Wheat Grain Development Is Characterized by Remarkable Trehalose 6-Phosphate Accumulation Pregrain Filling: Tissue Distribution and Relationship to SNF1-Related Protein Kinase1 Activity (original) (raw)
Abstract
Trehalose 6-phosphate (T6P) is a sugar signal that regulates metabolism, growth, and development and inhibits the central regulatory SNF1-related protein kinase1 (SnRK1; AKIN10/AKIN11). To better understand the mechanism in wheat (Triticum aestivum) grain, we analyze T6P content and SnRK1 activities. T6P levels changed 178-fold 1 to 45 d after anthesis (DAA), correlating with sucrose content. T6P ranged from 78 nmol g−1 fresh weight (FW) pregrain filling, around 100-fold higher than previously reported in plants, to 0.4 nmol g−1 FW during the desiccation stage. In contrast, maximum SnRK1 activity changed only 3-fold but was inhibited strongly by T6P in vitro. To assess SnRK1 activity in vivo, homologs of SnRK1 marker genes in the wheat transcriptome were identified using Wheat Estimated Transcript Server. SnRK1-induced and -repressed marker genes were expressed differently pregrain filling compared to grain filling consistent with changes in T6P. To investigate this further maternal and filial tissues were compared pre- (7 DAA) and during grain filling (17 DAA). Strikingly, in vitro SnRK1 activity was similar in all tissues in contrast to large changes in tissue distribution of T6P. At 7 DAA T6P was 49 to 119 nmol g−1 FW in filial and maternal tissues sufficient to inhibit SnRK1; at 17 DAA T6P accumulation was almost exclusively endospermal (43 nmol g−1 FW) with 0.6 to 0.8 nmol T6P g−1 FW in embryo and pericarp. The data show a correlation between T6P and sucrose overall that belies a marked effect of tissue type and developmental stage on T6P content, consistent with tissue-specific regulation of SnRK1 by T6P in wheat grain.
We recently identified a regulatory mechanism in growing tissues of Arabidopsis (Arabidopsis thaliana) and other species that regulates metabolism, end product synthesis, and growth and development (Zhang et al., 2009; Paul et al., 2010). This mechanism involves the pathway that synthesizes the nonreducing disaccharide trehalose. In Arabidopsis there are 21 genes that encode putative trehalose phosphate synthase (TPS) and trehalose phosphate phosphatase (TPP) genes (Avonce et al., 2006; Lunn, 2007). The trehalose pathway is ubiquitous in plants and it has been known that it is an indispensable component of plant development in seeds (Eastmond et al., 2002) and for vegetative development and transition to flowering (van Dijken et al., 2004) with trehalose 6-P (T6P) shown as the critical factor (Schluepmann et al., 2003). Significantly, when T6P levels are increased by genetic means, growth in response to Suc and expression of genes encoding enzymes for growth and biosynthetic pathways are increased (Zhang et al., 2009; Paul et al., 2010). Accumulation of important end products such as starch and cell walls can also be stimulated by T6P (Kolbe et al., 2005; Gómez et al., 2006).
We showed that a signaling partner of T6P in growing tissues is the SNF1-related protein kinase1, SnRK1, of the SNF1/AMPK group of protein kinases (Zhang et al., 2009; Paul et al., 2010). SNF1-related protein kinases perform a fundamental role in the physiological response of cells to energy limitation and starvation of carbon source through regulation of pathways and processes involved in metabolism, growth, and development (Hardie, 2007; Polge and Thomas, 2007; Halford and Hey, 2009). Active AMPK/SnRK1/SNF1 function to conserve energy and carbon supplies, switching off biosynthetic processes in response to carbon and energy limitation. In plants enzymes such as nitrate reductase and Suc phosphate synthase are phosphorylation targets of SnRK1 that inactivates them (Sugden et al., 1999). Recent work in Arabidopsis established 1,000 or so target genes of SnRK1 (AKIN10) involved in the response of metabolism and growth to starvation (Baena-González et al., 2007). It was shown that SnRK1 activates genes involved in degradation processes and photosynthesis and inhibits those involved in biosynthetic processes and, by so doing, regulates metabolism and growth in response to available carbon (Baena-González et al., 2007). In growing tissues inhibition of SnRK1 by T6P is a mechanism of switching from catabolism to anabolism by activation of biosynthetic processes by T6P in response to Suc supply (Zhang et al., 2009; Paul et al., 2010). Metabolic pathways such as those involved in cell wall and protein metabolism were up-regulated through T6P inhibition of SnRK1 in Arabidopsis seedlings (Zhang et al., 2009; Paul et al., 2010). Given that biosynthetic processes underpin growth and development this mechanism represents a potential means for the improvement of yield.
