Smac Mimetics Activate the E3 Ligase Activity of cIAP1 Protein by Promoting RING Domain Dimerization (original) (raw)
Abstract
The inhibitor of apoptosis (IAP) proteins are important ubiquitin E3 ligases that regulate cell survival and oncogenesis. The cIAP1 and cIAP2 paralogs bear three N-terminal baculoviral IAP repeat (BIR) domains and a C-terminal E3 ligase RING domain. IAP antagonist compounds, also known as Smac mimetics, bind the BIR domains of IAPs and trigger rapid RING-dependent autoubiquitylation, but the mechanism is unknown. We show that RING dimerization is essential for the E3 ligase activity of cIAP1 and cIAP2 because monomeric RING mutants could not interact with the ubiquitin-charged E2 enzyme and were resistant to Smac mimetic-induced autoubiquitylation. Unexpectedly, the BIR domains inhibited cIAP1 RING dimerization, and cIAP1 existed predominantly as an inactive monomer. However, addition of either mono- or bivalent Smac mimetics relieved this inhibition, thereby allowing dimer formation and promoting E3 ligase activation. In contrast, the cIAP2 dimer was more stable, had higher intrinsic E3 ligase activity, and was not highly activated by Smac mimetics. These results explain how Smac mimetics promote rapid destruction of cIAP1 and suggest mechanisms for activating cIAP1 in other pathways.
Keywords: Apoptosis, Protein-Protein Interactions, Signal Transduction, Ubiquitin Ligase, Ubiquitylation, Dimerization, IAP Antagonist Compounds, RING Domain, cIAP
Introduction
The attachment of ubiquitin and ubiquitin-like molecules to intracellular proteins is a key process that regulates many cellular events (1). A hierarchical multienzyme cascade brings about the attachment of ubiquitin to substrate proteins. Ubiquitin is first activated by the ubiquitin-activating enzyme (E1) and then is transferred to the active site cysteine of the ubiquitin-conjugating enzyme (E2). Subsequently, ubiquitin-protein ligases (E3s) promote the transfer of ubiquitin from the E2 to lysine residues in target proteins. The RING4 domain-containing E3 ligases, which are prevalent in mammals (>300), do not directly interact with ubiquitin; instead, the RING domain binds to the E2 and promotes the transfer of ubiquitin from the E2∼ubiquitin (E2∼Ub) thioester conjugate to the target protein (2).
The mechanism by which the RING domain promotes ubiquitin transfer is not obvious because the RING-binding site on the E2 is distant from the active site. It has been proposed that the RING domain simply brings the E2∼Ub conjugate into close proximity with the substrate and that the increased availability of E2s promotes transfer of ubiquitin to the substrate. However, interactions between RING domains and E2s are generally transient and have modest affinity, and tighter binding does not always correlate with increased activity. In addition, not all RING-E2 complexes promote transfer, demonstrating that proximity alone is insufficient (3). Notably, the RING domains of Brca1 and c-Cbl interact with UbcH7 and UbcH5b, but only interaction with UbcH5b results in ubiquitin transfer (3, 4). Others have suggested that an allosteric mechanism is important whereby interaction of the RING domain with the E2 leads to changes at its active site that promote release of ubiquitin (5, 6). Consistent with this, binding of the G2BR domain of the E3 gp78 induces allosteric changes in the E2 Ube2g2 that enhance ubiquitin loading (7).
Regulation of E3 ligase activity is important for cellular function, and different strategies have been adopted by various E3 ligases (2). For example, covalent modification of substrate proteins can regulate their interaction with E3 ligases so that they are appropriately targeted for ubiquitylation (8). In other cases, dimerization promotes the activity of RING E3 ligases (9, 10). However, it is unclear how dimerization of the RING domain increases activity because even when the non-E3 ligase RING domains from Bard1, mouse double minute X (MDMX), and Bmi1 form heterodimers with their active partners they promote E3 ligase activity (10–13). This indicates that RING dimerization does not increase ubiquitin transfer by simply increasing E2 availability but that dimerization stimulates the E3 ligase activity of the active partner.
Inhibitor of apoptosis (IAP) proteins are single subunit RING E3s that modulate apoptosis signaling pathways (14, 15). Some IAPs, such as X-linked IAP, can directly inhibit caspases and prevent proteolytic cleavage, whereas cIAP1 and cIAP2 are recruited to tumor necrosis factor (TNF) receptor complexes (16) and modulate the receptor-mediated apoptotic pathways that lead to NF-κB activation (17, 18). The C-terminal RING domains of cIAP1 and cIAP2 are critical (19, 20) with the RING domain required for dimerization, substrate ubiquitylation, and autoubiquitylation (21–23). In addition to the RING domain, all IAPs have N-terminal baculoviral IAP repeat (BIR) domains that can bind to other proteins, such as the IAP antagonist Smac/DIABLO (second mitochondrion-derived activator of caspase or direct IAP-binding protein with low pl) (24, 25). Some IAPs also contain a ubiquitin-associated (UBA) domain that binds to Lys-63-linked ubiquitin chains (26) and a less well characterized caspase recruitment domain (CARD) (see Fig. 1A).
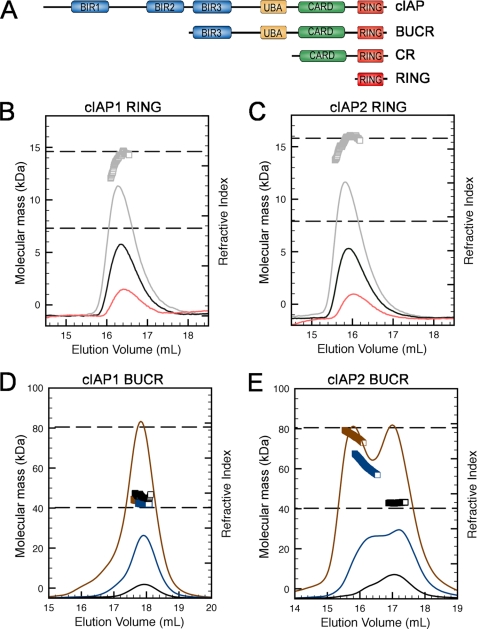
FIGURE 1.
N-terminal domains of cIAPs inhibit dimerization. A, schematic showing the domain organization of cIAPs and the constructs used in this study. Residues included in the human cIAP1 BUCR and RING constructs used were 266–618 and 553–618, respectively. The cIAP2 construct included BUCR (residues 255–604), CARD-RING (CR) (residues 435–604), and RING (residues 535–604). B, samples of cIAP1 RING at 10 (pink), 25 (black), and 50 μm (gray) were separated on an S75 column connected in line with a MALLS detector. The refractive index trace for each sample is shown, and the calculated mass is shown (gray squares) for the 50 μm sample. The dashed lines indicate the expected masses of monomeric and dimeric species. C, equivalent samples of cIAP2 RING were analyzed and displayed as in B. D, samples of cIAP1 BUCR at 25 (black), 100 (blue), and 300 μm (brown) were separated on an S200 column connected in line with a MALLS detector. The refractive index trace and the calculated mass are shown for each sample (color matches the trace). The dashed lines indicate the expected masses of monomeric and dimeric species. E, equivalent samples of cIAP2 BUCR were analyzed and displayed as in D.
IAP gene amplification and consequent increased expression of IAPs have been observed in several tumor types (27). This prompted the development of IAP antagonist compounds, which are commonly referred to as Smac mimetics (SMs). Like Smac/DIABLO, these compounds bind to the second and third BIR domains of a number of IAPs (28). However, because XIAP is the only bona fide caspase inhibitor (29), it was expected that cell death induced by these compounds would be a direct result of XIAP inhibition. Unexpectedly, however, SM compounds promoted rapid autoubiquitylation and proteasomal degradation of the cIAPs, which sensitized cells to TNF receptor-induced cell death in a variety of tumor types (17, 18). Several studies have now established that a key role of cIAPs is to regulate NF-κB signaling (30). Notably, cIAP-mediated ubiquitylation of RIPK1 has been shown to promote cancer cell survival, and loss of cIAPs due to addition of SMs leads to diminished ubiquitylation and stabilization of RIPK1, caspase-8 activation, and apoptotic cell death (31). Control of the E3 ligase activity of cIAPs is therefore critical for their antiapoptotic function, yet the mechanism by which their E3 ligase activity is stimulated and how SM compounds modulate it remain uncertain.
