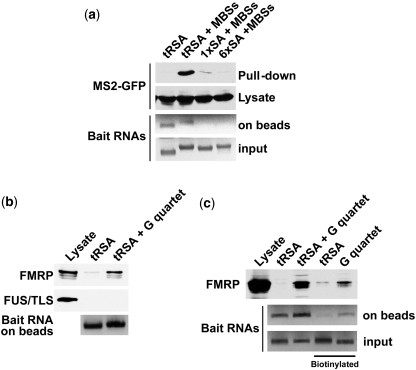
Figure 2.
The tRSA tag improves Streptavidin-binding efficiency, and can detect endogenous RBP. (a) RNA pull-down systems were tested using MS2 protein as a positive control. RNAs were transcribed in vitro and attached to streptavidin beads. RNA tags included tRSA, 1× SA and 6× SA aptamers, which were attached to 18× MBSs (MS2 binding sequences). Capture was performed with lysates of HEK 293 cells that expressed MS2-GFP. About 500 µg total protein were applied for each pull-down. Synthesized bait RNAs were quantified spectrometrically, and analyzed by gel electrophoresis in agarose containing 6.7% formaldehyde. (b) tRSA affinity tags can detect endogenously expressed protein. A single G quartet was attached to the tRSA. Pull-down was from 2 mg wild type HEK293 whole cell lysate protein. Proteins retained on the beads after washing were analyzed by immunoblot with anti-FMRP or anti-TLS/FUS antibodies. Half the beads then were kept aside to quantify tethered RNAs. Retained RNAs on the beads were quantified spectrometrically, and analyzed by gel electrophoresis in agarose containing 6.7% formaldehyde. (c) The tRSA method is more efficient than a biotinylated RNA pull-down. Pull-down was from 2 mg wild-type HEK293 whole cell lysate protein. Proteins retained on the beads after washing were analyzed by immunoblot with anti-FMRP RNA baits used in panels b and c were synthesized by in vitro transcription.
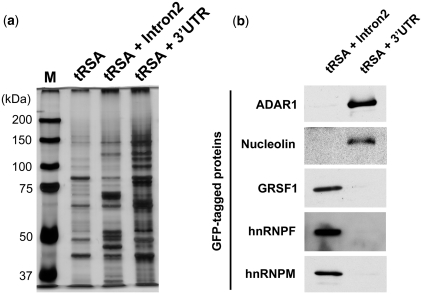
Figure 3.
Identification of specific RNA-interacting proteins by tRSA scaffold beads. (a) Pull-downs were performed from wild-type Caco-2 whole cell lysate, using Crb3 intron2 or 3′-UTR as baits. Captured proteins were analyzed by silver staining and unique bands were excised for analysis by mass spectrometry. (b) Proteins that associated specifically with either affinity matrix were GFP-tagged and tested for specific interaction with the tRSAs. Plasmids encoding GFP-tagged proteins were transfected into HEK293 cells, and pull-downs were performed from those lysates with tRSA-Intron2 or tRSA-3UTR synthetic RNA. About 1 mg total protein was applied for each pull-down. Proteins retained by the beads were analyzed by immunoblot using anti-GFP antibody.
Total cell lysates were lightly sonicated, and centrifuged for 10 min at 16 000_g_, 4°C. Supernatants were transferred to new tubes and protein was quantified by measuring absorbance at 280 nm. Egg white avidin (EMD chemicals, Cat#:189725, 10 µg/mg protein) and Yeast RNA (Sigma, Cat#R6750, ∼0.5 mg/mg protein) were added to block endogenous biotinylated proteins and non-specific RNPs, and incubated on a rotation shaker at 4°C, for 20 min. After blocking, the lysates were again centrifuged for 10 min at 16 000 g, 4°C, to prepare the final pre-cleared lysates. Supernatants were transferred to new tubes with 200 U/ml RNasin (Promega, cat#: N2115). In tandem with lysate preparation, 10 µg of synthetic RNAs were denatured at 65°C for 5 min then cooled to room temperature in the presence of 10 mM HEPES and 10 mM MgCl2. RNAs were applied to 30 µl streptavidin beads (Pierce, Cat#:20349). Streptavidin beads were washed 2× with lysis buffer before adding RNAs. The RNA tethering reaction was done in 300 µl lysis buffer including 200 U/ml RNasin, by incubating on a rotating shaker at 4°C for 20 min. Beads were then washed twice with fresh lysis buffer, and precleared lysates were applied to the RNA beads. After 1.5 h incubation, beads were washed ×5 with fresh lysis buffer. Captured proteins were analyzed by SDS–PAGE and immunoblotting.
