Shotgun proteomics aids discovery of novel protein-coding genes, alternative splicing, and “resurrected” pseudogenes in the mouse genome (original) (raw)
Abstract
Recent advances in proteomic mass spectrometry (MS) offer the chance to marry high-throughput peptide sequencing to transcript models, allowing the validation, refinement, and identification of new protein-coding loci. We present a novel pipeline that integrates highly sensitive and statistically robust peptide spectrum matching with genome-wide protein-coding predictions to perform large-scale gene validation and discovery in the mouse genome for the first time. In searching an excess of 10 million spectra, we have been able to validate 32%, 17%, and 7% of all protein-coding genes, exons, and splice boundaries, respectively. Moreover, we present strong evidence for the identification of multiple alternatively spliced translations from 53 genes and have uncovered 10 entirely novel protein-coding genes, which are not covered in any mouse annotation data sources. One such novel protein-coding gene is a fusion protein that spans the Ins2 and Igf2 loci to produce a transcript encoding the insulin II and the insulin-like growth factor 2–derived peptides. We also report nine processed pseudogenes that have unique peptide hits, demonstrating, for the first time, that they are not just transcribed but are translated and are therefore resurrected into new coding loci. This work not only highlights an important utility for MS data in genome annotation but also provides unique insights into the gene structure and propagation in the mouse genome. All these data have been subsequently used to improve the publicly available mouse annotation available in both the Vega and Ensembl genome browsers (http://vega.sanger.ac.uk).
The human genome sequence has been publicly available for 10 yr (Lander et al. 2001), but the exact protein-coding gene number is still under debate (Clamp et al. 2007). Automatic annotation systems such as Ensembl (Hubbard et al. 2002; Curwen et al. 2004) have been developed to generate gene sets by exploiting the power of integrating data from various sources, such as ab initio gene predictors (Kulp et al. 1996; Burge and Karlin 1997; Parra et al. 2000; Stanke and Waack 2003), comparative genomics (Roest Crollius et al. 2000; Korf et al. 2001; Miller 2001; Wiehe et al. 2001; Parra et al. 2003), and mapping of transcriptional (cDNA, EST) or translational evidence (protein sequence) to the DNA sequence (Gelfand et al. 1996; Birney and Durbin 1997).
However, manual annotation efforts, such as the Vertebrate Genome Annotation (VEGA) project (Ashurst et al. 2005; Wilming et al. 2008) or RefSeq (Pruitt et al. 2000; Pruitt and Maglott 2001), as well as quality assessment efforts (Guigo et al. 2006) still play a significant role in the validation and refinement of predicted gene models. The downside of manual investigation is that it its expensive and time consuming. Widespread use of DNA sequencing technologies will further accelerate the availability of new raw genomic sequences, all of which will require annotation.
New initiatives such as the International Mouse Phenotyping Consortium have the mammoth task of identifying the function of every mouse gene by gene knockout (Abbott 2010). However, the identification of protein-coding genes and the determination of their exact gene structure are not trivial tasks (Guigo et al. 2006). The recently completed clone-based assembly of the mouse strain C57BL/6J was reported to have 20,210 protein-coding genes, which was over 1000 more than human genes predicted at that time (Church et al. 2009). A high-throughput method, providing orthogonal data for validation and confirmation that accentuate the protein-coding potential, is required to complement these annotation efforts. A data source ideally suited for this purpose can be obtained from proteomics data in the form of peptides that can serve as translational evidence. State of the art tandem mass spectrometry (MS/MS) is the method of choice to identify peptides and proteins with high sensitivity and specificity in a high-throughput manner (Domon and Aebersold 2006). Efforts to combine genome annotation with protein MS led to the establishment of a new field, “proteogenomics,” a term first coined by Jaffe et al. (2004),which has subsequently been applied to other model organisms such as Drosophila melanogaster (Brunner et al. 2007; Tress et al. 2008) and Arabidopsis thaliana (Castellana et al. 2008).
In order to effectively use proteomic data for genome annotation, it is essential that peptide identification methods and significance measures are both sensitive and accurate. We have previously evaluated (Brosch et al. 2008) the standard database search engine Mascot (Perkins et al. 1999) and extended it with an improved semi-supervised machine learning algorithm, Percolator (Käll et al. 2007), to develop Mascot Percolator, which provides highly accurate significance measures and results in much improved sensitivity (Brosch et al. 2009). Moreover, Percolator provides two significance measures, the _q_-value (Storey and Tibshirani 2003; Käll et al. 2008a; 2008b) and the posterior error probability (PEP) (Käll et al. 2008b; 2008c). The former is an advanced notion of the false discovery rate (FDR) (Benjamini and Hochberg 1995; Chi et al. 2007) used in large-scale transcriptomics and proteomics studies as a standard metric to report the expected rate of wrong identifications among all accepted identifications. However, we believe that for genome annotation purposes, where the significance of each individual peptide should be known, the _q_-value or FDR as a global measure should be complemented with a peptide-level significance measure such as the PEP or the peptide probability as available through PeptideProphet (Keller et al. 2002; Ding et al. 2008).
MS data can be searched directly against a six-frame translation of the genome with the purpose of validating and refining existing gene annotation as well as the identification of novel genes (Yates et al. 1995; Choudhary et al. 2001; Kuster et al. 2001). However, searching a six-frame translation in higher eukaryotes is problematic; e.g., only 1%–2% of the human genome encodes for proteins (Claverie 2005; Birney et al. 2007), and therefore, most of the search space consists of translated noncoding sequence. The inflated search space increases the likelihood of false-positive identifications, and therefore, sensitivity decreases at a constant FDR. Moreover, this method does not account for splicing, which affects the majority of genes (Wang et al. 2008); nor does it account for the 20%–28% of tryptic peptides, depending on the number of allowed missed cleavages, whose coding regions span a splice site. The use of ab initio gene prediction algorithms, such as Augustus (Stanke and Waack 2003) or GeneID (Parra et al. 2000), offer a potential solution to this problem, since these algorithms report complete gene structures. A compact representation of the predicted proteome that removes redundancy of alternatively spliced transcripts can be achieved by an in silico digestion of the proteome into a peptide centric database that can be filtered and indexed to remove redundancy from alternatively spliced variants (Martens et al. 2005). Alternative approaches, such as the use of an exon splice graph database, have been developed to limit the search space (Tanner et al. 2007).
In this work, we build upon these efforts and apply a two-stage search strategy, aiming to validate and refine mouse genome annotation and to identify novel loci based on experimental translational evidence. First, MS data obtained from the PeptideAtlas project (Desiere et al. 2006) and data sets generated in-house were searched against a peptide-centric nonredundant superset of Ensembl, Vega, and IPI (Kersey et al. 2004) proteins. We expect that these databases comprise most of the proteome, and peptide identification sensitivity is maintained at a high level due to this limited search space. In a second stage, we incorporated protein predictions from Augustus that significantly inflate search space but enable refinement of existing gene annotations and the identification of novel protein-coding loci.
Results
Generation of high-confidence PSMs for genome annotation
We analyzed 10.5 million tandem mass spectra (downloaded from PeptideAtlas and from in-house experiments) using the genome annotation pipeline (Fig. 1; Methods). In total, 1,491,410 and 1,772,159 peptides were identified at a _q_-value (a more advanced notion of the FDR) of 1% and 5%, respectively. Application of a maximum allowed probability (PEP) of 1% and 5% of an individual peptide match to be incorrect reduced the number of identified peptides to 1,124,724 and 1,358,323, corresponding to a _q_-value of less than 0.14% and 0.59%, respectively.