Our previous structural studies showed that the RING domain of cIAP2 exists as a dimer and that the dimer bound directly to the E2 UbcH5b (20). To understand how IAP antagonists promote autoubiquitylation of cIAPs, we focused on identifying the essential features of the RING domain. By utilizing monomeric mutant proteins, we show that dimerization is required for E3 ligase activity because only dimeric cIAP RING domains could promote release of ubiquitin from the E2∼Ub conjugate. Our data also show that together the N-terminal BIR3, UBA, and CARD domains inhibited RING dimerization and that interaction of the monovalent and bivalent IAP antagonist compounds with the BIR3 domain overcame this. Thus, RING dimerization is the key regulatory trigger exploited by SM compounds to promote autoubiquitylation and degradation of cIAPs.
EXPERIMENTAL PROCEDURES
Plasmids and Mutagenesis
Various constructs of human cIAP1 and cIAP2 (GenBank accession numbers U37547 and U37546) and relevant mutants were cloned into pGEX-6p3 and confirmed by sequencing. Purified proteins have five additional N-terminal residues, GPLGS, as a result of cloning. The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate mutants. Ubiquitin was expressed in pQE80L with an N-terminal His6 fusion, and both UbcH5b and the C85S mutant were expressed as glutathione _S_-transferase (GST) fusion proteins. Inducible pF 5x UAS SV40 Puro mouse cIAP1 WT and mouse cIAP1 F610A (dimer tail mutant (DT)) cloned into pF 5x UAS SV40 Puro have been described previously (20). Mouse cIAP1 E2 binding mutants V567A/D570A (E2-m1) and L593A/I598A (E2-m2) and the cIAP1 dimerization interface mutant V576E (DI-m2) were generated by overlap PCR mutagenesis, cloned into the pF 5x UAS SV40 Puro vector using standard techniques, and confirmed by sequencing. The complete sequence of all constructs can be obtained upon request.
Transfections, Antibodies, and Reagents
Transient transfections typically using 1 μg of plasmid DNA/10-cm plate of cells were performed with EffecteneTM according to the manufacturer's instructions (Qiagen). Antibodies were sourced as follows: monoclonal anti-β-actin (Sigma), monoclonal anti-cIAP1 (1:500) (Alexis), monoclonal anti-FLAG (1:2000) (Sigma), monoclonal anti-monomeric Bcl-w (mbw) (1:1000) (in house), monoclonal anti-ubiquitin (Cell Signaling Technology), and polyclonal anti-cIAP2 (in house). MG132 (Boston Biochem) was used at 1 μm, and 4-hydroxytamoxifen (Sigma) was used at 5 nm (MEFs). Smac mimetic compounds (TetraLogic Pharmaceuticals) were used at 500 nm (bivalent Comps. A and F) or 1 μm monovalent (Comps. C, D, E, G, and H). All SM compounds are shown in supplemental Fig. 3. All reagents were used according to the manufacturer's protocols.
Protein Expression and Purification
All cIAP1 and cIAP2 proteins were expressed as GST fusions in Escherichia coli BL21(DE3). Protein expression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside, and cultures were incubated at 18 °C overnight. The recovered cell pellet was sonicated in lysis buffer (PBS, pH 7.4 containing 2 mm DTT), and the soluble GST fusion protein was bound to glutathione-Sepharose. Resin-bound GST-cIAP proteins were then washed and treated with PreScission protease (Amersham Biosciences). The soluble fraction was purified using either a Sephadex S75 column or a Superdex S200 column equilibrated in PBS. Fractions that contained purified protein were combined, concentrated, and quantified. Ubc5Hb was expressed in E. coli at 18 °C and purified from clarified lysate by affinity chromatography and size exclusion in a similar manner.
Multiple Angle Laser Light Scattering (MALLS) Analysis
Concentrated protein that had been fully reduced by addition of DTT was analyzed by MALLS (Wyatt Technology) when coupled to a Superdex 200 HR 10/30 column (GE Healthcare) equilibrated in 1× PBS, pH 7.4. Data were analyzed using ASTRA V (Wyatt Technology). SM compounds were added to protein samples at 2× the molar concentration of binding sites and left on ice for 30 min prior to their separation on the S200 column.
Ubiquitylation Assays
For ubiquitylation assays, ∼5 μm soluble cIAP proteins, which had been mixed with a 1.25 molar excess (based on the number of binding sites) of the indicated compounds for 15 min, was mixed with 7.5 μm UbcH5b and 100 nm E1 in 20 mm Tris, pH 7.5, 50 mm NaCl, 50 μm His-tagged ubiquitin, 5 mm ATP, 2 mm MgCl2, 2 mm DTT. Samples were incubated at 37 °C for the indicated times, and then following addition of 2× SDS-PAGE sample buffer, the reactions were resolved by SDS-PAGE on 4–12% BisTris gels and stained using Coomassie Blue. Stock solutions (10 mm) of bivalent Comp. A, its enantiomer Comp. B, and monovalent Comp. C were prepared and then diluted in water as required.
Conjugate Preparation and Discharge Assays
A stable E2∼Ub conjugate was prepared using UbcH5b with a Cys to Ser mutation at the active site (C85S) and either native ubiquitin or ubiquitin that had an N-terminal His tag. To prepare the conjugate, E2, ubiquitin, E1, ATP, creatine phosphate, and phosphocreatine were incubated at 37 °C until conjugate formation was complete (typically 6–10 h). Ubiquitin-charged C85S-UbcH5b (referred to as E2∼Ub conjugate) was stable and could be purified at pH 5 using an S75 column. To measure discharge, purified IAP proteins (E3) were added to the purified conjugate in 50 mm MES, pH 6.5, and the disappearance of the parent band and appearance of the ubiquitin and E2 bands were used to assess the efficiency of ubiquitin release.
Pulldowns
Maltose-binding protein (MBP) fusion proteins were used to directly measure interaction between cIAP2 CARD-RING proteins and the E2∼Ub conjugate. Soluble E2∼Ub conjugate and MBP-CARD-RING amylose resin-immobilized proteins were mixed for 60 min at 4 °C in PBS buffer containing 0.2% Tween 20 and 2 mm DTT and then washed with PBS containing 0.2% Triton X-100 and 2 mm DTT prior to addition of 2× SDS-PAGE sample loading buffer. Samples were then separated by 14.5% SDS-PAGE, and proteins were detected by immunoblotting and detection with anti-His antibodies.
Circular Dichroism (CD) Spectroscopy
The far-UV CD spectra of cIAP1 BIR3-UBA-CARD-RING (BUCR) were recorded between 195 and 260 nm using an Olis DCM-10 CD spectrophotometer. The measurements were taken in 20 mm NaP, 100 mm NaCl, pH 7.4 at a protein concentration of 12.5 μm in the absence of compounds or the presence of either Comp. A or Comp. C (final concentration of 25 μm) in a 1-mm cuvette at 20 °C. SM compounds were dissolved in methanol as 10 mm stocks because DMSO is known to interfere with CD measurements below 200 nm. The final methanol concentration was 5% (v/v). Measurements of buffer blank samples containing 5% (v/v) methanol were recorded and subtracted from the spectra of the compound alone or protein plus compound.
Cell Culture and Lentivirus Production
All cell lines were maintained in DMEM supplemented with 10% FCS, 2 mm l-glutamine, and penicillin/streptomycin and grown at 37 °C in 10% CO2. To generate lentiviral particles, 293T cells were transfected with packaging constructs pCMV ΔR8.2, pVSVg, and the relevant lentiviral plasmid in the ratio of 1:0.4:0.6. After 24 h, the virus-containing supernatants were harvested and filtered (0.8 μm). Polybrene was added (12 μg/ml), and target cells were infected with virus supernatant for 24 h. The medium was subsequently changed, and successful infection was selected for with puromycin (5 μg/ml; pF 5xUAS selection) or hygromycin B (300 μg/ml; GEV16 selection). pF 5x UAS inducible constructs were induced with 5 nm 4-hydroxytamoxifen for 16 h prior to harvesting lysates for Western blotting.
Generation of MEFs
Generation of MEFs and lentiviral particles has been described recently (18, 32). Briefly, MEFs were generated from embryos in accordance with standard procedures and were infected with SV40 large T antigen-expressing lentivirus. In vivo double knock-out cIAP1 and cIAP2 MEFs were obtained from LoxP/LoxP cIAP1 and FRT/FRT cIAP2 mice crossed first with a Cre transgenic mouse followed by a FlpE transgenic mouse. Double knock-out MEFs were generated at E10 instead of E15. In vivo cIAP1 MEFs were obtained from a LoxP/LoxP cIAP1 mouse crossed with a Cre transgenic mouse, generated at E15, and infected with SV40 large T antigen-expressing lentivirus.