Wheat (Triticum aestivum) is the dominant crop in temperate countries and is the most widely grown of the major crops worldwide. There is evidence that wheat yields have reached a plateau (http://energyfarms.wordpress.com/2009/09/03/crop-yield-projections-miss-biofuel-report-target/) and that to improve yields beyond this ceiling requires new insight and approaches. One means of doing this is to identify new regulatory processes as targets for manipulation. The trehalose pathway is present in cereals as in all other plants. TPP genes appear particularly numerous, 16 are present in rice (Oryza sativa), for example (Ge et al., 2008). A knockout of a TPP gene of maize (Zea mays) is the basis of the ramosa3 classical mutant of maize that affects inflorescence branching (Satoh-Nagasawa et al., 2006). As in dicotyledonous plants, overexpression of genes for the pathway has produced benefits such as stress tolerance in rice (Garg et al., 2002; Ge et al., 2008). Of the little that has been documented on the trehalose pathway in wheat, active enzymes are present in roots and shoots (El-Bashiti et al., 2005) and transcripts in roots (Mohammadi et al., 2007) and the transcripts of TPS and TPP enzymes in wheat grain (Weichert et al., 2010). SnRK1 plays an important role in the regulation of metabolism and development in crop sinks such as potato (Solanum tuberosum) tubers (McKibbin et al., 2006) and in seeds of legumes (Weber et al., 2005; Radchuk et al., 2010). Interestingly, it has been proposed that SnRK1 mediates responses to sugar signals required for early cotyledon establishment and patterning (Radchuk et al., 2010). To our knowledge, Radchuk et al. (2010) presented the first T6P measurements in a seed tissue in the embryos of pea (Pisum sativum). The significance of these data relative to the regulation of SnRK1 is not known, however. No combined analysis of T6P and SnRK1 has yet been performed in the seed of any major crop. In cereals, studies from cDNA from rye (Secale cereale) endosperm resulted in the isolation of plant SnRK1 through complementation of a yeast (Saccharomyces cerevisiae) snf1 mutant (Alderson et al., 1991). Subsequent studies have shown that the SnRK1 family in cereals has diverged into two separate groups, of which SnRK1a is more similar to the dicotyledonous form, whereas SnRK1b is unique to monocotyledonous plants with high expression in seed (Hannappel et al., 1995). SnRK1 of barley (Hordeum vulgare) is thought to regulate storage product accumulation during grain filling (Sreenivasulu et al., 2006). In rice and wheat SnRK1 enables seeds to respond to anoxic and starvation conditions (Laurie et al., 2003; Lee et al., 2009).
Here we measure T6P content and SnRK1 activities during wheat grain development. Wheat grain consists largely of endosperm, embryo, and maternal pericarp tissue that go through phases of development defined by events in the endosperm. Three phases are typically defined as a pregrain-filling phase for the first 10 d or so after anthesis when cell division, expansion, and differentiation give the characteristic structure of the endosperm before the storage or grain-filling stage. The transition to the grain-filling stage is marked by dramatic transcriptional and physiological changes (Wan et al., 2008). It is thought that sugars and sugar signals play a role in this transition and in seed maturation, at least in legume seeds (Weber et al., 2005). The desiccation phase then follows beyond 30 d after anthesis (DAA). We show unprecedented levels of T6P during early grain development before the transition to grain filling. In whole grains, T6P correlated strongly with sugar content. In vitro SnRK1 activity was strongly inhibited by T6P. As evidence that regulation of SnRK1 by T6P could operate in vivo, homologs of SnRK1 marker genes designated in Arabidopsis (Baena-González et al., 2007) were identified in wheat using Wheat Estimated Transcript Server (WhETS; Mitchell et al., 2007). Three hundred each of the most highly expressed SnRK1-induced and SnRK1-repressed marker genes showed different patterns of expression during the pregrain-filling period compared to the grain-filling period consistent with changes in T6P content. Grain was dissected into filial and maternal tissues during these two periods at 7 and 17 DAA to better understand the role of T6P. T6P was at high concentrations in filial and maternal tissue pregrain filling, sufficient to inhibit SnRK1, but during grain filling T6P became restricted at high concentrations to the endosperm and was at low concentrations in pericarp and embryo. The data show a relationship between T6P and Suc overall that belies a marked effect of tissue type and developmental stage on T6P content, consistent with tissue-specific regulation of SnRK1 by T6P in wheat grain.
RESULTS
Wheat Grain Development Is Characterized by Unprecedented Accumulation of T6P during Prestorage Phase and Wide Dynamic Range
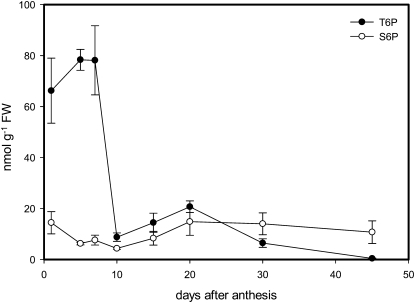
T6P levels changed 178-fold over the course of grain development between 1 and 45 DAA (Fig. 1). Between 1 and 7 DAA, pregrain-filling stage T6P levels were between 66 and 78 nmol g−1 fresh weight (FW), more than 100-fold higher than previously reported in Arabidopsis. Amounts then fell over 10 to 30 DAA to between 8 and 21 nmol g−1 FW during grain filling. By day 45 during the desiccation phase, amounts had decreased to 0.4 nmol g−1 FW, very similar to levels previously reported for T6P in Arabidopsis seedlings (Lunn et al., 2006; Paul et al., 2010). Amounts of Suc-6-P in comparison were stable between 4 and 14 nmol g−1 FW over the 45-d period.
Figure 1.
T6P and S6P contents in whole grains during wheat grain development 1 to 45 DAA. Means of four biological replicates with se of mean.