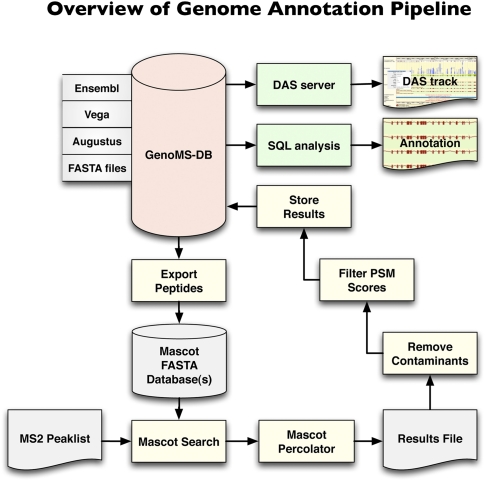
Figure 1.
Genome annotation pipeline. The database at the core of the system, GenoMS-DB, is built by integrating all peptides that are derived from an in silico digestion of available data sources (Ensembl, Vega, Augustus). Each peptide derived from these data sources is associated with its genomic locus and context (such as gene, transcript, exon, or splice site information). Peptides from FASTA protein databases can optionally be integrated but would lack genome mapping. A set of nonredundant in silico digested peptides is exported from GenoMS-DB to create the Mascot search database. Tandem MS spectra are searched with Mascot and post-processed with Mascot Percolator to derive accurate probabilities on a per PSM basis. A series of steps removes common contaminant sequences and low-scoring PSMs from the results, prior to storing the remaining identifications into the GenoMS-DB database. This integration of peptide-genome mapping together with peptide identifications enables streamlined analysis with standard SQL or visualization as a track in a genome browser via a DAS feature server. This is a flexible pipeline where alternative gene prediction tools could be used to provide source peptides, and alternative search engines and probability assessment algorithms could be integrated.
When data were searched against the database that was supplemented with the Augustus predictions (see Methods), 16% fewer identifications (1,253,074 and 1,490,020 at a _q_-value of 1% and 5%) were made due to the search space inflation of almost one order of magnitude (Fig. 2B). At a maximum PEP of 1% and 5%, we identified 967,131 and 1,171,060 peptides corresponding to _q_-values of 0.12% and 0.57%, respectively. It is interesting to note that Augustus predictions comprised 81% of all Ensembl peptides. This suggests good sensitivity for an ab initio gene predictor, but it should be noted that this is afforded by parameter settings that are tweaked to allow maximum sensitivity (see Methods).
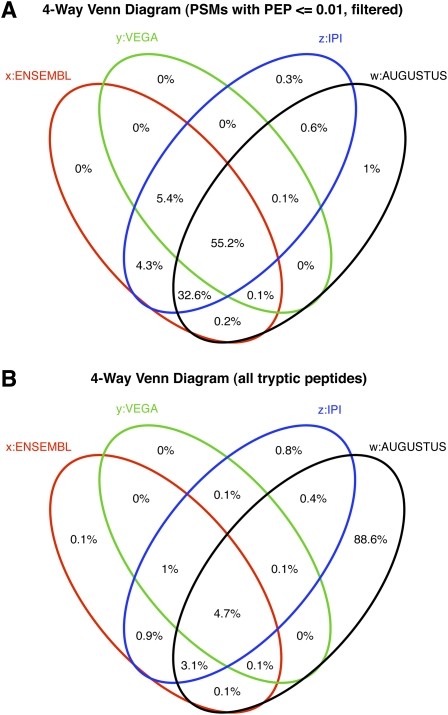
Figure 2.
Four-way Venn diagram showing distribution of origin of all identified peptides (A) and of all candidate peptides in the search database (B).
For subsequent analyses, only the best PEP and _q_-value score for each peptide sequence were considered, resulting in 95,606 distinct peptide identifications, 3260 of which matched common contaminants. Since isobaric amino acids, such as leucine/isoleucine as well as lysine/glutamine, cannot be discriminated in low energy collision induced dissociation data (Roepstorff and Fohlman 1984; Biemann 1988), all isoforms attributed to any of these residues were filtered out (1159 cases). Of the remaining peptides, 83% (76,029) mapped unambiguously to one genomic locus. Since only fully tryptic peptides were considered, it was further tested whether a semi-tryptic form of the peptide sequence mapped elsewhere in the genome (758 cases). As a last measure, the possibility that peptides with one residue substitution, insertion, or deletion could be identified elsewhere in the genome was tested, since coding SNPs were not considered in this study (6685 cases, mainly short peptide identifications). The total of 68,586 remaining distinct peptides formed the basis for subsequent genome annotation. However, PSMs with a PEP between 1%–5% were exclusively used as supplementing peptides, and only peptide identifications with a PEP of 1% or better (58,574 cases) were used as a primary annotation data source; meaning that the chance of a wrong peptide identification would be 1% in the worst-case scenario, which corresponds to a FDR of less than 0.14%. Most proteogenomics research studies to date have used a FDR of 1%–5% (Castellana et al. 2008; Tress et al. 2008), but we have adopted this conservative approach to avoid the propagation of erroneous identifications into genome annotation pipelines.
Validation of Ensembl/Vega gene annotation
We found that 98.1% of all identified peptides (PEP of 1% or better) matched the Ensembl/Vega database with only 1.9% attributed solely to IPI and Augustus (Fig. 2A). This is despite 88.6% of the candidate tryptic peptides in the search database originating solely from Augustus predictions (Fig. 2B). We therefore focus first on confirming Ensembl/Vega annotation at the level of gene translation and structure.
Verification of gene translation
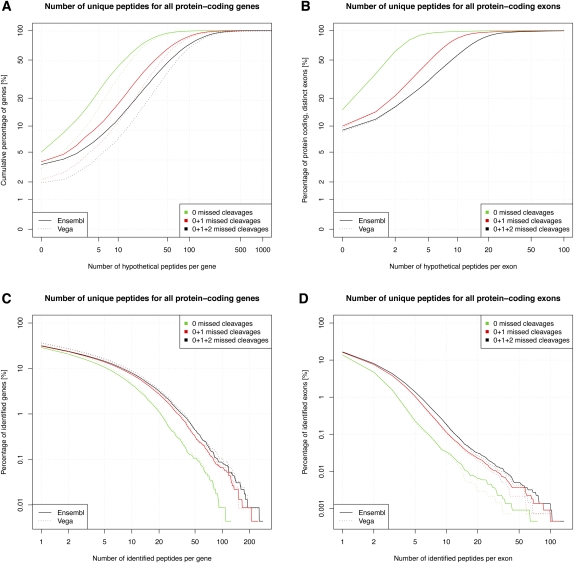
Figure 3 shows the cumulative percentage of genes that could be validated theoretically by tryptic peptides that map uniquely to a genomic locus and comprise between eight and 30 amino acids (Supplemental Fig. 1). Note that these are the default peptide parameters for all theoretical considerations in the remainder of this article. Interestingly, when zero, one, and two missed cleavages were allowed, only 5.0%, 3.8%, and 3.5% of protein-coding Ensembl gene products lack tryptic peptides, respectively. However, a large proportion of transcripts contain only a few tryptic peptides; e.g., 43.0% of transcripts comprise fewer than 10 peptides (no missed cleavages allowed), thereby potentially limiting the chances of validation.