Western Blotting and Immunoprecipitations
Samples were lysed in Death inducing signaling complex (DISC) lysis buffer containing 1% Triton X-100 supplemented with protease inhibitor mixture (Roche Applied Science) and _N_-ethylmaleimide on ice for 30 min and clarified by centrifugation. Samples were separated on precast 4–20% polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes for antibody detection. All membrane blocking steps and antibody dilutions were performed with 5% skim milk in PBS containing 0.1% Tween 20 (PTBS), and washing steps were performed with PTBS. Proteins on membranes were visualized using ECL (Amersham Biosciences) following incubation of membranes with HRP-coupled secondary antibodies.
RESULTS
Multidomain cIAP Proteins Form Less Stable Dimers than RING Domain Alone
Our previous studies showed that the RING domain of cIAP2 forms a dimer in solution, and the crystal structure revealed that dimerization was dependent on contacts between residues in the core of the RING domain as well as an N-terminal helix and a C-terminal extended strand, both of which flank the RING domain (20). Analysis of the RING domain from cIAP1 using multiple angle laser light scattering coupled in-line to size exclusion chromatography (MALLS-SEC) showed that it was also dimeric with a measured mass of 13.9 kDa (theoretical mass of the dimer is 14.6 kDa) (Fig. 1B). Both the cIAP1 and cIAP2 RING domain dimers appeared to be quite stable because samples at concentrations as low as 10 μm had an identical elution time (Fig. 1, B and C), and the average mass suggested that the proteins were dimeric, although at low protein concentrations the average mass measurement is not as accurate as at higher protein concentrations. However, when purifying longer cIAP2 proteins that contained domains in addition to the RING, the elution point varied depending on the concentration of the sample applied to the column, and often we observed two peaks suggestive of two oligomeric states (data not shown).
To explore this observation further, we purified cIAP1 and cIAP2 proteins that included the BUCR domains (Fig. 1A) and again used MALLS-SEC to characterize the oligomeric state of these proteins. The calculated molecular mass of the BUCR monomers is ∼40.0 kDa; therefore, a measured mass of ∼80 kDa would be expected for dimeric proteins. Surprisingly, the cIAP1 BUCR protein consistently eluted as a predominantly monomeric peak even when injected at concentrations up to ∼300 μm (Fig. 1D). In contrast, a monomer-dimer equilibrium was observed for the equivalent cIAP2 protein with the peak that corresponds to the dimeric species increased at higher protein concentrations (Fig. 1E). cIAP proteins that did not possess a RING domain behaved as monomers (data not shown).
This suggests that dimer formation depends on the RING domain of cIAPs, but the BIR3, CARD, and UBA domains modulate dimer stability. In addition, the oligomeric state of the longer proteins distinguishes cIAP1 from cIAP2 with the cIAP2 BUCR protein forming a more stable dimer than the corresponding cIAP1 protein.
Dimer Stability Correlates with E3 Ligase Activity and Distinguishes cIAP1 and cIAP2
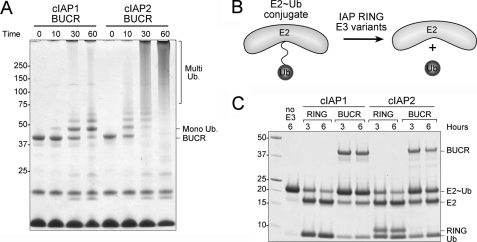
To evaluate the functional importance of cIAP dimerization, we assessed the ability of the different cIAP proteins to promote autoubiquitylation in vitro. As expected, both cIAP BUCR proteins efficiently promoted autoubiquitylation; however, the activity of the cIAP2 protein was much greater than for the analogous cIAP1 protein. For example, after incubation with ubiquitin, E1, and E2 for 30 min, most of the cIAP2 protein was highly ubiquitylated, whereas some of the cIAP1 protein was yet to be ubiquitylated, and most of it was only monoubiquitylated (Fig. 2A).
FIGURE 2.
E3 ligase activity correlates with dimer formation. A, purified cIAP1 and cIAP2 BUCR proteins were mixed with E1, UbcH5b, and ubiquitin for the indicated times and then separated by SDS-PAGE. B, schematic illustrating the discharge assay used to assess the ability of various RING domain-containing proteins to promote release of ubiquitin from the E2. C, purified RING and BUCR proteins were incubated with purified conjugate (E2∼Ub) for the indicated times at 37 °C and then separated by SDS-PAGE. The cIAP1 RING and untagged ubiquitin migrated to the same position. For reference, a sample of the conjugate that was incubated for 6 h without any E3 (no E3) is shown.
We next sought to determine whether the differences in dimerization propensity between cIAP1 and cIAP2 were responsible for their markedly different E3 ligase activity or whether the RING domains themselves were inherently different. To directly compare the ligase activity of BUCR and RING constructs, we developed a ubiquitin release assay as autoubiquitylation is considerably lower in RING-only constructs due to restricted lysine availability (supplemental Fig. 1). The active site cysteine mutant (C85S) of the promiscuous E2 UbcH5b, which mediates substrate ubiquitylation by cIAPs, was chosen for these studies (33, 34). The conjugate formed between this mutant and ubiquitin is relatively stable, allowing the E2∼Ub conjugate to be purified. We then compared the ability of different RING-containing proteins (at equimolar concentrations) to promote the conversion of the E2∼Ub conjugate (∼25 kDa) to free E2 (∼17 kDa) and ubiquitin (∼10 kDa) (Fig. 2B). Consistent with our autoubiquitylation assays (Fig. 2A), the cIAP2 BUCR protein more efficiently promoted discharge than the cIAP1 equivalent, which promoted only a modest increase in ubiquitin discharge. The RING domains of both cIAP1 and cIAP2 promoted disappearance of the conjugate to a similar extent and were more efficient than either of the longer constructs (Fig. 2C). Thus, without the inhibitory effects of their N-terminal domains, the cIAP1 RING domain is equivalent to that of cIAP2, and the ability to promote ubiquitin discharge correlates with the ability of the different proteins to form a stable dimer (Fig. 1).
Mutations That Disrupt RING Dimerization Disrupt Autoubiquitylation
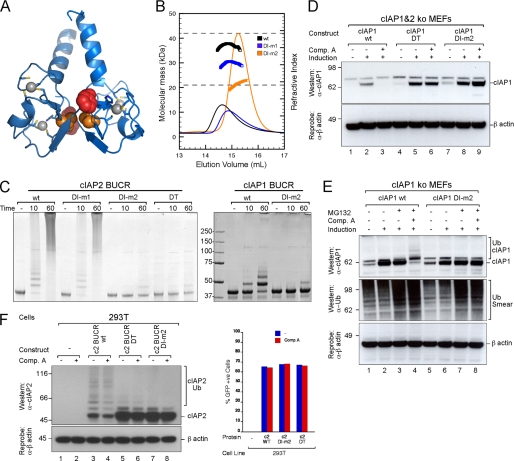
To further investigate the contribution of RING dimerization to cIAP protein function, we generated mutant cIAP proteins that had a reduced ability to form stable dimers. Previously, we showed that mutation of the aromatic residue three residues from the C terminus of the cIAP2 RING domain (F602A; subsequently referred to as DT) prevented dimer formation and abrogated the E3 ligase activity of the protein (20). Mutation of the analogous aromatic residue in the RING domain of MDM2 rendered the protein inactive, but the mutant MDM2 could still form heterodimers with MDM4, and the heterodimer was an active E3 ligase (13, 35), suggesting that the C-terminal residues might contribute to RING domain function in several ways. Therefore, to directly investigate the importance of RING dimerization for cIAP activity, we mutated Val-568, which is buried at the interface of the cIAP2 RING dimer (Fig. 3A). These mutants are referred to as dimer interface mutant 1 (DI-m1) (V568A) and DI-m2 (V568E) (see supplemental Table 1 for a summary of all mutants used in this study). Analysis of the purified proteins using MALLS-SEC showed that DI-m2 is largely monomeric in solution (Fig. 3B) because even at high concentrations (440 μm) it had an average molecular mass of 21.6 kDa, which is close to the calculated monomer mass of 21.19 kDa. In contrast, when samples of DI-m1 and wild-type protein at only ∼80–100 μm were loaded, the peaks eluted earlier, and the average molecular mass of the wild-type protein was close to that of the dimer, whereas the DI-m1 mutant had an average mass between that of the monomer and the dimer. This indicates that the conservative Val to Ala mutation slightly reduced the stability of the dimer but that the Val to Glu mutation, which would be predicted to place two similarly charged side chains opposite each other, was disfavored and destabilized the dimer, resulting in monomeric protein even at high concentrations. Analysis by circular dichroism spectroscopy indicated that the secondary structure of mutant proteins was comparable with that of the wild-type protein (supplemental Fig. 2A).
FIGURE 3.