Suc and Suc Catabolites Correlate Strongly with Amounts of T6P during Grain Development
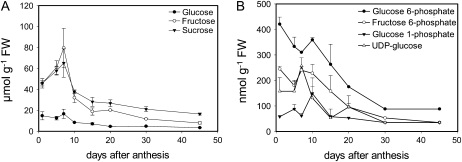
To establish why T6P levels could be so high in wheat grain and given that T6P levels are related to Suc supply in Arabidopsis (Lunn et al., 2006; Paul et al., 2010) we went on to measure sugars and closely related metabolites during grain development. The highest levels of Suc as for T6P were during early grain development between 40 and 65 μmol g−1 FW at 1 to 7 DAA (Fig. 2A). Amounts fell to between 20 and 40 μmol g−1 FW 10 to 45 DAA. Amounts of Fru were very similar to those of Suc; amounts of Glu were about 3-fold lower and showed a similar trend to Suc and Fru (Fig. 2A). Glc-6-P, UDP-Glc, Fru-6-P, and Glc-1-P, as for sugars, were highest between 1 and 10 DAA (Fig. 2B). After this period amounts of Glc-6-P, UDP-Glc, and Fru-6-P decreased to less than one-third of their values by day 45 compared to day 1. Amounts of Glc-1-P were relatively stable during grain development.
Figure 2.
Amounts of sugars, sugar phosphates, and UDP-Glc in whole grains during wheat grain development 1 to 45 DAA. A, Glc, Fru, and Suc. B, Glc 6-P, Fru 6-P, Glc 1-P, and UDP-Glc. Means of four biological replicates with se of mean.
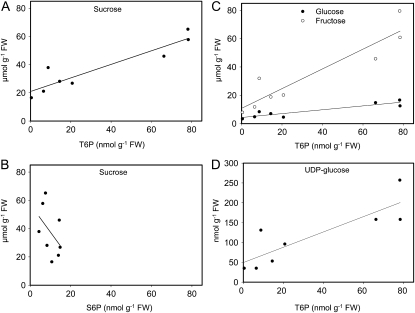
The interrelationship between Suc and T6P was determined during grain development. A strong positive relationship was obtained (Fig. 3A) with correlation coefficient 0.928 (P = 0.0009). The relationship of T6P and Suc is contrasted to that of Suc-6-P and Suc where a very different relationship was obtained (Fig. 3B, correlation coefficient −0.45). Correlation coefficients of T6P and Glc and T6P and Fru were also similar to Suc (0.928 and 0.919, respectively, P values 0.001; Fig. 3C). The correlation between Suc and UDP-Glc was also strong (correlation coefficient 0.853, P = 0.007; Fig. 3D).
Figure 3.
Correlation between T6P, sugars, and sugar phosphates and UDP-Glc. A, T6P and Suc. B, S6P and Suc. C, T6P and Glc and Fru. D, T6P and UDP-Glc.
Correlations between Glc-6-P, Fru-6-P, and T6P were less strong (correlation coefficients 0.665 and 0.676, respectively; Supplemental Fig. S1). There was no correlation between amounts of Glc-1-P and T6P (Supplemental Fig. S1B; correlation coefficient 0.071).
T6P Inhibition of SnRK1 Activity from Extracts at Different Stages of Grain Development
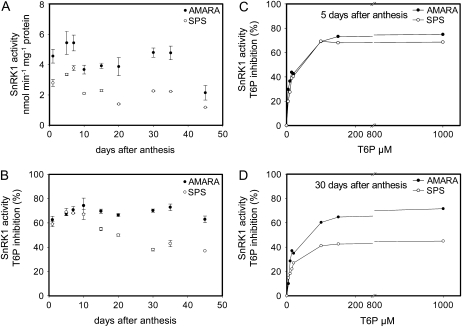
To determine if wheat grain SnRK1 is inhibited by T6P in vitro, SnRK1 activities were measured during grain development using both AMARA and SPS peptides as substrates. Desalting was carried out to remove endogenous T6P so that it could be added to the SnRK1 assay in defined amounts. Without T6P in the assay SnRK1 activity profiles were similar using AMARA or SPS; SnRK1 activities measured with AMARA were approximately 2-fold higher compared to activities with SPS as substrate (Fig. 4A). SnRK1 activities in desalted extracts with no T6P in the assay changed less than 3-fold over the course of development with highest values during 1 to 7 DAA (Fig. 4A). When T6P was included in SnRK1 assays with AMARA as substrate, SnRK1 activity was inhibited by between 62% and 74% at 1 mm T6P (Fig. 4B). This amount of inhibition by T6P was similar to that when SPS was used as substrate between 1 and 7 DAA. However, after 10 DAA the amount of inhibition by T6P in assays using the SPS peptide fell to about 40% (Fig. 4B). This would indicate a change in the nature of SnRK1 beyond 10 DAA detected by the SPS peptide. These measurements were confirmed at a wider range of T6P levels in grain harvested at 5 DAA and at 30 DAA (Fig. 4, C and D). SnRK1 was inhibited by 50% between 50 and 60 μm T6P at 5 DAA using both AMARA and SPS peptide and at 30 DAA using AMARA peptide, but inhibition was much less at 30 DAA using the SPS peptide.
Figure 4.