Figure 3.
(A) Cumulative gene identification rate as a function of the number of potential identifiable (hypothetical) peptides per protein-coding gene. (B) As before, but analysis for protein-coding exons. Note that considered peptides were fully tryptic, ranged from eight to 30 residues and were unique to a genomic locus. (C) Inverse cumulative validation rate of all protein-coding genes as a function of the number of peptides identified per gene. (D) As before, but for protein-coding exons.
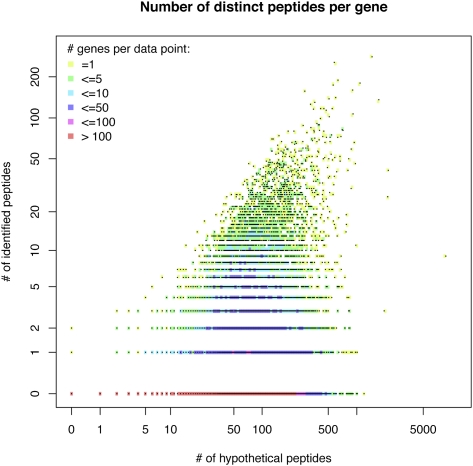
We report translational validation of 7221 (4463) protein-coding Ensembl (Vega) genes, corresponding to 31.6% (36.7%) of all protein-coding genes. However, peptide coverage was limited, with only 7.9% (9.0%) of the genes being validated by more than 10 peptides and 0.08% (0.09%) by more than 100 peptides (Fig. 3C). In order to further study the relationship between identified and potentially identifiable peptides, we tested whether a linear model could be fitted (Fig. 4). A perfect fit would mean that the MS instrument would sample more peptides from gene products with more potential peptides. However, we found that there is no correlation (_R_2 = 0.10), and this is consistent with studies that show that peptide sampling is mainly determined by relative protein abundance (Ishihama et al. 2005; Lu et al. 2007). Furthermore, genes that are only expressed in specific tissues would not be identified if the tissue of interest was not analyzed. For example, obscurin (ENSMUSG00000061462) is among the top 10 genes with most potentially identifiable peptides (1192), and yet none of the peptides were identified (see also http://tinyurl.com/Obscurin). In contrast, plectin (ENSMUSG00000022565), a cytoskeletal protein that is more widely expressed, has a similar number of potential peptides (1447) but has the highest number of identified peptides in this study (280).
Figure 4.
Correlation analysis between the number of identified peptides and the number of potential identifiable peptides per gene. Since many data points have the same _x_–_y_-values, the number of overlaying data points (genes) is encoded with the color gradient available from the legend.
It is important to note that the consideration of missed cleavages makes a significant difference to this analysis; even though trypsin is a very specific enzyme, it is not always 100% efficient. In fact, 31.7% of all peptides identified in this study had one missed cleavage site, 9.9% had two missed, and only 58.4% had none. Therefore more than 90% of the peptides have none or one missed tryptic cleavage site.
Gene structure validation
Theoretical calculations, using the same peptide properties as described previously, revealed that, when zero, one, or two missed cleavages are allowed, 15.1%, 10.0%, and 9.0% of all Ensembl protein-coding exons do not contain detectable peptides, respectively. In addition, 93.6%, 47.8%, and 30.4% of the protein-coding Ensembl exons contained five or fewer peptides (Fig. 3C). The lower peptide coverage of exons compared with complete genes can be explained by the fact that the average protein-coding exon count per gene in mouse is around 9.7. Nevertheless, a total of 16.7% of the total 222,378 Ensembl protein-coding exons could be validated by peptide identifications. We validated 8.0% and 1.4% of Ensembl exons by at least two and five peptides (Fig. 3D).
A more difficult challenge is to validate annotation of introns, since this requires a fully tryptic, and unique, peptide spanning splice boundaries. Defining splice donor and acceptor sites is not trivial, and a peptide spanning these sites not only validates them but also implicitly validates the joined exons and thereby significantly contributes to gene structure validation. Of the 202,205 (131,336) introns in Ensembl (Vega) that span a protein-coding splice boundary, up to 70.9% and 86.2% could theoretically be confirmed by peptides, allowing for one or two missed cleavages, respectively. However, when zero missed cleavages are considered, the theoretical validation rate drops to 46%. Using the subset of identified peptides that span a splice site, a total of 14,426 (9347) Ensembl (Vega) introns could be confirmed, corresponding to 7.1% of all splice sites that join protein-coding exons, 1.3% of which were validated with two or more distinct peptides.
Clearly, the translational evidence is valuable for independent gene structure validation. Up to 91.0% of all protein-coding exons and 86.2% of all introns could theoretically be confirmed with peptides obtained in typical proteomics experiments. Applying the peptides identified in this study, 16.7% of all exons and 7.1% of all introns could be confirmed, highlighting that with relatively moderate efforts a significant proportion of gene structures can be validated.
Validating evidence for alternative translation
Until recently, only limited evidence of expression of alternatively spliced transcripts was available at the protein level (Tress et al. 2008). The detection of these variants by standard MS proteomics experiments is hindered by the fact that the majority of protein sequence is shared between the variant transcripts, differing only in small parts of the translation products. Validation of alternative translation requires identification of at least one “signature” peptide for each protein isoform. While 8877 (40%) protein-coding Ensembl genes code for alternative products, only 16,664 transcripts from 1542 genes could theoretically be discriminated by 168,726 “signature” peptides. For example, Catenin (cadherin associated protein), delta-1 (ENSMUSG00000034101), has 25 alternative transcripts annotated as coding, but only nine “signature” peptides could theoretically distinguish the alternative translation of three protein isoforms.
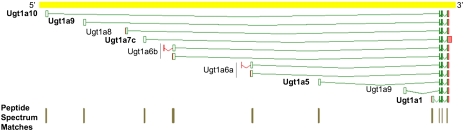
Nevertheless, protein evidence for alternatively translated genes from tryptic digests was shown recently; Tanner et al. (2007) found evidence for 16 human genes, Castellana et al. (2008) found evidence for 47 A. thaliana genes, and Tress et al. (2008) identified 130 D. melanogaster genes that express at least two protein isoforms. Here, a total of 370 peptides enabled discrimination of 112 Ensembl transcripts in 53 genes, corresponding to 3.4% of all protein-coding genes with annotated multiple protein-coding isoforms that can be discriminated by a peptide. The UDP-glucuronosyltransferase family, polypeptide A6A (ENSMUSG00000054545), which has 12 alternative coding transcripts within one locus, is unusual as all variants have an alternative 5′ exon spliced to a common set of downstream constant exons. These variable first exons confer diverse functional mRNAs with different, tissue specific expression profiles (Zhang et al. 2004). Figure 5 shows the overview of this complex locus with evidence for expression of five alternative protein isoforms from 27 “signature” peptides. Other examples with evidence for three alternative gene products include the following: ankyrin 2 brain isoform 2 (ENSMUSG00000032826), synaptotagminVII (ENSMUSG00000024743), and H2A histone family member Y (ENSMUSG00000015937). Two alternative isoforms were validated for each of the remaining 49 genes.