Mutations that disrupt dimerization disrupt autoubiquitylation. A, ribbon representation of the cIAP2 RING dimer showing the zinc-binding sites. Residues that when mutated resulted in a monomeric protein, Phe-602 (red) and Val-568 (orange), are shown as spheres. B, samples of wild-type (black), DI-m1 (blue), and DI-m2 (orange) cIAP2 CARD-RING protein at either ∼80–100 (DI-m1 and WT) or ∼440 μm (DI-m2) were analyzed using an S200 column coupled in line to a MALLS detector. The refractive index profiles and average masses for the indicated peaks are shown. The dashed lines indicate the expected masses of the monomeric and dimeric proteins. C, the ability of wild-type and mutant BUCR proteins to promote autoubiquitylation was assessed. Equivalent samples of WT cIAP2 and mutants DI-m1, DI-m2, and DT as well as WT cIAP1 and the DI-m2 mutant were incubated with purified E1, E2, and ubiquitin for 10 and 60 min at 37 °C. Samples were then separated by SDS-PAGE and stained with Coomassie Blue. E1 was omitted from the controls (−). D, cIAP1 cIAP2 double knock-out MEFs were immortalized with SV40 large T and infected with a lentivirus containing inducible mouse cIAP1 that was either WT or contained the mutations DI-m2 and DT. For each construct, single clones were induced with 5 nm 4-hydroxytamoxifen for 16 h and treated with or without 500 nm bivalent Comp. A for 16 h. DISC lysates were separated by SDS-PAGE, and proteins were detected using antibodies for cIAP1 and β-actin. E, cIAP1−/− MEFs were immortalized with SV40 large T and infected with a lentivirus containing WT or DI-m2 inducible mouse cIAP1 construct. Following selection of single clones, MG132 was added for 2 h prior to incubation with or without 500 nm bivalent Comp. A for 1 h. DISC lysates were separated by SDS-PAGE, and proteins were detected using antibodies for cIAP1, ubiquitin, and β-actin. F, 293T cells were transfected with cIAP2 BUCR WT, DT and DI-m2 mutants, and GFP. Cells were treated with or without 500 nm bivalent Comp. A for 1 h. DISC lysates were prepared, separated by SDS-PAGE, and analyzed by Western blot for cIAP2 and β-actin. Cells from each sample were also analyzed in parallel by flow cytometry for expression of GFP. +ve, positive.
Autoubiquitylation assays using both the cIAP2 CARD-RING (supplemental Fig. 2B) and BUCR (Fig. 3C, left panel) proteins showed that the wild-type protein and DI-m1 efficiently promoted ladder formation. In contrast, the monomeric DI-m2 and DT mutants did not promote efficient autoubiquitylation (Fig. 3C, left panel). Mutation of the equivalent Val to Glu (V576E; referred to as DI-m2) at the dimer interface in cIAP1 also resulted in an E3 ligase with diminished activity (Fig. 3C, right panel). Because disruption of the dimer interface suppressed the E3 ligase activity of cIAP1, this suggests that, although difficult to detect, RING dimerization is required for activity.
To investigate the effect of specifically disrupting dimer formation on the E3 ligase activity of IAPs in a cellular setting, we made the DI-m2 mutation in full-length cIAP1 and expressed it in cIAP1−/− cIAP2−/− cells. Consistent with our previous results (20), mutation of the C-terminal aromatic residue to alanine in cIAP1 (F610A; DT) that rendered the protein an inactive E3 ligase resulted in accumulation of the protein in the absence of SM compounds and inhibited degradation induced by the bivalent SM (Comp. A; Fig. 3D). All compounds used in this study are described in supplemental Fig. 3. Likewise, cIAP1 containing the DI-m2 mutation was more stable than the wild-type protein, and it was not degraded when Comp. A was added (Fig. 3D, cf. lanes 2 and 3, lanes 5 and 6, and lanes 8 and 9). Because RING domains that contain DT and DI-m2 are monomers, this indicates that RING domain-mediated dimerization is required for SM compounds to cause the rapid degradation of cIAP1. This is consistent with our previous results that showed that the DT mutant inhibits the TNF-induced activation of NF-κB (36). Others have also shown that the DT mutation disrupts ubiquitylation of RIPK1 (19).
To confirm that degradation of cIAP1 was due to autoubiquitylation and targeting to the proteasome for degradation and that this process was dependent on RING dimer formation, we treated cells with the proteasome inhibitor MG132 to prevent degradation of ubiquitylated products. Comp. A promoted ubiquitylation of wild-type cIAP1 but not the DI-m2 mutant (Fig. 3E, cf. lanes 4 and 8). We also analyzed cIAP2 to determine whether the greater stability of the dimer and higher E3 ligase activity in vitro were mirrored within a cellular setting. For these experiments, we co-transfected 293T cells with wild-type cIAP2 or monomeric cIAP2 mutants together with green fluorescent protein (GFP) to ensure that the transfection efficiency was equivalent. Consistent with our in vitro results, wild-type cIAP2 was extensively modified when overexpressed, whereas the cIAP2 dimerization mutants had a significantly reduced level of modification and appeared to be more stable compared with the wild-type protein (Fig. 3F, cf. lane 3 with lanes 5 and 7). However, unlike cIAP1, cIAP2 levels remained relatively high upon treatment with IAP antagonist compounds (Fig. 3F, lane 4). This suggests that RING dimerization is required for the E3 ligase activity of cIAP2, but because the basal E3 ligase activity is relatively high, it is not highly stimulated upon SM compound addition (discussed further below).
RING Dimerization Is Required for Interaction with E2∼Ub Conjugate and to Promote Ubiquitin Discharge
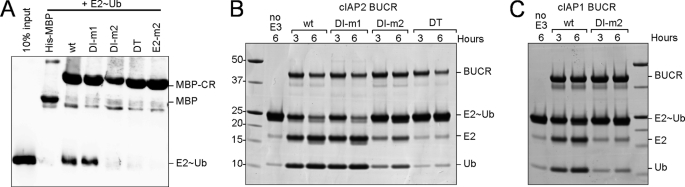
RING domains must interact with an E2, such as UbcH5b, to promote ubiquitylation (2). We therefore sought to test whether the monomeric cIAP proteins were inefficient E3 ligases because they could no longer interact with the E2. To assess E2 recruitment by the different proteins, we compared the ability of immobilized mutant and wild-type cIAP2 MBP-CARD-RING fusion proteins to interact with the E2∼Ub conjugate formed between the E2 (UbcH5b) and ubiquitin. In pulldown assays, wild-type cIAP2 and the DI-m1 mutant bound comparable amounts of the E2∼Ub conjugate, suggesting that interaction of the conjugate with the DI-m1 mutant was not significantly diminished (Fig. 4A). Neither the two monomeric mutant proteins DI-m2 and DT nor the control cIAP2 E2 interface mutant E2-m2 (R592A and V559A) bound to the conjugate, showing that dimerization of the cIAP2 RING domain and the integrity of the E2 binding surface are required for interaction of the RING domain with the UbcH5b∼ubiquitin conjugate.
FIGURE 4.
Monomeric cIAP proteins do not interact with E2∼Ub conjugate and promote discharge. A, wild-type or the indicated RING mutants within the context of cIAP2 CARD-RING (CR) protein were expressed as His-MBP fusions and immobilized on resin. Soluble His-tagged E2∼Ub conjugate was added to resin-bound proteins, incubated at 4 °C for 1 h, and then washed before separation by SDS-PAGE and transfer to nitrocellulose. The cIAP2-MBP fusions and the E2∼Ub conjugate were both detected using α-His antibodies. B, for discharge assays, wild-type cIAP2 or the mutant BUCR proteins were mixed with the E2∼Ub conjugate for the indicated times at 37 °C, and then samples were removed. A sample of conjugate that had been incubated in the absence of any E3 is included as a control (lane 2). All samples were separated by SDS-PAGE and stained with Coomassie Blue. C, wild-type and DI-m2 cIAP1 BUCR proteins were incubated with conjugate and analyzed as described in B.
Although interaction with the E2∼Ub conjugate was significantly reduced for the monomeric mutants, some RING E3s have been observed to efficiently promote ubiquitin transfer even though their interaction with the E2 is transient and difficult to detect (13, 35). Therefore, we also utilized the ubiquitin discharge assay to evaluate the ability of the mutant cIAP proteins to functionally interact with the E2 (Fig. 2B). As expected, the inactive E2 binding mutant E2-m2 (supplemental Fig. 4A) could not promote discharge of ubiquitin, whereas the wild-type cIAP2 protein promoted breakdown of the conjugate into free E2 and ubiquitin (supplemental Fig. 4B). Furthermore, the stable active DI-m1 mutant and wild-type protein promoted comparable amounts of ubiquitin discharge (Fig. 4B). In contrast, the conjugate was stable in the presence of the monomeric mutant cIAP2 proteins (DI-m2 and DT). Similarly, the equivalent wild-type cIAP1 protein promoted discharge, but the DI-m2 mutant did not (Fig. 4C).