SnRK1 activity in whole grains during wheat grain development. A, Between 1 and 45 DAA using AMARA and SPS peptide as substrate. B, Between 1 and 45 DAA using AMARA and SPS peptide as substrate in the presence of 1 mm T6P in the assay expressed as percent inhibition. C, At 5 DAA using AMARA and SPS peptide as substrate in the presence of between 5 μm and 1 mm T6P. D, At 30 DAA using AMARA and SPS peptide as substrate in the presence of between 5 μm and 1 mm T6P. Means of four biological replicates with se of mean.
SnRK1 activities were also measured in fully expanded flag leaves over the same course of grain development for comparison (Supplemental Fig. S2A). Activities were up to 6-fold lower than in wheat grain and tended to increase during the post-anthesis period. When T6P was included in the SnRK1 assays, the amount of inhibition was far lower than in the grain (Supplemental Fig. S2B).
SnRK1 Marker Gene Expression Indicates Differential SnRK1 Activity during Wheat Grain Development
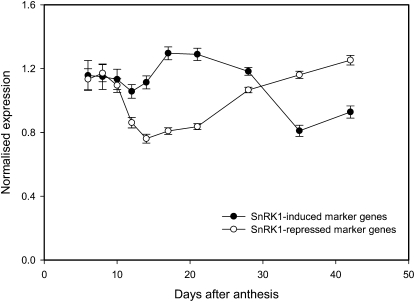
As a measure of SnRK1 activity in vivo, SnRK1 marker genes from Arabidopsis for which expression is repressed or induced by SnRK1 were taken from Baena-González et al. (2007). Corresponding wheat probesets were selected using the WhETS tool (Supplemental Table S1). For each list of 600 repressed or induced SnRK1 marker genes the top 300 most abundantly expressed in grain were selected. Figure 5 shows the average of the normalized expression for these sets during grain development. SnRK1-induced and SnRK1-repressed marker gene expression changed beyond the pregrain-filling period 10 DAA, indicating inhibition of SnRK1 activity before 10 DAA, but greater SnRK1 activity after 10 DAA coincident with changes in T6P levels (Fig. 1).
Figure 5.
SnRK1 target gene transcript abundance. Transcript abundance of wheat Affymetrix probesets corresponding to sets of 600 SnRK1-induced and 600 SnRK1-repressed Arabidopsis genes according to Baena-González et al. (2007) determined using WhETS tool (Mitchell et al., 2007). Log of normalized expression (i.e. divided by the median expression for that probeset) was averaged and se calculated. The resulting averages with se are displayed, back transformed to a linear scale. The 300 most abundantly expressed SnRK1-induced and 300 most abundantly SnRK1-repressed genes are presented.
Marked changes in grain development characterize the transition to grain filling. To increase understanding of the events pregrain-filling compared to grain-filling grain tissues were dissected at 7 DAA (pregrain filling) and 17 DAA (grain filling) and T6P levels and SnRK1 activities determined.
Analysis of T6P, SnRK1 Activity, and Suc in Dissected Grain
Grain tissue was separated into filial and maternal tissue. At 7 DAA pregrain-filling T6P concentrations were high in both filial tissue (endosperm, 119 nmol g−1 FW) and maternal tissue (outer pericarp, 49 nmol g−1 FW and inner chlorophyll-containing pericarp, 117 nmol g−1 FW; Table I). In contrast, at 17 DAA during grain filling T6P was largely restricted to the endosperm (43 nmol g−1 FW) with small amounts in the pericarp and embryo (0.6–0.8 nmol g−1 FW).
Table I. T6P levels in dissected maternal and filial grain tissues at 7 DAA compared with 17 DAA.
Data are expressed as nmol g−1 FW with se of mean of four replicates. ND, Not determined.
| Wheat Grain Tissue | 7 DAA | 17 DAA |
|---|---|---|
| Maternal tissue | ||
| Outer pericarp (white) | 49 ± 9.0 | 0.8 ± 0.4 |
| Inner pericarp (green) | 117 ± 8.1 | 0.7 ± 0.5 |
| Filial tissue | ||
| Endosperm | 119 ± 15 | 43 ± 4.2 |
| Embryo | ND | 0.6 ± 0.03 |
In contrast to the marked changes in T6P SnRK1 activities in vitro were similar at both 7 and 17 DAA (Table II). Activities in maternal tissue decreased from 7 to 17 DAA; whereas in endosperm SnRK1 activities were stable and embryo had the highest activities of all tissues at 17 DAA. SnRK1 activity in all tissues was inhibited in vitro by 1 mm T6P, although outer pericarp SnRK1 was only little affected by T6P at 17 DAA.
Table II. SnRK1 activity in dissected maternal and filial grain tissues at 7 DAA compared with 17 DAA.
Data are expressed as nmol min−1 mg−1 protein with se of mean of three replicates. Percentage inhibition with inclusion of 1 mm T6P in the assay in parentheses. ND, Not determined.
| Wheat Grain Tissue | 7 DAA | 17 DAA |
|---|---|---|
| Maternal tissue | ||
| Outer pericarp (white) | 3.7 ± 0.3 (60%) | 1.1 ± 0.1 (17%) |
| Inner pericarp (green) | 3.6 ± 0.4 (59%) | 2.1 ± 0.2 (42%) |
| Filial tissue | ||
| Endosperm | 3.2 ± 0.5 (65%) | 3.2 ± 0.5 (43%) |
| Embryo | ND | 4.5 ± 0.6 (47%) |
Suc was present in maternal and filial tissues pregrain filling and during grain filling (Table III), with 49 to 105 μmol g−1 FW at 7 DAA compared to 22 to 64 μmol g−1 FW at 17 DAA.