Figure 5.
MS PSMs confirm the protein-coding potential of five alternatively translated products of the UDP-glucuronosyltransferase 1 family, polypeptide A6 (highlighted in bold). Ambiguous PSMs are shown for the two alternatively spliced transcripts of the Ugt1a6a and Ugt1a6b genes, respectively; and as clusters for each of the 3′ exons.
Furthermore, we have identified an additional 31 novel alternative splice isoforms of known mouse genes based on single high-stringency peptides (Supplemental Table 1). Of these, 29 putatively represent splice variants, of which eight are variants with extensions to the N terminus of the CDS, resulting from the use of an ATG upstream of that currently annotated, and one utilizes a novel termination codon. Both remaining transcripts represent translated upstream open reading frames (upORFs) lying in the 5′ UTR sequence of annotated splice variants. Although each of these alternatively spliced isoforms is supported by only one MS peptide, validation is shown as 23 of the 31 supporting peptides were recorded in more than one data set and none were recorded once in only one data set (Supplemental Table 2). However, these putative objects have been tagged for the addition of experimental validation before they become persistent annotations included in the Ensembl/Vega gene sets.
Manual identification of protein-coding novel loci and alternative splice variants
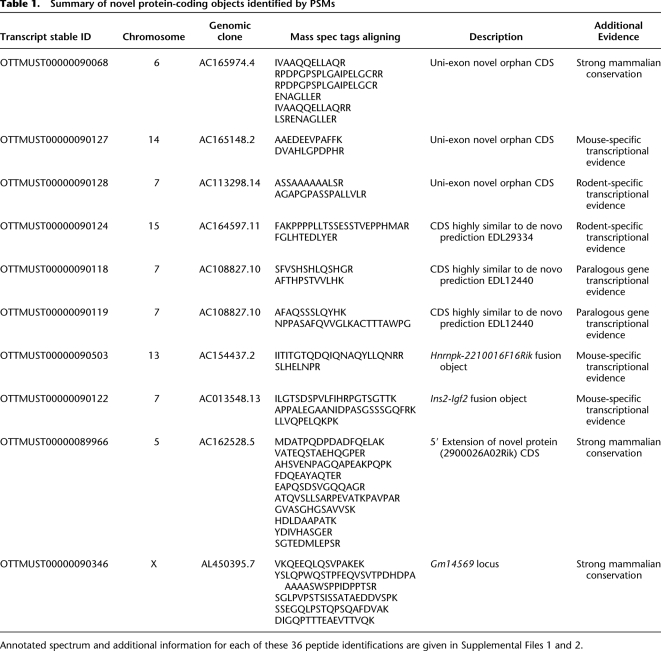
We used the gene finding algorithm Augustus to over-predict protein-coding genes on the genome ab initio and to populate the search database (see Methods). Assuming that the Ensembl gene list is close to complete, the Augustus database contains 90% previously unannotated sequence (Fig. 2B). Therefore, reliable and stringent peptide scoring, together with subsequent filtering to exclude ambiguous matches, is crucial to minimize any false-positive identifications. To reiterate, the least significant peptide match considered in this study had a 1% probability to be incorrect, corresponding to a FDR of less than 0.14%. For subsequent analysis, where peptides were not supported by any existing annotation, this was further constrained in that at least two peptides (one of which with a PEP of less than 0.01; the second, less than 0.05) had to be identified prior to investigation. We found that 1.9% of all peptide identifications matched neither Ensembl nor Vega but were present in either the IPI database or Augustus gene predictions. These peptides represent a significant number of identifications that contribute to refinements of gene structure or annotation of novel genes (Fig. 2A). Using this approach, 36 MS PSMs were identified that provide clear support for the translation of 10 novel protein-coding loci. Transcripts at each of these loci were manually curated using current HAVANA annotation guidelines (http://www.sanger.ac.uk/research/projects/vertebrategenome/havana/assets/guidelines.pdf), and the details of such (and the additional supporting evidence for each) are given in Table 1. These 10 loci fall into four categories.
Table 1.
Summary of novel protein-coding objects identified by PSMs
Single-exon loci
Three of these 10 novel objects have a single exon. Such objects are known to exist within mammalian genomes and, as in these cases, are often difficult to identify due to a paucity of nonsplicing transcriptional support. Paralogous gene family members are often used as annotation aids for such genes (e.g., defensin genes) (Amid et al. 2009). However, orphan genes, which lack sufficient paralogous evidence to be annotated as protein-coding loci, remain a significant annotation problem (J. Mudge, pers. comm.). MS proteomic support is of obvious benefit in the annotation of these genes.
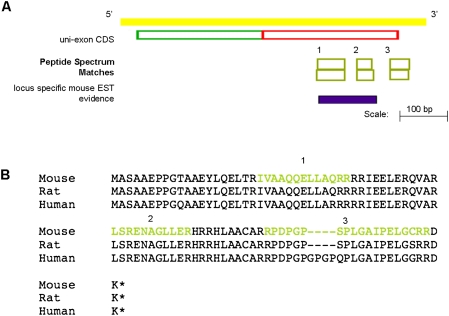
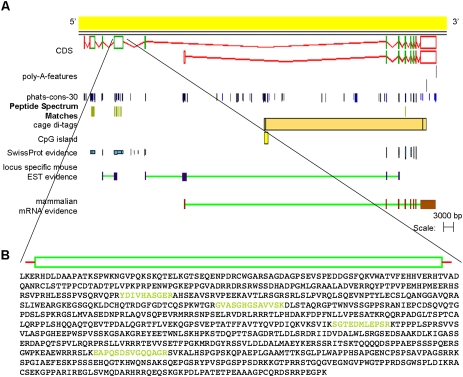
Figure 6A shows one such example of a single-exon protein-coding transcript (OTTMUST00000090068). Interestingly, the CDS region of this model is well conserved across a number of mammalian genomes (Fig. 6B), although it has no identifiable protein domains. This conservation allows us to speculate that this locus is likely to be functional. Further validation for the translation of this novel gene is found in the tissues that the supporting MS PSMs were detected. Each of the six peptides that validate the protein-coding potential of this locus was detected from brain tissue data sets (data not shown). In addition, three of these six matches were recorded from within the same data set. This correlation in tissue expression adds validation to our claim that this locus represents a novel translated gene from the mouse genome.
Figure 6.
(A) Species-specific EST and peptide evidence supports the annotation of a protein-coding object at this locus. (B) Clustal alignment of the polypeptide sequences of this novel single-exon object with the orthologous objects from the rat and human genomes, respectively. There is no transcriptional support for either the rat or human model; however, the CDS frame is highly conserved and intact. The residues of the mouse translation that are covered by PSMs are colored yellow.
Confirmation of ab initio predicted protein-coding loci
Three of our 10 novel protein-coding objects confirm the annotation of ab initio coding gene predictions. None of these genes are supported by species-specific transcriptional support; however, the proteomics data support their annotation. Each of these three transcripts is supported by two MS peptides, with expression of each detected in the liver (OTTMUST00000090124), liver and brain (OTTMUST00000090118), and brain (OTTMUST00000090119), respectively. The putative translations from each of these loci contain no detectable domains. However, each peptide supporting each transcript was recorded at least five times in two different data sets, adding considerable validation to each model. Again, these gene transcripts will be experimentally validated to test their transcription and confirm gene models prior to their being made available as part of the Vega data set.