Together, these results indicate that RING dimerization is required for recruitment of the E2∼Ub conjugate and for efficient discharge of ubiquitin. Dimerization has been reported to increase the E3 ligase activity of a number of RING domains. Our results suggest that this may be because the RING dimer is required for a productive interaction with the E2∼Ub conjugate so that the oxyester (or thioester) bond between the E2 and ubiquitin is destabilized in preparation for catalysis. These data support a model whereby RING dimerization has an important role in regulating the E3 ligase activity of the cIAPs.
Mono- and Bivalent IAP Antagonist Compounds Promote RING-dependent Dimerization and Autoubiquitylation
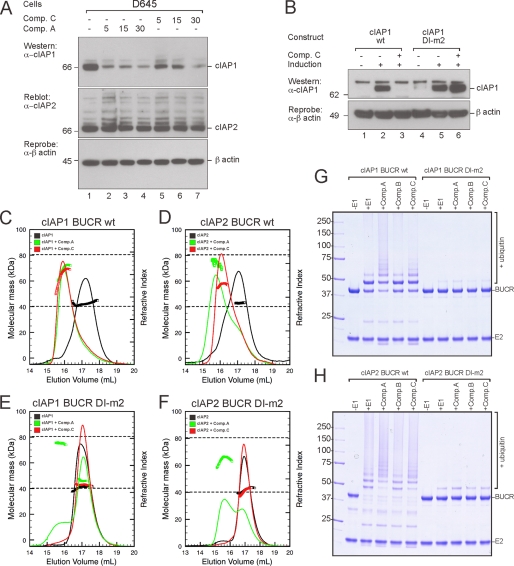
The ability of IAP antagonist compounds to promote IAP-dependent cell death is now well established (17, 18), but the mechanism by which the SM compounds promote ubiquitylation remains uncertain. Given that dimerization of the cIAP proteins is required for E3 ligase activity and is modulated by the N-terminal domains, we wondered whether IAP antagonist compounds promote autoubiquitylation by stabilizing the cIAP dimer. For these studies, we used the bivalent SM (Comp. A) and the monovalent SM (Comp. C) (supplemental Fig. 3) with monovalent compounds added at twice the concentration of the bivalent compounds to maintain the same number of mimetic moieties. As observed previously when cIAP1 was overexpressed (Fig. 3D), the bivalent compound also promoted the rapid degradation of endogenous cIAP1 in D645 cells (Fig. 5A). In contrast to cIAP1 and consistent with our results when we overexpressed cIAP2 (Fig. 3F), endogenous cIAP2 appeared to have a significant level of basal modification in the untreated sample, and the levels of cIAP2 were not significantly decreased upon SM treatment. Endogenous cIAP1 was also efficiently degraded upon treatment with Comp. C (Fig. 5A), indicating that the SM compounds stimulate autoubiquitylation in the absence of BIR domain cross-linking by bivalent compounds. However, the DI-m2 mutant was resistant to Comp. C-induced degradation (Fig. 5B), indicating that SM-induced cIAP1 degradation requires the formation of a RING dimer.
FIGURE 5.
Mono- and bivalent IAP antagonist compounds promote RING-dependent dimerization and ubiquitylation. A, D645 cells were treated with 500 nm bivalent Comp. A or 1 μm monovalent Comp. C for 5, 15, and 30 min. Lysates were prepared, and cIAP1 and cIAP2 were detected as described previously. B, cIAP1−/− MEFs were immortalized with SV40 large T and infected with a lentivirus containing inducible wild-type mouse cIAP1 or the monomeric DI-m2 mutant. Single clones were induced with 5 nm 4-hydroxytamoxifen for 16 h and 1 μm monovalent Comp. C for 16 h. DISC lysates were separated by SDS-PAGE and immunoblotted for cIAP1 and β-actin. C, MALLS analysis of wild-type cIAP1 BUCR protein in the absence of compound (black) and in the presence of either the monovalent (red) or bivalent (green) compounds is shown. Samples contained 25 μm BUCR protein and either 62.5 μm bivalent Comp. A or 125 μm monovalent Comp. C. The dashed lines indicate the expected masses of the monomeric and dimeric proteins. D, samples of cIAP2 BUCR were analyzed as described for cIAP1 BUCR. E and F, samples of the cIAP1 and cIAP2 DI-m2 proteins were also analyzed. G, purified wild-type and DI-m2 cIAP1 BUCR proteins were used in autoubiquitylation assays in the absence or presence of the indicated compounds. Samples were incubated at 37 °C for 30 min, then separated by SDS-PAGE, and detected by Coomassie staining. H, similar assays were carried out using wild-type and DI-m2 cIAP2 BUCR proteins.
Next we used MALLS-SEC to evaluate the ability of these compounds to promote dimerization of the BUCR proteins. As observed previously, at ∼25 μm, both BUCR proteins were largely monomeric (Fig. 5, C and D), although the greater stability of cIAP2 was apparent as a broad leading edge on the main peak. Addition of the mono- and bivalent compounds to the cIAP1 protein readily promoted dimer formation, and the two samples appeared to have comparable stability (Fig. 5C). For cIAP2, bivalent Comp. A also promoted dimerization, although the stability of the dimer was reduced when monovalent Comp. C was added because the average mass was 58.7 kDa, and the peak eluted between that of the monomer and dimer peaks (Fig. 5D). For both proteins, no shift in mass or peak position was observed when the monovalent compound was added to the DI-m2 mutant (Fig. 5, E and F). Dimerization of the DI-m2 mutant was also reduced upon addition of the bivalent compound, although some cross-linking due to interaction of the compound with BIR domains from two IAP molecules occurred in the absence of a stable RING dimer (see Fig. 6C).
FIGURE 6.
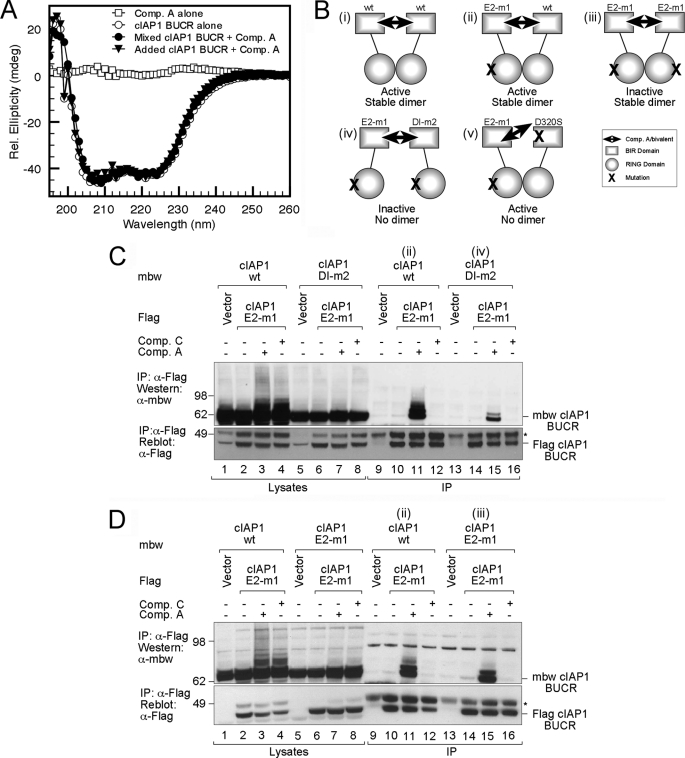
E2 binding is not required for compound-induced dimerization. A, CD spectra of Comp. A alone and cIAP1 BUCR in the absence or presence of Comp. A as indicated. The theoretical spectra obtained by adding that for the protein alone and Comp. A alone is also shown. B, schematic detailing the cIAP1 BUCR WT and mutants and the active and inactive combinations (labeled i–v) used in the immunoprecipitation (IP) assays. C, 293T cells were transfected with FLAG-tagged cIAP1 BUCR containing the E2-m1 mutation or an empty FLAG vector as a control, and either mbw-tagged wild-type cIAP1 BUCR or a similar construct with DI-m2 mutation (as indicated in B ii and iv). Cells were treated for 1 h with or without 500 nm bivalent Comp. A or 1 μm monovalent Comp. C SM compound. DISC lysates were immunoprecipitated with anti-FLAG beads for 1 h. Proteins were eluted by boiling, separated by SDS-PAGE, and immunoblotted for mbw and FLAG. D, experiments similar to those described in C were carried out except that that FLAG-tagged constructs were incubated together with either mbw-tagged wild-type cIAP1 BUCR or an equivalent E2-m1 construct (as indicated in B ii and iii). Cells were treated for 1 h with or without bivalent Comp. A or monovalent Comp. C SM compound. Samples were prepared as before and separated by SDS-PAGE before immunoblotting for mbw and FLAG. rel., relative; mdeg, millidegrees. In C and D, the * indicates the heavy chain of the antibody used for immunoprecipitation.