In some experiments, 60 µl RNA beads were prepared by adding 30 µg synthetic RNAs for each samples. Subsequently, half were applied for RNA pull-down, and half were treated with 200 µl Trizol (Invitrogen) to recover retained RNAs without adding protein samples. About 7.5 µg of each RNA was recovered and 1.5 µg of these RNAs were analyzed by formaldehyde agarose gel electrophoresis.
For LC/MS analysis, Caco-2 cells (24 × 15 cm dish, approximately 108 cells) were maintained in DMEM supplemented with 10% FBS and penicillin–streptomycin at 37°C in 5% CO2. Cells were harvested after the cells reached confluence by adding 10 ml lysis buffer. After harvesting cells, the detergent concentration was corrected by adding 10% Triton ×-100. To prepare the RNA beads, 50 µg of RNAs (tRSA, tRSA + intron 2 and tRSA + 3′-UTR) were added on 100 µl bed volume Streptavidin beads. About 8 mg protein was applied for each pull-down. After precipitation, the final sample volume was adjusted to 100 µl by adding sample buffer. Samples (20 µl) were analyzed by SDS–PAGE followed by silver staining, and unique protein bands were cut for mass spectrometry analyses.
Mass spectrometry
Protein gel bands were excised and in-gel digested with trypsin (0.6 µg), and the tryptic peptides were subjected to matrix-assisted laser desorption-ionization mass spectrometry (MALDI-MS) on an Applied Biosystems 4700 Proteomic Analyzer® time of flight (TOFTOF®) mass spectrometer. Positive mode time of flight was used to identify peptides, and individual peptides were sequenced by MS/MS using collision-induced dissociation. All sequence and peptide fingerprint data was searched using the NCBI database and Mascot search engine.
RESULTS
tRSA tag improves the efficiency of RNA pull-down assay
As a starting point we employed the S1 SA for RNA affinity capture of RNPs. We attached the SA via a linker to the 5′-side of the bait RNA (15). However, this construct bound only weakly to the Streptavidin beads. To improve the efficiency, we tested two strategies (Figure 2a). First, we created tandemly repeated aptamers (6×SA), which we reasoned would increase both the affinity and specificity for the affinity matrix. Second, because aptamers are likely to be flexible and may adopt structures that do not recognize their target, we constructed tRNA-scaffolded aptamers.
To test the efficiency of these systems for the purification of RBPs, we employed the MS2 phage coat protein and its cognate RNA binding sequence (MBS). The MBS was attached to the 3′-ends of the aptamer constructs (see schematic in Figure 1a). Initially, chimeras were expressed in HEK293 cells together with GFP-tagged MS2, and lysates were incubated with Streptavidin beads. However, we were unable to capture any detectable GFP-MS2 under these conditions (data not shown). Therefore, we expressed the RNA chimeras in vitro, and attached them to beads prior to adding cell lysate (see schematic of method in Figure 1b). Single SA-, 6× SA-, and tRSA-tagged MBS were synthesized in vitro as bait RNAs (Figure 2a). Surprisingly, tRSA-tagged MBS captured the GFP-MS2 protein from cell lysates with 10-fold higher efficiency than did the 1× SA tag, while the 6× SA tag showed much lower efficiency than the single SA tag (Figure 2a). To evaluate how efficiently various RNAs were tethered, we treated the beads with Trizol to release the RNAs, and the extracts were analyzed by formaldehyde gel electrophoresis. Single SA and 6× SA tagged MBS RNAs were almost undetectable, whereas tRSA tagged RNAs showed clear bands (Figure 2a). We conclude that the tRNA scaffold significantly improves SA function. The failure of the 6× SA may be due to interference of one aptamer sequence with another, leading to misfolded structures that cannot recognize Streptavidin.