Protein-coding fusion loci
An increasing number of locus-spanning transcripts have been identified in EST and cDNA sequence databases in recent years (Gingeras 2009). The advent of new sequencing technologies has expanded this transcript category in terms of both numbers and transcript complexity (Ruan et al. 2007; Gingeras 2009). However, there remains considerable uncertainty over the function of such transcripts, and very little evidence for their ability to encode stable proteins exists (Gingeras 2009).
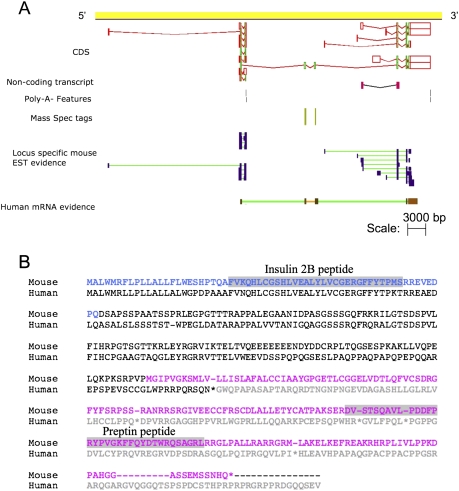
Two of our 10 novel protein-coding objects support the annotation of coding splice variants that contain exons from more than one coding locus. Object OTTMUST00000090122 is evidence of a protein-coding fusion transcript linking the mouse Igf2 (insulin-like growth factor 2) and Ins2 (insulin II) loci (Fig. 7A). Our proteomic data support the annotation of this read-through transcript as a coding variant. Comparative genomic analysis of the orthologous human loci shows the existence of an equivalent fusion transcript. However, the human transcript (OTTHUMT00000026061) is likely to be a target for the nonsense-mediated decay (NMD) pathway due to the presence of a premature STOP codon in the putatively translated sequence (Fig. 7B). This STOP is absent from the mouse fusion transcript, allowing this object to be annotated as fully protein-coding. The PSMs that support the annotation of both of these fusion loci were detected in multiple tissues (data not shown). Therefore, although the _Ins2_-Igf2 and _Hnrnpk_-2210016F16Rik fusion transcripts are supported by only two and three peptides, respectively, the fact that each were detected multiple times across a number of tissues gives supporting validation to the annotation of these protein-coding fusion transcripts.
Figure 7.
(A) Mouse Ins2-Igf2 fusion object contains a valid CDS, supported by human cDNA and species-specific peptide evidence. (B) Clustal alignment of the translation of the mouse and human fusion transcripts. This human translation would be a target for the NMD pathway due to a frame-shift mutation, caused by the inclusion of an additional exon not present in the mouse transcript. The residues of the Ins2 and Igf2 polypeptides are colored blue and pink; with the known domains within each highlighted in gray.
Interestingly, the Igf2 and Ins2 genes are functionally linked and are co-regulated (Buchanan et al. 2001). Ins2 codes for a protein that is processed to give rise to two active peptides (insulin 2A and 2B) that form heterodimers that regulate blood glucose concentration. Igf2 also codes for a protein that is processed to give rise to an active peptide, preptin, which is an insulin-like growth factor (Buchanan et al. 2001). Preptin is co-secreted with insulin and is regulated by glucose levels and in turn stimulates further insulin secretion, thereby amplifying the effect of increased glucose levels (Buchanan et al. 2001). The _Ins2_-Igf2 fusion protein that we have validated contains the insulin 2B and preptin but not the insulin 2A peptide. Whether this fusion protein is functional and can actually be processed to produce insulin 2B and preptin remains to be determined, but it raises the possibility of an ultimate form of co-regulation of these two products expressed in a single protein precursor.
Merging of annotated noncoding transcript objects
Incomplete transcriptional support can lead to protein-coding genes being annotated as fragments (i.e., as multiple noncoding transcripts due to uncertainty over the complete gene model and/or frame of translation). In such cases, the addition of proteomic data is of obvious benefit. Two of our 10 novel protein-coding objects support the merging of previously annotated transcripts. An example of such a locus in the mouse genome is that of Gm14569 (Supplemental Fig. 2). This locus was represented as two noncontiguous noncoding transcripts based on the available species-specific EST support. Our filtered MS data aligned five peptides to this locus that, in conjunction with the annotated human locus (KIAA1210, OTTHUMT00000058020), provide sufficient support to build a full-length protein-coding gene model. A second example is that of model OTTMUST0000008966 (Fig. 8). We have typed this object as a novel protein-coding locus as, although the proteomic evidence lies close to model OTTMUST00000063646, our data allow the annotation of an object which more than doubles the CDS region of this locus.
Figure 8.
(A) MS PSMs allow the annotation of OTTMUST00000089966, for which there is no full-length transcriptional support. (B) Focus on the 2664-bp exon of this transcript. Exons of this length are uncommon and are problematic for manual annotation. Translation of this exon is shown, with the positions of four PSMs that cover this exon highlighted in yellow.
Interestingly, both of these newly annotated objects contain a large (>2 kb) coding exon. Such exons are notoriously difficult for transcript-based computational or manual annotation methods to identify. However our proteomic data have been of considerable benefit where limitations of transcriptional evidence are found.
Resurrected pseudogenes
While some duplicated pseudogenes have been shown to gain novel function despite a loss of protein-coding potential (e.g., XIST) (Duret et al. 2006); retrotransposed or processed pseudogenes have generally been considered as “dead on arrival”; i.e., they lose functional activity as a corollary of their creation (Zheng et al. 2007). While there is increasing evidence for the transcription of retrotransposed pseudogenes (Khachane and Harrison 2009), there is very little evidence for gain of novel function at the transcript level. Mouse makorin (Hirotsune et al. 2003; Kaneko et al. 2006) was identified as encoding a transcript that regulated the expression of the parent locus in trans (however these findings have been contradicted by Gray et al. 2006), and a NOS pseudogene performs a similar function in the snail Lymnaea stagnalis (Korneev et al. 1999). At the level of protein-coding function, retrotransposition has been identified as a minor but important mechanism for the creation of novel combinations of functional domains (Babushok et al. 2007) and has also been identified as having the potential to create protein-coding loci de novo (Kaessmann et al. 2009). However, while it has been estimated that more than 100 human protein-coding loci might have arisen by this route (Vinckenbosch et al. 2006), very few retrotransposed loci have evidence of function at the protein level; notable exceptions include human GLUD2 (Shashidharan et al. 1994) and Drosophila Prat (Malmanche et al. 2003). While the increasing number of transcribed retrotransposed genes creates additional candidate protein-coding loci (Baertsch et al. 2008), there is no evidence that proteins originate from such loci.