In parallel with these experiments, we assessed the ability of the compounds to promote autoubiquitylation. For cIAP1 BUCR, we observed a significant increase in autoubiquitylation upon addition of the mono- and bivalent compounds (Fig. 5G) but not with the inactive enantiomer (Comp. B is the enantiomer of Comp. A). In contrast, although the active compounds promoted autoubiquitylation of cIAP2 (Fig. 5H), the increase over the basal activity was less obvious. This is consistent with the diminished ability of the compounds to promote cIAP2 dimerization (Fig. 5D) and the modest effect of the compounds on cIAP2 levels in cells (Figs. 2F and 5A). For both cIAP1 and cIAP2, the monomeric DI-m2 mutant was largely inactive in the presence or absence of the compounds (Fig. 5, G and H).
These results indicate that both mono- and bivalent compounds can efficiently promote cIAP dimerization that largely depends on the ability of the RING domains to interact. In addition, it appears that upon SM treatment dimerization is more efficiently promoted for cIAP1, but cIAP2 forms a more stable dimer in the absence of compounds. The ability of the monovalent compounds that bind to the BIR3 domain to increase RING-dependent dimer formation strongly suggests that SM compounds antagonize an interdomain interaction that inhibits RING dimerization. As a consequence, the monomer-dimer equilibrium shifts in favor of dimer formation.
Dimeric IAP Antagonists Induce RING-dependent Dimerization in Cells That Is Independent of E2 Binding
Given the critical role for RING dimerization in generating an active E3 ligase and the ability of mono- or bivalent SM compounds to promote this, we sought to further examine how cIAP dimerization was regulated. As discussed above, we noted that the RING domain alone formed a more stable dimer than that formed by longer proteins with the cIAP1 BUCR protein eluting as a monomer even at 300 μm, whereas the cIAP1 RING domain behaved as a stable dimer at 10 μm. This suggested that the longer proteins contained sequences that restricted dimer formation. Because attempts to crystallize the longer proteins were unsuccessful, we used CD spectroscopy to investigate the nature of the changes that occur upon addition of the SM compounds to cIAP1 BUCR. For both Comp. A (Fig. 6A) and Comp. C (supplemental Fig. 5A), the complex spectra overlay with the theoretical spectra obtained by adding the spectra of the proteins and compounds alone. This indicates that compound addition does not result in a significant change in the secondary structure. These results are most consistent with the SM compounds stimulating domain rearrangement that allows two RING domains to interact.
To identify additional features that influenced dimer stability, we utilized a cell-based system and assessed dimer formation by analyzing the ability of different mutant BUCR proteins to be co-immunoprecipitated from cells (Fig. 6B). Consistent with the modest affinity of the cIAP1 dimers, no interaction of tagged proteins could be detected despite abundant protein in the lysates (Fig. 6C, cf. lanes 2 and 10). However, we utilized the ability of bivalent Comp. A to interact tightly with the BIR domain from two BUCR molecules and effectively cross-link cIAP proteins that have formed a stable RING-mediated dimer (Fig. 6B, i, ii, and iii). Initially, we assessed the ability of wild-type cIAP1 tagged with mbw, a previously described inert carrier protein that has approximately the same size and shape as an IAP BIR repeat (22), and FLAG-tagged cIAP1 to interact (Fig. 6B, i). In the presence of bivalent Comp. A, FLAG-tagged wild-type cIAP1 and mbw-tagged wild-type cIAP1 co-immunoprecipitated but not when the monovalent Comp. C was added (supplemental Fig. 5B). In a comparable experiment, FLAG-tagged cIAP1 that contained the E2 interface mutation V573A/D576A (referred to as E2-m1) was co-immunoprecipitated (Fig. 6B, ii). The interaction with the E2-m1 protein was easier to detect (supplemental Fig. 5B) probably because of decreased autoubiquitylation and degradation; we therefore used this mutant for subsequent co-immunoprecipitation assays.
To evaluate the possibility that the bivalent compound cross-linked cIAP proteins irrespective of whether the RING domains interacted, we utilized the dimer interface mutant DI-m2 (Fig. 6B, iv). When this mutant was co-expressed together with the E2-m1 mutant and Comp. A was added, considerably less protein was immunoprecipitated (Fig. 6C, cf. lanes 11 and 15). As before, no interaction was detected with the monovalent compound (Comp. C). However, addition of either the mono- or bivalent compound resulted in increased smearing above the dominant IAP band when wild-type cIAP1 was present, indicating that both compounds can promote RING dimer-dependent autoubiquitylation of cIAPs (Fig. 6C, cf. lanes 3 and 4 with lanes 7 and 8). Furthermore, a D320S mutation that disrupts the SM-binding pocket on BIR3 prevented co-immunoprecipitation when the bivalent compound was added (supplemental Fig. 5C). However, autoubiquitylation was still increased, indicating that the RING dimer forms when just one molecule in the dimer can bind to the SM compound (Fig. 6B, v). These results demonstrate that the RING domain-mediated dimer forms in the absence of cross-linking, but the bivalent compound is required for it to be detected.
Next we investigated the contribution E2 binding made to the stability of the RING dimer by using constructs where both E2-binding sites were disrupted (Fig. 6B, iii). We co-expressed mbw-tagged cIAP1 BUCR constructs of wild type or the E2-m1 mutant together with the FLAG-tagged E2-m1 mutant. Upon addition of the bivalent compound (Comp. A), a comparable amount of protein was immunoprecipitated by both the wild-type and E2-m1 proteins (Fig. 6D, cf. lanes 11 and 15), indicating that interaction with the E2 does not stabilize the cIAP1 dimer. Taken together, these results suggest that both E2 recruitment and RING dimerization are required for autoubiquitylation, but interaction with the E2 does not increase formation of the cIAP1 dimer.
All Active Smac Mimetic Compounds Promote Dimerization
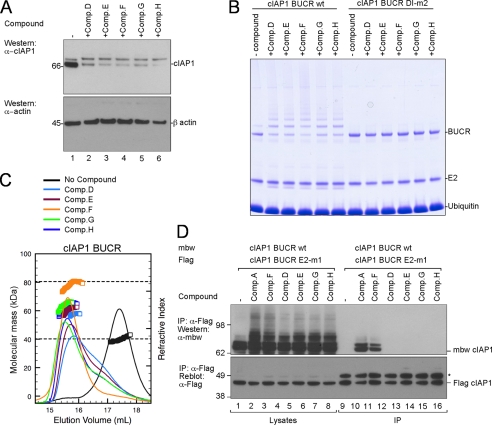
A number of IAP antagonist compounds have been developed independently (37), and we next investigated whether other SM compounds promoted cIAP autoubiquitylation in a similar manner. We were particularly interested to see whether other monovalent compounds also promoted dimerization and whether this was associated with autoubiquitylation. We therefore synthesized a selection of published structurally distinct compounds (supplemental Fig. 3) and confirmed that each compound promoted rapid loss of cIAP1 in a cellular setting as reported (Fig. 7A) (31, 38). These compounds also promoted efficient autoubiquitylation of wild-type cIAP1 BUCR in vitro but not of the monomeric cIAP1 BUCR DI-m2, indicating that the E3 ligase activity induced by all the tested SMs requires RING dimerization (Fig. 7B). MALLS-SEC analysis was then used to evaluate the oligomeric state of cIAP1 BUCR following addition of the compounds (Fig. 7C). As observed for Comp. A and Comp. C, every SM compound that promoted cIAP1 autoubiquitylation and degradation also efficiently promoted dimerization of the BUCR protein in vitro, although bivalent Comp. F resulted in the most stable cIAP1 dimer (Fig. 7C).
FIGURE 7.
All active compounds from patent literature tested promote dimerization. A, wild-type MEFs were incubated with the indicated compounds for 30 min. DISC lysates were separated by SDS-PAGE, and then cIAP1 and β-actin were detected by immunoblotting. All compounds are monovalent except Comp. F. B, purified wild-type and DI-m2 cIAP1 BUCR proteins were mixed with E1, UbcH5b, ubiquitin, and the indicated compounds for 10 min, and then samples were separated by SDS-PAGE. C, MALLS analysis of wild-type cIAP1 BUCR protein in the absence of compound (black) and in the presence of the indicated compounds is shown. Samples contained 25 μm BUCR protein and a 125 μm concentration of each compound, except 62.5 μm bivalent compound F was added. The dashed lines indicate the expected masses of the monomeric and dimeric proteins. D, 293T cells were transfected with FLAG-tagged cIAP1 BUCR containing an E2 binding mutation (E2-m1) and mbw-tagged wild-type cIAP1 BUCR. Cells were treated for 1 h with or without 500 nm bivalent (Comps. A and F) or 1 μm monovalent (Comps. D, E, G, and H) SM compounds. DISC lysates were immunoprecipitated (IP) with anti-FLAG beads for 1 h. Proteins were eluted by boiling and separated by SDS-PAGE prior to immunoblotting for mbw and FLAG. The * indicates the heavy chain of the antibody used for immunoprecipitation.