The tRSA tag is applicable to detect endogenously expressed RBP by RNA pull-down assay
To evaluate whether the tRSA system is applicable to the detection of endogenously expressed RBPs, we chose Fragile X Mental Retardation Protein (FMRP, also called FMR1), which interacts with a specific sequence motif, called the G quartet, through its RGG box. The G quartet was first identified as a FMRP binding RNA element by an in vitro selection strategy, and is frequently found in FMRP target mRNAs (16). We prepared tRSA-tagged G quartet RNA in vitro, and performed pull-down assays from HEK293 lysates. About 60 pmols each of synthetic RNAs were added to 30 µl bed volume SA beads to prepare the RNA beads. Lysate containing 2 mg protein was applied for each pull-down. Beads were washed 5× with lysis buffer, and analyzed by immunoblot. Notably, the G quartet affinity tag specifically captured endogenous FMRP, but did not bind to a different RBP, TLS/FUS (Figure 2b). These data demonstrate that the tRSA system can specifically capture endogenous RBPs. We further tested the tRSA system by comparison with a standard biotinylated-RNA pull-down assay (Figure 2c). Biotinylated tRSA, and a G quartet lacking the tRSA scaffold sequence were synthesized, and pull-down assays were performed from the same whole cell lysate as was used for the tRSA pull-downs. The tRSA sequence itself did not bind to FMRP with either method (Figure 2c). However, RNA tethering efficiency and FMRP pull-down efficiency were both higher with the tRSA-G quartet than with the biotinylated G-quartet (Figure 2c). The affinity of tRSA tag to beads was reduced by biotinylation (Figure 2c), possibly because biotinylation affects formation of secondary structures in RNAs. These data indicate that our tRSA system is able to capture RBPs with higher efficiency than a traditional biotinylated RNA pull-down method.
Identification of novel transcript-specific RNP by tRSA system
To test the ability of the tRSA system to identify novel, transcript-specific RBPs, we focused on the Crumbs3 (Crb3) gene. The Drosophila genome contains a single Crb gene that is essential for epithelial polarity, and its mRNA localizes to the apical surface (4). The 3′-UTR is required for Crb mRNA localization, but the zipcode within this region has not been mapped, and RBPs required for transport and anchoring of the transcript have not been identified (17).
We recently found that the mammalian Crb3 mRNA is also apically localized in epithelial cells (H. Iioka and I.G. Macara, unpublished data). To screen for RBPs that recognize specific elements within the Crb3 mRNA, we created tRSA chimeras with the 192-nt 3′-UTR and the 761-nt intron2 of the human Crb3 transcript. Empty tRSA RNA was used as a negative control. Affinity selection was performed using cell lysate from human intestinal epithelial Caco-2 cells. Lysate was pre-blocked with 10 µg white egg avidin/mg protein. RNA beads were prepared by coupling 50 µg of RNAs (tRSA, tRSA + intron2 or tRSA + 3′-UTR) to 100 µl streptavidin beads, and ∼8 mg of protein was applied for each pull-down.
Proteins retained on the beads were analyzed by silver staining, and the patterns were strikingly different for the Intron2 and 3′-UTR RNA tags as compared with the empty tRSA (Figure 3a). We identified the major bands by LC/MS, and almost all were RBPs (Table 1). Encouragingly, one of the bands specific to the 3′-UTR-tRSA pull-down was Staufen1, which is known as a regulator of mRNA localization and interacts with the 3′-UTRs of target mRNAs. In addition, hnRNPs and GRSF1, which may be involved in pre-mRNA processing, were specifically captured by Crb3 intron2 (Table 1).
Table 1.
Crb3 intron2 or 3′-UTR interacting proteins
| Protein name | Accession No. | No. of unique peptides |
|---|---|---|
| Crb3 intron2 interecting proteins | ||
| Heterogeneous nuclear ribonucleoprotein M | gi|14141154 | 34 |
| Heterogeneous nuclear ribonucleoprotein F | gi|4826760 | 28 |
| Heterogeneous nuclear ribonucleoprotein H1 | gi|48145673 | 19 |
| G-rich sequence factor 1 | gi|55977848 | 10 |
| Heterogeneous nuclear ribonucleoprotein A2/B1 | gi|4504447 | 8 |
| Small nuclear ribonucleoprotein polypeptide A | gi|4759156 | 7 |
| Crb3 3′-UTR interecting proteins | ||
| Polyribonucleotide nucleotidyltransferase 1 | gi|115502437 | 24 |
| Nucleolin | gi|55956788 | 23 |
| Eukaryotic translation initiation factor 2-alpha kinase 2 (PKR) | gi|4506103 | 22 |
| NOL1/NOP2/Sun domain family 2 protein | gi|39995082 | 19 |
| NSAP1 protein | gi|5031512 | 19 |
| Splicing factor proline/glutamine-rich | gi|29881667 | 17 |
| Staufen isoform a | gi|82659083 | 16 |
| IFI-4 (ADAR1) | gi|2326524 | 12 |
| BAIAP2 | gi|21619132 | 11 |
To validate the specificities of these RBPs, we constructed N-terminally GFP-tagged versions and expressed them transiently in HEK293 cells. Lysates from 10 cm dishes were divided between different RNA beads for affinity capture. Most of the GFP-proteins showed specific interaction with either the intron2 or 3′-UTR RNA tags (Figure 3b).