Our MS data provide support for the translation of nine processed pseudogenes in the reference mouse genome. Each pseudogene is supported by at least two peptides, and all aligning PSMs are locus specific, showing exact similarity to only one translated locus of the mouse genome. More specifically, each PSM shows at least two amino acid substitutions compared with the translated parent protein sequence. As an additional validation step, each supporting PSM for each translated pseudogene needed to be detected in at least two different tissue data sets. Great care was taken in assigning parents to each of these pseudogenes to ensure high confidence that these MS PSMs do indeed represent translations of these pseudogenic loci and not polymorphisms of the parent locus; therefore, we require that:
- The residues substituted in our PSMs in comparison with the parent polypeptide are conserved in the amino acid sequences of the 1:1 rat and human orthologs; and
- There is no evidence of single nucleotide polymorphism/deletion-insertion polymorphism (SNP/DIP) at these codon positions of the parent mouse locus within the available sequence data of 44 mouse strains undergoing genome sequencing (http://www.ensembl.org/Mus_musculus).
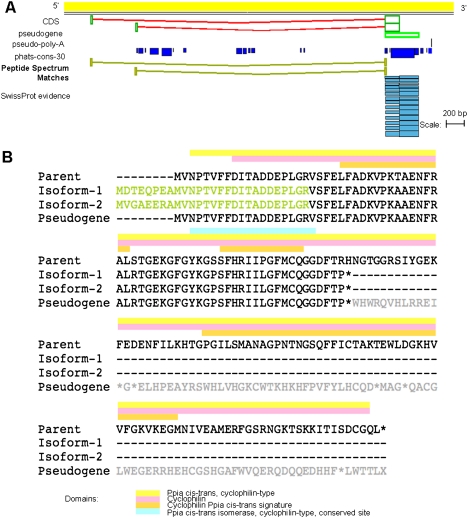
Figure 9 shows one example where proteomic data have allowed the annotation of two protein-coding variants of a mouse peptidylprolyl isomerase A (Ppia) pseudogene (OTTMUST00000018507). Both peptides aligning to this locus identify exons 5′ to the main body of this pseudogene object. Each PSM commences with a methionine residue, suggesting that we have captured the translational start sites of both of these CDS objects within our proteomic data set. Furthermore, both peptides possess canonical sites in splicing to the main body of this Ppia processed pseudogene. The translated sequence of these 5′ exons of both CDS objects could not be found within 300 kb upstream of the 5′ end of the parental Ppia locus (OTTMUSG00000000783), confirming that they do not represent unannotated splice variants of the parent locus.
Figure 9.
(A) Two canonically splicing MS PSMs support the annotation of coding isoforms of a mouse Ppia processed pseudogene locus (OTTMUST00000018507). (B) Clustal alignment of parent PPIA protein (SWISS-PROT P17742), translations of both coding isoforms of the mouse Ppia processed pseudogene, and the putative translation of the pseudogene object. Residues of the coding isoforms that are covered by MS evidence are highlighted yellow. Residues of the parent polypeptide that are part of known domains are shown by colored boxes above the alignment.
The Ppia parent gene of this translated pseudogene is known to be involved in the acceleration in the folding of proteins (Colgan et al. 2000). As to be expected from this ubiquitous role, expression of this gene has been detected in a number of diverse tissues, including the kidney, lung, heart, and 11-d-old embryo (http://www.informatics.jax.org/searches/estclone_report.cgi?_Marker_key=12618&sort=Tissue). The MS PSMs supporting both isoforms of this translated pseudogene were both recorded in these same tissues. In addition, we have been able to confirm both of these translated pseudogene isoforms by RT-PCR in using template extracted from 11-d-old embryo (Supplemental Text 2). Homozygous “knock-out” mutation analysis of the parent Ppia gene has been associated with a variety of effects to the cardiovascular, endocrine, hematopoietic, immune, renal, and optic systems (http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=alleleDetail&key=33820). We have added the translated Ppia pseudogene to our internal pipeline for similar “knock-out” analysis in order to investigate the potential function of this locus.
Interestingly, of these nine translated pseudogenes, only two show a syntentic ortholog in the reference rat genome, and none possess human orthologs. This is in spite of the genes surrounding each translated mouse pseudogene showing strong syntentic conservation with the equivalent rat and human genomic loci (data not shown). This suggests that seven of these pseudogenes have arisen since the divergence of the rat and mouse lineages some 25–30 Myr ago (Church et al. 2009). We propose two hypotheses to explain the detection of translated polypeptide sequences from these nine mouse loci:
- The polypeptides detected from these pseudogenic loci are simply relics of translation being generated until the locus has accrued sufficient mutations that allow all translations generated to be targets for the NMD pathway.
- Of all retrotransposition events that change a genome, only a fraction will insert at a position that is permissive of translation. It is likely that of these processed pseudogenes, again only a fraction result in translations that provide a selective advantage to the organism and are therefore positively selected for across generations, and it is therefore translation from such loci that we have detected.
It is unlikely that only one of these hypotheses are able to fully explain the translated polypeptide sequences that we have detected from all nine pseudogenic loci. We plan to investigate these translations further through knock-out mutation analysis.
Discussion
We have described the construction of a novel genome annotation pipeline for tandem MS data, which provides highly sensitive and accurate peptide identification, efficient peptide-genome mapping, and automated data analysis for gene structure validation and correction. We have evaluated the implications and limitations of this approach and have shown that, theoretically, peptide evidence could validate up to 97% of all protein-coding genes, 91% of all protein-coding exons, and 86% of all protein-coding exon–exon junctions of the reference mouse genome if all tryptic peptides could be detected. However, the mouse proteome is far from being saturated by MS-based peptide identifications. Even if every organ with all its regions, cell types, and organelles could be isolated and analyzed, there would probably be a significant set of genes that would be missed because expression occurs only under specific and transient cellular and developmental conditions. There have not been systematic analyses at these levels of complexity, but if we compare studies from 10 yr ago with today, it is clear that MS data have become a richer and more valuable resource for genome annotation. By applying our proteogenomics pipeline, we report the first systematic validation of the mouse genome annotation with tandem MS data. Analysis of a collection of 10 million tandem MS spectra, available from the PeptideAtlas and our own data, provides translational evidence for a third of all protein-coding genes, over a sixth of all exons and in excess of 14,000 splice boundaries. In addition, we have uncovered strong evidence for 53 genes with alternative translations.
Moreover, using ab initio gene predictions to populate the search database, our approach can also be used for refining and discovery of new genes. We highlight the value of proteogenomics to refine gene structures in reporting experimental validation for the translation of nine processed pseudogenes from the reference mouse genome. Although locus-specific evidence of transcription is available for a number of mouse pseudogenes, these nine loci are the first examples of putatively translated pseudogenes from the mouse genome. It remains unclear whether these loci are able to produce functional proteins, and further experimental validation is required to identify their functional roles. In addition, we have been able to identify 10 novel protein-coding loci at high confidence. Interestingly, although orthologous protein-coding loci for eight of these 10 can be found in the reference human genome, these mouse loci were not identified by either the RefSeq or Ensembl annotation pipelines. Instead, the identification of these loci has been mediated only by the application of our proteomic data and manual investigation.
Overall, we demonstrate that translational evidence in the form of proteomic data, available through tandem MS, could significantly enhance genome validation and annotation efforts. Coverage of proteogenomics data is set to increase as continually improving methods and instrumentation allow for deeper proteome sequencing, offering the validation and discovery of more genes and splice isoforms.