Lastly, all the unrelated SMs also increased autoubiquitylation in cell-based assays, but only the bivalent Comp. F stabilized the RING dimer and allowed co-immunoprecipitation (Fig. 7D). This parallels the results obtained using Comps. A and C and suggests that all SM compounds promote RING dimerization, and this is required for their mechanism of action and subsequent therapeutic effects.
DISCUSSION
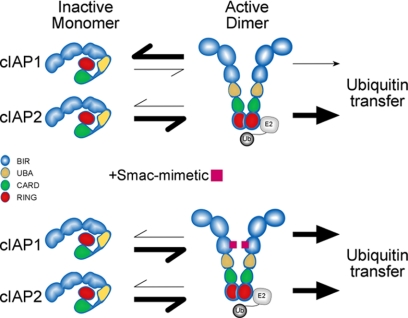
cIAP1 and cIAP2 are important components of the TNF receptor signaling complex, and a functional RING domain is required for TNF-induced activation of NF-κB (19, 31, 36). Here we report that cIAP1 and cIAP2 require RING domain-mediated dimerization for E3 ligase activity and that in the absence of a stimulus cIAP1 is largely monomeric and inactive. This suggests that molecules that promote dimer formation activate the E3 ligase activity of cIAP1. Consistent with this, SM compounds that promoted cIAP dimerization increased the E3 ligase activity of cIAP1. In contrast, cIAP2 formed a more stable dimer and had higher intrinsic E3 ligase activity that was not dramatically induced upon SM compound addition (Fig. 8). Our results suggest that the N-terminal domains of cIAPs inhibit RING dimerization to regulate the E3 ligase activity and that IAP antagonists modulate this inhibition.
FIGURE 8.
Model indicating key role of RING dimerization. A monomer-dimer equilibrium is present for both cIAP1 and cIAP2 although cIAP1 predominantly exists as a monomer, whereas cIAP2 exists primarily as an active dimer. cIAP RING domain-mediated dimers must form before a productive interaction with the E2∼Ub conjugate can occur, and this interaction is required for autoubiquitylation. Because cIAP2 forms a stable dimer in the absence of stimulus, the E2∼Ub conjugate is more readily destabilized by cIAP2, and ubiquitin is efficiently transferred. When IAP antagonist compounds are added, RING-mediated dimerization is favored for both cIAP1 and cIAP2, resulting in efficient E2∼Ub recruitment and increased autoubiquitylation.
Using cIAP RING domain mutant proteins that can no longer dimerize, we show that E3 ligase activity correlated with dimer formation and that monomeric IAPs had a significantly reduced ability to promote ubiquitin release from UbcH5b. Dimerization is used by many proteins to alter their activity, and this has often been observed for RING domain E3 ligases (2). For example, recent work shows that dimerization of MDM2 and polyubiquitylation of its target p53 are inhibited by phosphorylation (39). Dimerization of the RNF4 RING domain is also required for E3 ligase activity (40), and only dimeric RNF4 proteins can effectively complement yeast strains in which the homologous proteins Rfp1 and Rfp2 have been inactivated. The RING domains of MDM2, RNF4, and cIAPs form similar dimers, suggesting that other C-terminal RING domains, such as those found in caspase-associated RING proteins and mind bomb proteins (41, 42), will also form comparable dimers, and this might be important for controlling the E3 ligase activity of this class of RING domains. However, it remains to be investigated whether dimerization of these proteins is regulated in a manner similar to that observed for cIAP proteins.
Although RING dimerization is essential for E3 ligase function, it is still not clear why. Structures of RING domains, even dimeric RING domains, in complex with their cognate E2 indicate that E2s bind to RING surfaces far from the dimer interface; therefore, dimerization was not thought to be required for E2 interaction (2). In contrast to this, our data suggest that RING dimerization is required for interaction with the ubiquitin-charged E2 and that interaction of the E2∼Ub thioester intermediate with a RING domain dimer destabilizes the thioester linkage. The nature of this change is uncertain, but it is likely that interaction of the E2∼Ub conjugate with the RING dimer leads to rearrangements in the E2 that impact the stability of the E2∼Ub thioester (or oxyester) bond so that catalysis and attack by the target lysine residue is favored. An allosteric mechanism of E2 activation by E3s has been proposed (6), and interaction with the E3 is clearly important for discharge and can lead to rearrangements near the active site of the E2 (7, 43). However, further structural studies are required because the E2 does not undergo a significant structural change upon binding to its cognate RING dimer (20, 44, 45). However, ubiquitin is not present in these structures. An important role for ubiquitin is possible because structural analysis of E2s in the presence of ubiquitin indicates that as well as interacting with surface-exposed residues in the vicinity of the catalytic cysteine, conjugation with ubiquitin has an indirect effect on other residues (46), and extended chains can form (47).
Notwithstanding the mechanistic details, it is clear that cIAP RING domains can only promote ubiquitin transfer when the RING domain simultaneously contacts its cognate E2 and forms a RING homodimer. In this way, RING function is regulated by a combination of a relatively indiscriminate E2 binding event and a highly specific dimerization event, both of which are relatively low affinity and transient. As a consequence, it is likely that inappropriate ubiquitylation of proteins is diminished. The importance of modulating the rate of ubiquitin transfer from the E2 was highlighted previously with Deshaies and colleagues (48, 49) showing that for Cullin RING ligases the dynamics of substrate-E3 and E3-E2 encounters ensure that substrate proteins are either not ubiquitylated or are polyubiquitylated and degraded.
The requirement for RING dimerization means that regulating protein self-association can control the E3 ligase activity of cIAPs, especially cIAP1 (Fig. 8). The mechanism by which this is achieved is uncertain, but a role for the BIR3 domain in inhibiting RING dimerization is suggested because the SM compounds that bind to a surface-exposed pocket on this domain promote dimerization, rapid autoubiquitylation, and degradation of cIAPs (17, 18, 38). Previous studies have concluded that dimerization is not important for cIAP function because monovalent SM compounds promote autoubiquitylation and degradation (17). These studies assumed that compounds promoted IAP dimerization only by cross-linking two IAPs via their BIR domains, and because the monovalent compounds could not mediate this interaction, they concluded that dimerization was not essential. However, although bivalent compounds are able to stabilize an IAP dimer, mutation of the RING dimer interface results in substantially less IAP dimer in vivo. The bivalent BIR binding compounds therefore promote RING dimer formation but also trap this weak RING-RING interaction in cells by interacting with the BIR3 domain from the two molecules. Because structurally distinct monovalent SMs are all able to activate cIAP E3 ligase activity, cross-linking of BIRs is unlikely to be a significant element of IAP antagonist function. The inability of longer cIAP1 constructs to form stable dimers suggests that the UBA, CARD, or BIR3 domain restrains the RING domain in an inactive monomeric conformation, and we propose that IAP antagonists promote a structural rearrangement that stabilizes RING dimerization. Whatever the mechanism, it is clear that the functional IAP dimer is a RING-mediated dimer and that both monovalent and bivalent compounds are able to promote this modality.
For TNFR1 to activate p65/RelA NF-κB following TNF stimulation, cIAP1 must interact with TRAFs and bear a RING domain that is capable of forming dimers (36, 50). This suggests that upon TNF stimulation cIAP1 is recruited to receptor complexes by TRAF2, and cIAP1 dimerization is promoted. Recently, it has been shown that the TRAF2 trimer only binds to a single cIAP protein (51, 52); therefore, cIAP dimer formation is likely to require the interaction of cIAP molecules bound to two TRAF2 trimers. Whether two separate TNFR1-TRADD complexes recruit two TRAF2 trimers or whether the cIAP RING dimer is sufficient to facilitate cIAP1/TRAF2 recruitment without the involvement of another receptor is an interesting question. If two TRAF2 trimers must be recruited by two TNFR1-TRADD complexes, then this suggests at least two TNF/TNFR1 receptors must be engaged and in close proximity for TNFR1 signaling to occur. The stoichiometry of the TNF receptor signaling complex is therefore uncertain; however, the essential role for cIAP RING dimerization is clear because unlike wild-type cIAP1 (31) the monomeric cIAP1 RING mutant neither promotes RIPK1 ubiquitylation following TNF receptor stimulation in cIAP1−/− cIAP2−/− cells nor does it recover normal canonical NF-κB activation (19, 36).