DISCUSSION
RNA aptamers are potentially ideal RNA tags for the analysis of RNP interactions. Yet in practice, aptamers have not worked well to isolate RNPs from cell lysates, especially for proteins that interact with mRNAs or pre-mRNA complexes (11). RNA folding is a critical factor not only for aptamer function but also for RNA–protein interactions. However, there is no generalized strategy to design bait RNAs that will fold correctly and maintain a stable conformation. Here we have shown the effectiveness of a scaffolding strategy to stabilize the aptamer RNA conformation. Using this approach, we successfully demonstrated the identification of novel RNA–RNP interactions using Crb3 mRNA. Strikingly, different sets of proteins bound to the 3′-UTR versus the second intron of this transcript.
We note that one candidate 3′-UTR binding protein interacted also with the tRSA negative control, even though there was no detectable interaction with the tRSA-Intron2. Such associations might arise if the protein participates in translation regulation and the translation complex interacts with the tRNA segment of the tRSA tag. For this reason, we recommend the use of another negative control rather than the empty tRSA tag.
Our tRSA pull-down is simple and effective, special reagents are not required, and it can be applied flexibly from small-scale functional analyses to high throughput screens. These advantages should make the isolation of RBPs almost as simple as protein immunoprecipitations.
FUNDING
Japanese Society for the Promotion of Science fellowship and grant (to H.I.); Human Frontier Science Program Grant (to I.G.M.); Excellent Young Researchers Overseas Visit Program (to H.I.); National Institutes of Health grant GM070902 (to I.G.M.). Funding for open access charge: National Institutes of Health (grant GM070902).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Stavroula Mili to providing plasmids (pcNMS2-GFP-NLS and β-globin-24bs/-). We also thank Prof. Hiroshi Sakamoto and members of the science department (especially Ms Yae Kamisako and Ms Miyako Fujiwara) in Kobe University for their continuous encouragement for H.I. through this study.
REFERENCES
- 1.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Prokipcak RD, Herrick DJ, Ross J. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J. Biol. Chem. 1994;269:9261–9269. [PubMed] [Google Scholar]
- 6.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta A, Driscoll DM. A sequence-specific RNA-binding protein complements apobec-1 to edit apolipoprotein B mRNA. Mol. Cell. Biol. 1998;18:4426–4432. doi: 10.1128/mcb.18.8.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Said N, Rieder R, Hurwitz R, Deckert J, Urlaub H, Vogel J. In vivo expression and purification of aptamer-tagged small RNA regulators. Nucleic Acids Res. 2009;37:e133. doi: 10.1093/nar/gkp719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Reed R. Purification of functional RNA-protein complexes using MS2-MBP. Curr. Protoc. Mol. Biol. 2003 doi: 10.1002/0471142727.mb2703s63. Chapter 27, Unit 27 23. [DOI] [PubMed] [Google Scholar]
- 10.Windbichler N, Schroeder R. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat. Protoc. 2006;1:637–640. doi: 10.1038/nprot.2006.95. [DOI] [PubMed] [Google Scholar]
- 11.Walker SC, Scott FH, Srisawat C, Engelke DR. RNA affinity tags for the rapid purification and investigation of RNAs and RNA-protein complexes. Methods Mol. Biol. 2008;488:23–40. doi: 10.1007/978-1-60327-475-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat. Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 14.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 15.Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Wang L, Hays TS, Cai Y. Dynein-mediated apical localization of crumbs transcripts is required for Crumbs activity in epithelial polarity. J. Cell. Biol. 2008;180:31–38. doi: 10.1083/jcb.200707007. [DOI] [PMC free article] [PubMed] [Google Scholar]