Table III. Suc in dissected maternal and filial grain tissues at 7 DAA compared with 17 DAA.
Data are expressed as μmol g−1 FW with se of mean of four replicates. ND, Not determined.
| Wheat Grain Tissue | 7 DAA | 17 DAA |
|---|---|---|
| Maternal tissue | ||
| Outer pericarp (white) | 49 ± 3.2 | 22 ± 2.3 |
| Inner pericarp (green) | 105 ± 6.5 | 64 ± 3.4 |
| Filial tissue | ||
| Endosperm | 74 ± 4.1 | 23 ± 1.9 |
| Embryo | ND | 41 ± 2.0 |
To better understand trehalose pathway and SnRK1 genes in wheat responsible for T6P levels and SnRK1 activities, respectively, probesets corresponding to Arabidopsis class I and class II TPS and TPP genes and SnRK1 genes were identified using WhETS.
Trehalose Pathway and SnRK1 Gene Expression during Wheat Grain Development
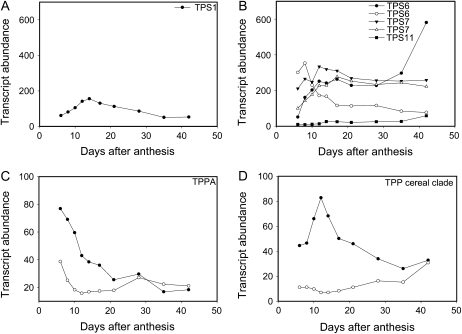
Contrasting expression patterns for TPS and TPP genes were obtained over the course of grain development (Fig. 6, A–D), including representative transcripts of the distinctive clade of TPP genes found only in grasses (Satoh-Nagasawa et al., 2006; Supplemental Fig. S3, A and B). A class I TPS ortholog to AtTPS1 known to be catalytically active in Arabidopsis (Blázquez et al., 1998) was expressed at low levels over the whole course of grain development (Fig. 6A). Class II TPS genes displayed contrasting patterns of expression (Fig. 6B). TPP genes most similar to Arabidopsis TPPA displayed highest patterns of expression during early grain development (Supplemental Fig. S3B; Fig. 6C). TPPs of the distinct clade of grass TPPs displayed distinctive patterns of expression (Fig. 6D).
Figure 6.
Transcript abundance of trehalose pathway genes during grain development. A, Class I TPS, Bradi2g19640. B, Class II TPS, Bradi1g69420 (○) and Bradi4g29730 (•; TPS6), Bradi2g49870 (▾) and Bradi2g19710 (△; TPS7), and Bradi3g53790 (▪; TPS11). C, TPP genes most closely related to Arabidopsis TPPA, Bradi3g50810 (•), Bradi3g32970 (○). D, TPP from the distinct clade of TPP genes found in grasses, Bradi1g21420 (•), Bradi4g29030 (○).
Transcripts for a number of SnRK1 genes were also present (Supplemental Fig. S4). Expression of an AKIN10-like gene was particularly prominent (Supplemental Fig. S4A) as previously observed, termed SnRK1b (Hannappel et al., 1995). Transcripts of genes of other subunits of the SnRK1 complex, β1, β3, γ, and βγ were also found (Supplemental Fig. S4, B–E).
DISCUSSION
T6P is an important regulatory molecule in plants with a large impact on metabolism, growth, and development (Eastmond et al., 2002; Schluepmann et al., 2003; Paul et al., 2008; Smeekens et al., 2010). We recently established a mechanistic basis for the signaling function of T6P in growing tissues through inhibition of SnRK1 of the SNF1/AMPK group of protein kinases (Zhang et al., 2009; Paul et al., 2010). A working model for the function of T6P in Arabidopsis is that it signals Suc availability to regulate SnRK1. In this article we establish, to our knowledge, the first link between T6P, Suc, and SnRK1 activities during grain development of a major cereal crop. Furthermore, we present data for T6P levels in different seed tissues at contrasting developmental stages and show large tissue specificity of T6P levels during grain filling.