Our results establish a critical role for RING dimerization in cIAP-mediated ubiquitylation and show that efficient discharge of ubiquitin from the E2 depends on interaction with a RING domain dimer. RING dimerization is critical for the action of the IAP antagonist compounds, and these studies suggest that there might be other ways to promote cIAP dimerization and degradation. This study also suggests that cIAP1 and cIAP2 may have distinct roles with cIAP1 primarily targeted by SM compounds. It also remains unclear how the balance between substrate and autoubiquitylation by cIAPs is altered, and it will be interesting to see, in a cellular context, whether SM addition alters the nature of the complex formed.
Supplementary Material
Supplemental Data
*
M. McKinlay, S. M. Condon, and S. K. Chunduru are employees of TetraLogic Pharmaceuticals; J. Silke is a consultant; and D. L. Vaux is on the scientific advisory board.
4
The abbreviations used are:
RING
really interesting new gene
cIAP
cellular inhibitor of apoptosis
IAP
inhibitor of apoptosis
MALLS
multiple angle laser light scattering
NF-κB
nuclear factor-κ light chain enhancer of activated B cells
SEC
size exclusion chromatography
Smac
second mitochondrion-derived activator of caspase
SM
Smac mimetic
TRAF
TNF receptor-associated factor
RIPK1
receptor-interacting protein kinase-1
Comp.
compound
BIR
baculoviral IAP repeat
Ub
ubiquitin
UBA
ubiquitin-associated
CARD
caspase recruitment domain
MEF
mouse embryonic fibroblast
BisTris
2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
MBP
maltose-binding protein
BUCR
BIR3-UBA-CARD-RING
E
embryonic day
DT
dimer tail mutant
DI-m
dimer interface mutant
E2-m
E2 binding mutant
mbw
monomeric Bcl-w
FRT
Flippase Recognition Target
DISC
Death Inducing Signaling Complex
TRADD
TNF Receptor Associated Death Domain.
REFERENCES
- 1.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 2.Deshaies R. J., Joazeiro C. A. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 3.Huang A., de Jong R. N., Wienk H., Winkler G. S., Timmers H. T., Boelens R. (2009) J. Mol. Biol. 385, 507–519 [DOI] [PubMed] [Google Scholar]
- 4.Christensen D. E., Brzovic P. S., Klevit R. E. (2007) Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 5.Ye Y., Rape M. (2009) Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozkan E., Yu H., Deisenhofer J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18890–18895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 9.Barbash O., Zamfirova P., Lin D. I., Chen X., Yang K., Nakagawa H., Lu F., Rustgi A. K., Diehl J. A. (2008) Cancer Cell 14, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knipscheer P., Sixma T. K. (2007) Curr. Opin. Struct. Biol. 17, 665–673 [DOI] [PubMed] [Google Scholar]
- 11.Brzovic P. S., Keeffe J. R., Nishikawa H., Miyamoto K., Fox D., 3rd, Fukuda M., Ohta T., Klevit R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 14.Salvesen G. S., Duckett C. S. (2002) Nat. Rev. Mol. Cell Biol. 3, 401–410 [DOI] [PubMed] [Google Scholar]
- 15.Vaux D. L., Silke J. (2005) Nat. Rev. Mol. Cell Biol. 6, 287–297 [DOI] [PubMed] [Google Scholar]
- 16.Rothe M., Pan M. G., Henzel W. J., Ayres T. M., Goeddel D. V. (1995) Cell 83, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 17.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 18.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 19.Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., Walczak H. (2009) Mol. Cell 36, 831–844 [DOI] [PubMed] [Google Scholar]
- 20.Mace P. D., Linke K., Feltham R., Schumacher F. R., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) J. Biol. Chem. 283, 31633–31640 [DOI] [PubMed] [Google Scholar]
- 21.Hu S., Yang X. (2003) J. Biol. Chem. 278, 10055–10060 [DOI] [PubMed] [Google Scholar]
- 22.Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung H. H., Plenchette S., Kern C. J., Mahoney D. J., Korneluk R. G. (2008) Mol. Biol. Cell 19, 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du C., Fang M., Li Y., Li L., Wang X. (2000) Cell 102, 33–42 [DOI] [PubMed] [Google Scholar]
- 25.Verhagen A. M., Ekert P. G., Pakusch M., Silke J., Connolly L. M., Reid G. E., Moritz R. L., Simpson R. J., Vaux D. L. (2000) Cell 102, 43–53 [DOI] [PubMed] [Google Scholar]
- 26.Gyrd-Hansen M., Darding M., Miasari M., Santoro M. M., Zender L., Xue W., Tenev T., da Fonseca P. C., Zvelebil M., Bujnicki J. M., Lowe S., Silke J., Meier P. (2008) Nat. Cell Biol. 10, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter A. M., LaCasse E. C., Korneluk R. G. (2007) Apoptosis 12, 1543–1568 [DOI] [PubMed] [Google Scholar]
- 28.Vucic D., Fairbrother W. J. (2007) Clin. Cancer Res. 13, 5995–6000 [DOI] [PubMed] [Google Scholar]
- 29.Eckelman B. P., Salvesen G. S. (2006) J. Biol. Chem. 281, 3254–3260 [DOI] [PubMed] [Google Scholar]
- 30.Gyrd-Hansen M., Meier P. (2010) Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 31.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 32.Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dynek J. N., Goncharov T., Dueber E. C., Fedorova A. V., Izrael-Tomasevic A., Phu L., Helgason E., Fairbrother W. J., Deshayes K., Kirkpatrick D. S., Vucic D. (2010) EMBO J. 29, 4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankenship J. W., Varfolomeev E., Goncharov T., Fedorova A. V., Kirkpatrick D. S., Izrael-Tomasevic A., Phu L., Arnott D., Aghajan M., Zobel K., Bazan J. F., Fairbrother W. J., Deshayes K., Vucic D. (2009) Biochem. J. 417, 149–160 [DOI] [PubMed] [Google Scholar]
- 35.Uldrijan S., Pannekoek W. J., Vousden K. H. (2007) EMBO J. 26, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feltham R., Moulin M., Vince J. E., Mace P. D., Wong W. W., Anderton H., Day C. L., Vaux D. L., Silke J. (2010) J. Biol. Chem. 285, 17525–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flygare J. A., Fairbrother W. J. (2010) Expert Opin. Ther. Pat. 20, 251–267 [DOI] [PubMed] [Google Scholar]
- 38.Gaither A., Porter D., Yao Y., Borawski J., Yang G., Donovan J., Sage D., Slisz J., Tran M., Straub C., Ramsey T., Iourgenko V., Huang A., Chen Y., Schlegel R., Labow M., Fawell S., Sellers W. R., Zawel L. (2007) Cancer Res. 67, 11493–11498 [DOI] [PubMed] [Google Scholar]
- 39.Cheng Q., Chen L., Li Z., Lane W. S., Chen J. (2009) EMBO J. 28, 3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liew C. W., Sun H., Hunter T., Day C. L. (2010) Biochem. J. 431, 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald E. R., 3rd, El-Deiry W. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6170–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C., Li Q., Jiang Y. J. (2007) J. Mol. Biol. 366, 1115–1128 [DOI] [PubMed] [Google Scholar]
- 43.Petroski M. D., Deshaies R. J. (2005) Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 44.Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 45.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 46.Serniwka S. A., Shaw G. S. (2009) Biochemistry 48, 12169–12179 [DOI] [PubMed] [Google Scholar]
- 47.Sakata E., Satoh T., Yamamoto S., Yamaguchi Y., Yagi-Utsumi M., Kurimoto E., Tanaka K., Wakatsuki S., Kato K. (2010) Structure 18, 138–147 [DOI] [PubMed] [Google Scholar]
- 48.Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierce N. W., Kleiger G., Shan S. O., Deshaies R. J. (2009) Nature 462, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vince J. E., Pantaki D., Feltham R., Mace P. D., Cordier S. M., Schmukle A. C., Davidson A. J., Callus B. A., Wong W. W., Gentle I. E., Carter H., Lee E. F., Walczak H., Day C. L., Vaux D. L., Silke J. (2009) J. Biol. Chem. 284, 35906–35915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mace P. D., Shirley S., Day C. L. (2010) Cell Death Differ. 17, 46–53 [DOI] [PubMed] [Google Scholar]
- 52.Zheng C., Kabaleeswaran V., Wang Y., Cheng G., Wu H. (2010) Mol. Cell 38, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data