Methods
MS/MS data
This study is based on 10,465,149 tandem MS spectra, where 729,583 spectra were obtained from in-house experiments on nuclear protein extracts of murine embryonic stem cells and murine brain membrane fractions. These data sets have been submitted to the PRIDE database with accession numbers 15297, 15298, and 15299. We obtained 9,735,566 spectra from the PeptideAtlas project (Mus musculus, Feb. 2009 data snapshot, http://www.peptideatlas.org/repository/). Data were not associated with any publication records, but associated metadata show the sampling of various tissues of the mouse such as the brain, liver, lung, heart, kidney, testes, and placenta (Supplemental Table 2).
GenoMS-DB database construction
Details of search database construction are given in Figure 1 and Supplemental Text 1. Briefly, gene products from Ensembl, Vega, and IPI were digested in silico, stored into the GenoMS-DB database, and exported for Mascot analysis. The Ensembl Perl API was utilized to capture the peptide-genome mapping into the database during this process to enable subsequent analysis. Additionally, gene product predictions from Augustus predictions were similarly processed through the database to provide a second Mascot database for use in this two-stage search strategy.
Data processing and database searching with Mascot and Mascot Percolator
In-house LTQ-FT- and LTQ-FT Ultra (Thermo Fisher Scientific)–generated MS raw data files were processed to peak lists with BioWorks (version 3.2 and 3.3; Thermo Fisher Scientific). Processing parameters were identical to those used by Brosch et al. (2009).
All MS peaklist data (in-house and PeptideAltas) were searched with Mascot and post-processed with Mascot Percolator (1.09, default settings) using Percolator version 1.12. For this, each peaklist file was searched against both target and decoy databases using an enzyme setting that is compatible with the custom-made peptide-centric search databases; therefore, the artificial amino acid J was introduced under the Mascot config file that defines the amino acid masses. J was set to a mass that does not correspond to a naturally occurring amino acid, e.g., 300 Da. The enzyme was set to cut at the N- and C-terminal of the peptide, thereby only fully tryptic peptides that were separated by “J” were searched with Mascot.
Distributed Annotation System
Using the Perl-based Proserver (Finn et al. 2007), a Distributed Annotation System (Dowell et al. 2001) (DAS) feature server was implemented that allows the identified peptides stored in the GenoMS-DB database (Fig. 1) to be visualized as tracks in various genome browsers and curation tools. Meta-information for each peptide is provided in the form of, not exhaustively, scoring statistics (_q_-value, log transformed PEP value), uniqueness of the peptide within the genome, experiment, Mascot search log ID, etc. The uniqueness, together with the PEP value, is color-coded, so it is very easy to visually validate whether a peptide is unique to a genomic location and is also significant. The DAS data source can be accessed at http://www.sanger.ac.uk/research/publications/supp-info/ms-data/.
Manual annotation
MS PSMs overlapping annotated loci were annotated based on current HAVANA annotation guidelines (http://www.sanger.ac.uk/research/projects/vertebrategenome/havana/assets/guidelines.pdf). For loci unsupported by existing annotation, an annotation hierarchy of RT-PCR > species-specific transcriptional support > rodent specific transcriptional support > strong mammalian conservation > paralogous gene transcriptional evidence was used for aiding annotation (Table 1).
Translated pseudogenes
The parent of each mouse translated pseudogene identified was selected in a two-step process. First, parents were assigned by homology scoring of the putative translation of the processed pseudogene object against the SWISS-PROT data set using the current HAVANA annotation guidelines. Second, as an additional check, each of the PSMs aligning to the pseudogenic loci were individually assigned to a parent mouse protein by aligning to the complete UniProt database using the hidden Markov model–based program HMMER. In all cases, the same parent mouse protein was assigned to each of our translated pseudogene loci using both strategies.
Genes orthologous to these parent mouse proteins were identified using the orthologous gene identifying application of the Ensembl website (http://www.ensembl.org/Mus_musculus/Gene/Compara_Ortholog?g=). Parent proteins were aligned with the putative translations of the translated pseudogene loci using the online ClustalW2 application available at the website of the European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/Tools/clustalw2/index.html). The identification of domains within these translations and parent proteins was aided by the InterProScan application, another tool of the EBI website (http://www.ebi.ac.uk/Tools/InterProScan/).
Acknowledgments
We thank Eric Deutsch and coworkers for the mgf data export of the PeptideAtlas data and Mario Stanke for help and support with Augustus. We further thank Michael Tress and Laurens Wilming for comments on the manuscript, Felix Kokocinski and Jonathan Warren for help with the DAS annotation server setup, James Gilbert for the compilation of the NMD mouse transcripts, and the Ensembl and Havana team for their ongoing support. This work was funded by the Wellcome Trust. D.J.A. is funded by Cancer Research-UK.
Footnotes
References
- Abbott A 2010. Mouse project to find each gene's role: International Mouse Phenotyping Consortium launches with a massive funding commitment. Nature 465: 410 doi: 10.1038/465410a [DOI] [PubMed] [Google Scholar]
- Amid C, Rehaume LM, Brown KL, Gilbert JG, Dougan G, Hancock RE, Harrow JL 2009. Manual annotation and analysis of the defensin gene cluster in the C57BL/6J mouse reference genome. BMC Genomics 10: 606 doi: 10.1186/1471-2164-10-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashurst J, Chen C, Gilbert J, Jekosch K, Keenan S, Meidl P, Searle S, Stalker J, Storey R, Trevanion S, et al. 2005. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res 33: D459–D465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Ostertag EM, Kazazian HH Jr 2007. Current topics in genome evolution: Molecular mechanisms of new gene formation. Cell Mol Life Sci 64: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J 2008. Retrocopy contributions to the evolution of the human genome. BMC Genomics 9: 466 doi: 10.1186/1471-2164-9-466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Biemann K 1988. Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom 16: 99–111 [DOI] [PubMed] [Google Scholar]
- Birney E, Durbin R 1997. Dynamite: A flexible code generating language for dynamic programming methods used in sequence comparison. Proc Int Conf Intell Syst Mol Biol 5: 56–64 [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos J, Dutta A, Guigo R, Gingeras T, Margulies E, Weng Z, Snyder M, Dermitzakis E, Thurman R, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch M, Swamy S, Hubbard T, Choudhary J 2008. Comparison of Mascot and X!Tandem performance for low and high accuracy mass spectrometry and the development of an adjusted Mascot threshold. Mol Cell Proteomics 7: 962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch M, Yu L, Hubbard T, Choudhary J 2009. Accurate and sensitive peptide identification with mascot percolator. J Proteome Res 8: 3176–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Ahrens C, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch E, Panse C, de Lichtenberg U, Rinner O, et al. 2007. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol 25: 576–583 [DOI] [PubMed] [Google Scholar]
- Buchanan C, Phillips A, Cooper G 2001. Preptin derived from proinsulin-like growth factor II (proIGF-II) is secreted from pancreatic islet β-cells and enhances insulin secretion. J Biochem 360: 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Castellana N, Payne S, Shen Z, Stanke M, Bafna V, Briggs S 2008. Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A, Bai D, Geer L, Shabanowitz J, Hunt D 2007. Analysis of intact proteins on a chromatographic time scale by electron transfer dissociation tandem mass spectrometry. Int J Mass Spectrom 259: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary J, Blackstock W, Creasy D, Cottrell J 2001. Matching peptide mass spectra to EST and genomic DNA databases. Trends Biotechnol 19: S17–S22 [DOI] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin M, Kellis M, Lindblad-Toh K, Lander E 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J 2005. Fewer genes, more noncoding RNA. Science 309: 1529–1530 [DOI] [PubMed] [Google Scholar]