All published data on amounts of T6P in plants are from Arabidopsis seedlings and leaf material with other data from pea embryos (Radchuk et al., 2010). But there has been no detailed and comprehensive analysis of T6P in the harvested parts of any major crop such as wheat. Given the important function of T6P in affecting growth processes and the high Suc content of sink tissues such as grain, knowledge of T6P content in such tissues is an important step for crop improvement through modification of the trehalose pathway. To our knowledge, we present the first data of T6P measurements in wheat grain. Remarkably, during most of the course of grain development amounts of T6P in whole grain far exceeded values reported for Arabidopsis in the region of 20- to 100-fold higher than previously observed. Between 1 and 7 DAA at pregrain-filling stage amounts of T6P were particularly high (66–78 nmol g−1 FW; Fig. 1). There was also a wide dynamic range of T6P over the course of grain development from 78 nmol g−1 FW at 5 DAA to 0.4 nmol g−1 FW at 45 DAA. The latter value at desiccation stage is comparable to levels normally found in Arabidopsis seedlings (Lunn et al., 2006). The dynamic range of T6P has been shown to be related to Suc supply (Lunn et al., 2006). To test the possibility that Suc availability could account for the levels of T6P we compared the pattern of T6P with that of sugars and sugar metabolites. First, we found that Suc levels in wheat grain were far higher (50-fold) than in Arabidopsis seedlings. Second, we found a strong correlation between amounts of Suc and amounts of T6P. Amounts of Suc during grain development changed in a pattern similar to T6P, over a smaller dynamic range (Fig. 2A). It is interesting to compare this with the interrelationship between Suc and Suc-6-P. Here the relationship was completely different (Fig. 3B), indicative of the different respective functions of the two compounds, Suc-6-P as a substrate for Suc and T6P as a signal of Suc availability. Good interrelationships were also found between T6P and the direct catabolites of Suc (hexoses and UDP-Glc; Fig. 3, C and D), indicating a relationship between T6P, Suc, and Suc breakdown and that T6P likely occurs where Suc is being consumed consistent with a role in biosynthetic processes. We then determined if SnRK1 could be regulated by T6P in wheat grain.
Activities of SnRK1 that reflect the maximum catalytic potential of the enzyme (without T6P in the assay) were similar to those previously reported for Arabidopsis seedlings (Fig. 4A; Zhang et al., 2009). Activities changed 3-fold over grain development. This is a relatively small change in catalytic activity compared to the changes in T6P content. Hence T6P is potentially a significant component of the regulation of SnRK1 in wheat grain. To establish this possibility, SnRK1 assays were performed including T6P in the assay. Inhibition of SnRK1 by T6P was seen in wheat grain extracts as observed in Arabidopsis (Fig. 4B; Zhang et al., 2009). Given the higher levels of T6P observed in wheat grain than in Arabidopsis the potential for inhibition of SnRK1 by T6P in vivo is significant. To gain insight into SnRK1 activity in vivo in wheat grain we took an approach analogous to that in Zhang et al. (2009) and analyzed the expression of genes known to be regulated by SnRK1 (Baena-González et al., 2007). Using the WhETS tool (Mitchell et al., 2007) we identified wheat transcripts that correspond most closely to genes induced by SnRK1 and repressed by SnRK1 in Arabidopsis and looked in the developing grain transcriptome from Wan et al. (2008). SnRK1-induced and SnRK1-repressed marker gene expression changed beyond the pregrain-filling period 10 DAA (Fig. 5), indicating inhibition of SnRK1 activity before 10 DAA, but greater SnRK1 activity after 10 DAA coincident with changes in T6P levels (Fig. 1).
The transition from pregrain filling to grain filling is characterized by large developmental changes. To gain better insight into the interaction between T6P and SnRK1 over this time T6P levels and SnRK1 activities were measured in filial and maternal grain tissues in dissected grain at 7 and 17 DAA coincident with pregrain-filling and grain-filling phases. At 7 DAA T6P levels were high in filial and maternal tissue at concentrations sufficient to inhibit SnRK1 in all tissues (Table I). SnRK1 activity was similar in all these tissues (Table II). In marked contrast at 17 DAA high concentrations of T6P were confined to the endosperm with very low amounts in embryos and pericarp. T6P levels in wheat embryos (0.6 nmol g−1 FW) are of a similar order to those in pea embryos (2 nmol g−1 FW; Radchuk et al., 2010) and more than 70-fold lower than amounts measured in endosperm and maternal tissues. SnRK1 was again active in all tissues, but it is likely that SnRK1 activity in pericarp and embryo is not inhibited by T6P at 17 DAA. The expression profile of SnRK1 marker genes at 7 and 17 DAA can be explained by differences in T6P concentrations at these times. Active SnRK1 is clearly important for embryo development (Radchuk et al., 2010) as is T6P in Arabidopsis embryos (Eastmond et al., 2002). However, T6P may not be sufficiently high to inhibit SnRK1 in embryos and regulation of SnRK1 by T6P may not be a primary function of T6P in this tissue. Interestingly, Suc is still present in all tissues at 17 DAA albeit 2- to 3-fold lower than at 7 DAA (Table III). While T6P and Suc correlate well when determined at the whole-grain level, this belies large tissue-specific changes that develop during the grain-filling period and where this relationship may become restricted to the endosperm. Further detailed analysis of the interrelationship between T6P and Suc contents in endosperm would confirm this. In contrast during pregrain filling T6P may be necessary for the early development of all grain tissues. Transcripts for a number of TPS and TPP genes known to be catalytically active in Arabidopsis to regulate T6P levels were present in developing grain including representatives from a distinct clade of TPPs found in grasses (Satoh-Nagasawa et al., 2006; Fig. 6; Supplemental Fig. S3B). TPS1 is constitutively expressed at low levels in Arabidopsis and catalytically active and a similar function may be retained in wheat. Most TPPs are thought to possess catalytic activity to regulate T6P content and display much greater tissue and cell specificity and developmental control (Schluepmann and Paul, 2009) that may regulate the large tissue-specific differences in T6P content during grain filling where T6P becomes restricted to the endosperm. The exact nature of the interrelationship between T6P, Suc, and trehalose pathway gene expression in the determination of the early grain development and yield of endosperm is the subject of further work.
In summary, we link T6P and SnRK1 in an important cereal crop and present data for T6P levels in different seed tissues at contrasting developmental stages. Unprecedented levels of T6P are reported for a plant species. The data show a relationship between T6P and Suc overall, which belies a marked effect of tissue type and developmental stage on T6P content, consistent with tissue-specific regulation of SnRK1 by T6P in wheat grain.
MATERIALS AND METHODS
Wheat Plants
Wheat (Triticum aestivum var. Cadenza) plants were grown in pots of soil containing Rothamsted standard compost mix and full nutrition in a glasshouse during summer with supplementary lighting to give a 16-h photoperiod and day/night temperature of 18°C/15°C. Samples were taken during the middle of the photoperiod and material was snap frozen immediately in liquid nitrogen and stored at −80°C until analysis. Ears were tagged at anthesis. The two outermost grains of the eight middle spikelets from an ear were sampled. These were combined together with four other similar samples from different ears that constituted one biological replicate. Data are means of four biological replicates. Experiments were repeated at least twice and representative data from one experiment are presented. Grain from 7 and 17 DAA were dissected under the growing conditions. Each grain was cut in half through the central groove and separated under a light microscope into maternal and filial tissues. At 7 DAA maternal tissue was separated into an outer white layer here called outer pericarp and the inner green chlorenchyma (chlorophyll-containing layers) of the pericarp. Embryo tissue was too small to dissect at this stage and filial tissue at 7 DAA is just endosperm. At 17 DAA embryo was dissected from the grain. Endosperm consisted mainly of the bulk of starchy endosperm and the aleurone layer. No clear observation was made concerning the fate of nucellar epidermis and integuments because they were too small. Data are means of four biological replicates. The two outermost grains of the eight middle spikelets from an ear were sampled. These were combined together with at least four other similar samples from different ears until enough material had been obtained for one biological replicate of at least 50 mg FW in the case of embryos and at least 100 mg FW in the case of other tissues. Whole grains and separated tissue were ground in liquid nitrogen in a pestle and mortar. Flag leaf samples are four biological replicates of individual flag leaves.
T6P Determinations
T6P was quantified in wheat grain extracts using anion-exchange HPLC coupled with electrospray ionization mass spectrometry (Delatte et al., 2009). The method achieved baseline resolution of all compounds from wheat grains with a mass-to-charge ratio ranging from 418 to 423 that eluted in the vicinity of T6P (Supplemental Fig. S5). Exogenous addition of the standards lactose-1-P (Sigma-Aldrich), Suc-6-P (Sigma-Aldrich), or maltose-6-P (Professor Jack Thompson, National Institutes of Health) showed that these compounds do not coelute with T6P.
Sugar, Sugar Phosphate, and UDP-Glc Determinations
Suc and hexoses, sugar phosphates, and UDP-Glc were measured using spectrophotometric assays as in Pellny et al. (2004).
SnRK1 Assays
SnRK1 activity was extracted from developing grain and from flag leaves, spin desalted, and assayed as in Zhang et al. (2009) using AMARA or SPS peptides as substrates. AMARA is as defined in Zhang et al. (2009); the SPS peptide, RDHMPRIRSEMQIWSED (Baena-González et al., 2007).
Bioinformatics
Transcript profiles were derived from the dataset reported by Wan et al. (2008) with the normalization protocols reported there. The full transcriptome set is available in the ArrayExpress database (accession no. E-MEXP-1193). Transcripts were identified by blasting Affymetrix probeset target sequences to find the closest known nucleotide sequences present in EMBL/GenBank/DDBJ. For analysis of SnRK1 marker genes sets of 600 Arabidopsis (Arabidopsis thaliana) genes for which expression is repressed or induced by SnRK1 were taken from Baena-González et al. (2007). Wheat Affymetrix probesets that corresponded most closely to these were identified using the WhETS tool (Mitchell et al., 2007). For each list, the top 300 most abundantly expressed in grain of these probesets were selected. Quantitative reverse transcription-PCR profiling of the expression of genes including SnRK1 markers myb3, Yab2 confirmed induction and repression, respectively, over the grain developmental period (Wan et al., 2008).
Phylogenetic Analysis
Brachypodium distachyon is a model grass that is the closest fully sequenced organism to wheat (International Brachypodium Initiative, 2010) and transcripts are labeled according to the closest Brachypodium gene model. From analysis of wheat ESTs, one TPP or TPS gene in Brachypodium corresponds to one gene on each of the three genomes of hexaploid wheat (data not shown). Phylogenetic trees of Arabidopsis (TAIR9) and B. distachyon (Bd21_v1 gene models) protein sequences were generated for the TPP and TPS gene families (Fig. 6, A and B). Trees were created with the PHYML program (Guindon et al., 2005), implementing the WAG model of amino acid substitution (Whelan and Goldman, 2001) from a full-length alignment (excluding gaps). Consensus trees from 100 bootstrap runs are shown and agree well with those previously published for Arabidopsis; trees from Brachypodium proteins closely resemble those published for rice (Oryza sativa; Lunn, 2007).
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure S1. Correlation between T6P and sugar phosphates.
- Supplemental Figure S2. SnRK1 activity in most recently fully expanded flag leaves.