- Colgan J, Asmal M, Luban J 2000. Isolation, characterization and targeted disruption of mouse ppia: Cyclophilin A is not essential for mammalian cell viability. Genomics 68: 167–178 [DOI] [PubMed] [Google Scholar]
- Curwen V, Eyras E, Andrews T, Clarke L, Mongin E, Searle S, Clamp M 2004. The Ensembl automatic gene annotation system. Genome Res 14: 942–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiere F, Deutsch E, King N, Nesvizhskii A, Mallick P, Eng J, Chen S, Eddes J, Loevenich S, Aebersold R 2006. The PeptideAtlas project. Nucleic Acids Res 34: D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Choi H, Nesvizhskii A 2008. Adaptive discriminant function analysis and reranking of MS/MS database search results for improved peptide identification in shotgun proteomics. J Proteome Res 7: 4878–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon B, Aebersold R 2006. Mass spectrometry and protein analysis. Science 312: 212–217 [DOI] [PubMed] [Google Scholar]
- Dowell R, Jokerst R, Day A, Eddy S, Stein L 2001. The distributed annotation system. BMC Bioinformatics 2: 7 doi: 10.1186/1471-2105-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Finn R, Stalker J, Jackson D, Kulesha E, Clements J, Pettett R 2007. ProServer: A simple, extensible Perl DAS server. Bioinformatics 23: 1568–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand M, Mironov A, Pevzner P 1996. Gene recognition via spliced sequence alignment. Proc Natl Acad Sci 93: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR 2009. Implications of chimaeric non-co-linear transcripts. Nature 461: 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Wilson A, Fortin PJ, Nicholls RD 2006. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proc Natl Acad Sci 103: 12039–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigo R, Flicek P, Abril J, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic V, Birney E, et al. 2006. EGASP: The human ENCODE Genome Annotation Assessment Project. Genome Biol 7: S2.1–S2.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91–96 [DOI] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. 2002. The Ensembl genome database project. Nucleic Acids Res 30: 38–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Jaffe J, Berg H, Church G 2004. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4: 59–77 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M 2009. RNA-based gene duplication: Mechanistic and evolutionary insights. Nat Rev Genet 10: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Canterbury J, Weston J, Noble W, MacCoss M 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4: 923–925 [DOI] [PubMed] [Google Scholar]
- Käll L, Storey J, MacCoss M, Noble W 2008a. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 7: 29–34 [DOI] [PubMed] [Google Scholar]
- Käll L, Storey J, MacCoss M, Noble W 2008b. Posterior error probabilities and false discovery rates: Two sides of the same coin. J Proteome Res 7: 40–44 [DOI] [PubMed] [Google Scholar]
- Käll L, Storey J, Noble W 2008c. Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics 24: i42–i48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Aki I, Tsuda K, Mekada K, Moriwaki K, Takahata N, Satta Y 2006. Origin and evolution of processed pseudogenes that stabilize functional Makorin1 mRNAs in mice, primates and other mammals. Genetics 172: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii A, Kolker E, Aebersold R 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kersey P, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R 2004. The International Protein Index: An integrated database for proteomics experiments. Proteomics 4: 1985–1988 [DOI] [PubMed] [Google Scholar]
- Khachane AN, Harrison PM 2009. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC Genomics 10: 435 doi: 10.1186/1471-2164-10-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Flicek P, Duan D, Brent M 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: S140–S148 [DOI] [PubMed] [Google Scholar]
- Korneev SA, Park JH, O'Shea M 1999. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci 19: 7711–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese M, Eeckman F 1996. A generalized hidden Markov model for the recognition of human genes in DNA. Proc Int Conf Intell Syst Mol Biol 4: 134–142 [PubMed] [Google Scholar]
- Kuster B, Mortensen P, Andersen J, Mann M 2001. Mass spectrometry allows direct identification of proteins in large genomes. Proteomics 1: 641–650 [DOI] [PubMed] [Google Scholar]
- Lander E, Linton L, Birren B, Nusbaum C, Zody M, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lu B, Ruse C, Xu T, Park S, Yates JR 2007. Automatic validation of phosphopeptide identifications from tandem mass spectra. Anal Chem 79: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanche N, Drapeau D, Cafferty P, Ji Y, Clark DV 2003. The PRAT purine synthesis gene duplication in Drosophila melanogaster and Drosophila virilis is associated with a retrotransposition event and diversification of expression patterns. J Mol Evol 56: 630–642 [DOI] [PubMed] [Google Scholar]
- Martens L, Vandekerckhove J, Gevaert K 2005. DBToolkit: Processing protein databases for peptide-centric proteomics. Bioinformatics 21: 3584–3585 [DOI] [PubMed] [Google Scholar]
- Miller W 2001. Comparison of genomic DNA sequences: Solved and unsolved problems. Bioinformatics 17: 391–397 [DOI] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigo R 2000. GeneID in Drosophila. Genome Res 10: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Agarwal P, Abril J, Wiehe T, Fickett J, Guigo R 2003. Comparative gene prediction in human and mouse. Genome Res 13: 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D, Pappin D, Creasy D, Cottrell J 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pruitt K, Maglott D 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res 29: 137–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Katz K, Sicotte H, Maglott D 2000. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet 16: 44–47 [DOI] [PubMed] [Google Scholar]
- Roepstorff P, Fohlman J 1984. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom 11: 601. [DOI] [PubMed] [Google Scholar]
- Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fischer C, Fizames C, Wincker P, Brottier P, Quetier F, et al. 2000. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat Genet 25: 235–238 [DOI] [PubMed] [Google Scholar]
- Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, et al. 2007. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs). Genome Res 17: 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidharan P, Michaelidis TM, Robakis NK, Kresovali A, Papamatheakis J, Plaitakis A 1994. Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J Biol Chem 269: 16971–16976 [PubMed] [Google Scholar]
- Stanke M, Waack S 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics-Oxford 19: 215–225 [DOI] [PubMed] [Google Scholar]
- Storey J, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S, Shen Z, Ng J, Florea L, Guigo R, Briggs S, Bafna V 2007. Improving gene annotation using peptide mass spectrometry. Genome Res 17: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress M, Bodenmiller B, Aebersold R, Valencia A 2008. Proteomics studies confirm the presence of alternative protein isoforms on a large scale. Genome Biol 9: R162 doi: 10.1186/gb-2008-9-11-r162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci 103: 3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore S, Schroth G, Burge C 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe T, Gebauer-Jung S, Mitchell-Olds T, Guigo R 2001. SGP-1: Prediction and validation of homologous genes based on sequence alignments. Genome Res 11: 1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming L, Gilbert J, Howe K, Trevanion S, Hubbard T, Harrow J 2008. The vertebrate genome annotation (Vega) database. Nucleic Acids Res 36: D753–D760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Eng J, McCormack A 1995. Mining genomes: Correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem 67: 3202–3210 [DOI] [PubMed] [Google Scholar]
- Zhang T, Haws P, Wu Q 2004. Multiple variable first exons: A mechanism for cell- and tissue-specific gene regulation. Genome Res 14: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, Lu Y, Denoeud F, Antonarakis SE, Snyder M, et al. 2007. Pseudogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Res 17: 839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]