- Supplemental Figure S3. Phylogenetic trees of Arabidopsis (TAIR9) and B. distachyon (Bd21_v1 gene models) protein sequences for the TPS and TPP gene families.
- Supplemental Figure S4. SnRK1 gene transcript abundance during grain development.
- Supplemental Figure S5. Separation of phosphodisaccharides with identical mass as T6P in wheat extracts using the LC/MS method described by Delatte et al. (2009).
- Supplemental Table S1. SnRK1 Arabidopsis marker genes taken from Baena-González et al. (2007) identified in wheat Affymetrix probesets using the WhETS tool (Mitchell et al., 2007).
Acknowledgments
The analysis of transcripts during grain development was made possible through funding to Professor Peter Shewry and Dr. Yongfang Wan by the Biotechnology and Biological Sciences Research Council (grant nos. EGA17694 and D/10608). We thank Dr. Sabine Gubatz and Dr. Till Pellny for showing us how to dissect and separate wheat grain into different tissue fractions. We thank Professor Peter Shewry and Professor Maurice Moloney for comments on a draft of the manuscript. We thank Dr. Stephen Powers for advice on statistical methods. We further thank Professor Nigel Halford for facilitating the visit of Dr. Eleazar Martínez-Barajas and for discussions.
References
- Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreis M, Shewry PR, Halford NG. (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci USA 88: 8602–8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. (2006) Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Santos E, Flores CL, Martínez-Zapater JM, Salinas J, Gancedo C. (1998) Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 13: 685–689 [DOI] [PubMed] [Google Scholar]
- Delatte TL, Selman MHJ, Schluepmann H, Somsen GW, Smeekens SC, de Jong GJ. (2009) Determination of trehalose-6-phosphate in Arabidopsis seedlings by successive extractions followed by anion exchange chromatography-mass spectrometry. Anal Biochem 389: 12–17 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA. (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- El-Bashiti T-E, Hamamci H, Oktem HA, Yucel M. (2005) Biochemical analysis of trehalose and its metabolising enzymes in wheat under abiotic stress conditions. Plant Sci 169: 47–54 [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L-F, Chao D-Y, Shi M, Zhu M-Z, Gao J-P, Lin H-X. (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228: 191–201 [DOI] [PubMed] [Google Scholar]
- Gómez LD, Baud S, Gilday A, Li Y, Graham IA. (2006) Delayed embryo development in the Arabidopsis trehalose 6-phosphate synthase 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46: 69–84 [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. (2005) PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res (Web Server issue) 33: W557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Hannappel U, Vicente-Carbajosa J, Barker JHA, Shewry PR, Halford NG. (1995) Differential expression of two barley SNF1-related protein kinase genes. Plant Mol Biol 27: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Hardie DG. (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P. (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, McKibbin RS, Halford NG. (2003) Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an α-amylase (α-Amy2) gene promoter in cultured wheat embryos. J Exp Bot 54: 739–747 [DOI] [PubMed] [Google Scholar]
- Lee K-W, Chen P-W, Lu C-A, Chen S, Ho T-HD, Yu S-M. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lunn JE. (2007) Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol 34: 550–563 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JH, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKibbin RS, Muttucumaru N, Paul MJ, Powers SJ, Burrell MM, Coates S, Purcell PC, Tiessen A, Geigenberger P, Halford NG. (2006) Production of high-starch, low-glucose potatoes through over-expression of the metabolic regulator SnRK1. Plant Biotechnol J 4: 409–418 [DOI] [PubMed] [Google Scholar]
- Mitchell RAC, Castells-Brooke N, Taubert J, Verrier PJ, Leader DJ, Rawlings CJ. (2007) Wheat estimated transcript server (WhETS): a tool to provide best estimate of hexaploid wheat transcript sequence. Nucleic Acids Res (Web Server issue) 35: W148–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Kav NNV, Deyholos MK. (2007) Transcriptional profiling of hexaploid wheat (Triticum aestivum L.) roots identifies novel, dehydration-responsive genes. Plant Cell Environ 30: 630–645 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Jhurreea D, Zhang Y, Primavesi LF, Delatte T, Schluepmann H, Wingler A. (2010) Upregulation of biosynthetic processes associated with growth by trehalose 6-phosphate. Plant Signal Behav 5: 386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59: 417–441 [DOI] [PubMed] [Google Scholar]
- Pellny TK, Ghannoum O, Conroy JP, Schluepmann H, Smeekens S, Andralojc J, Krause KP, Goddijn O, Paul MJ. (2004) Genetic modification of photosynthesis with E. coli genes for trehalose synthesis. Plant Biotechnol J 2: 71–82 [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M. (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschke W, Weber H. (2010) Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J 61: 324–338 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Paul M. (2009) Trehalose metabolism in Arabidopsis: elusive, active and central. The Arabidopsis Book 7: e0122, doi/10.1199/tab.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U. (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant J 47: 310–327 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. (1999) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken AJH, Schluepmann H, Smeekens SC. (2004) Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Poole RL, Huttly AK, Toscano-Underwood C, Feeney K, Welham S, Gooding MJ, Mills C, Edwards KJ, Shewry PR, et al. (2008) Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Weichert N, Saalbach I, Weichert H, Kohl S, Erban A, Kopka J, Hause B, Varshney A, Sreenivasulu N, Strickert M, et al. (2010) Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiol 152: 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RAC, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]