The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1–TRAF6 pathway (original) (raw)
Abstract
The atypical protein kinase C (aPKC)-interacting protein, p62, has previously been shown to interact with RIP, linking these kinases to NF–κB activation by tumor necrosis factor α (TNFα). The aPKCs have been implicated in the activation of IKKβ in TNFα-stimulated cells and have been shown to be activated in response to interleukin–1 (IL–1). Here we demon– strate that the inhibition of the aPKCs or the down-regulation of p62 severely abrogates NF–κB activation by IL–1 and TRAF6, suggesting that both proteins are critical intermediaries in this pathway. Consistent with this we show that p62 selectively interacts with the TRAF domain of TRAF6 but not that of TRAF5 or TRAF2 in co-transfection experiments. The binding of endogenous p62 to TRAF6 is stimulus dependent, reinforcing the notion that this is a physiologically relevant interaction. Furthermore, we demonstrate that the N–terminal domain of TRAF6, which is required for signaling, interacts with ζPKC in a dimerization-dependent manner. Together, these results indicate that p62 is an important intermediary not only in TNFα but also in IL–1 signaling to NF–κB through the specific adapters RIP and TRAF6.
Keywords: IL-1–TRAF6 pathway/NF–κB activation/p62/ζPKC
Introduction
The two members of the atypical protein kinase C (aPKC) subfamily of isozymes, ζPKC and λ/ιPKC, have been implicated in the control of important cellular functions such as cell proliferation and survival (Dominguez et al., 1992; Berra et al., 1993, 1997; Akimoto et al., 1996; Diaz–Meco et al., 1996a; Murray and Fields, 1997), probably through the regulation of critical signaling pathways such as those that activate the AP-1 and NF–κB transcription factors (Diaz-Meco et al., 1993, 1994a,b; Berra et al., 1995; Bjorkoy et al., 1995, 1997; Akimoto et al., 1996; Folgueira et al., 1996; Liao et al., 1997; Sontag et al., 1997; Schonwasser et al., 1998; Wang et al., 1999; Wooten, 1999; Wooten et al., 1999). In this regard, the inhibition of the aPKCs severely impairs the activation of the MEK–ERK cascade by mitogens (Berra et al., 1995; Liao et al., 1997) and the IKK system by tumor necrosis factor α (TNFα) (Lallena et al., 1999). The mechanisms whereby one given kinase can be involved in different cascades are not completely understood but could be related to the existence of specific anchors/scaffolds that confer selectivity to the kinase's effects (Mochly–Rosen, 1995; Mochly–Rosen and Gordon, 1998; Colledge and Scott, 1999). The aPKCs can be modulated by protein–protein interactions (Diaz–Meco et al., 1994b, 1996a,b; Puls et al., 1997; Izumi et al., 1998; Sanchez et al., 1998; Kuroda et al., 1999). Thus, both aPKCs, but not the classical or the novel isoforms, bind selectively to p62 (Sanchez et al., 1998), which is neither a regulator nor a substrate of these kinases but has a number of motifs that suggest a role as an adapter, potentially linking the aPKCs to receptor signaling complexes (Puls et al., 1997; Sanchez et al., 1998). In fact, this laboratory has recently found that p62 binds the adapter protein RIP connecting the aPKCs to the TNFα receptor (Sanz et al., 1999). RIP, a death domain kinase that associates with the TNF receptor 1 (TNF–R1) through its interaction with the adapter molecule TRADD (Hsu et al., 1996a,b), is a very important molecule in the activation of NF–κB by TNFα, since cells from RIP-deficient mice are severely impaired in the activation of NF–κB by this cytokine (Kelliher et al., 1998). On the other hand, the down-regulation of p62 with an antisense plasmid or the use of dominant-negative mutants dramatically inhibits the activation of NF–κB through the TNFα pathway (Sanz et al., 1999), strongly suggesting that the RIP–p62 interaction is critical for that function. TRAF2 is another protein that interacts with TRADD and RIP (Hsu et al., 1996b), but its role during NF–κB activation is not fully understood. Thus, although the overexpression of TRAF2 activates NF–κB, TRAF2-deficient cells display only minor alterations in the activation of this transcription factor by TNFα but show a completely impaired activation of the JNK/SAPK cascade (Lee et al., 1997; Yeh et al., 1997). Interestingly, TRAF2 did not show any interaction with p62 (Sanz et al., 1999).
NF–κB is also triggered in response to other inflam– matory cytokines such as interleukin–1 (IL–1). Previous results from other groups have demonstrated that the aPKCs are activated in IL–1-treated cells, which suggests that these kinases may play a critical role during IL–1 signaling, in a manner similar to that reported for the TNFα pathway (Rzymkiewicz et al., 1996; Herrera-Velit et al., 1997). In this regard, there is a notable parallel between both signaling cascades. Thus, the IL–1 receptor, together with the receptor accessory protein, interacts with MyD88, a functional analog of TRADD, which recruits the kinase IRAK, whose enzymatic activity, like that of RIP, is dispensable for the activation of NF–κB (Cao et al., 1996a). IRAK plays a critical role in NF–κB activation (Thomas et al., 1999) through its interaction with TRAF6, which is a required intermediary for IL–1 signaling (Cao et al., 1996b; Lomaga et al., 1999). Here, we demonstrate that p62 selectively interacts with TRAF6 and that it is an important intermediary in IL–1 signaling toward NF–κB activation.
Results
Role of the aPKCs and p62 in the activation of NF–κB by IL–1 signaling
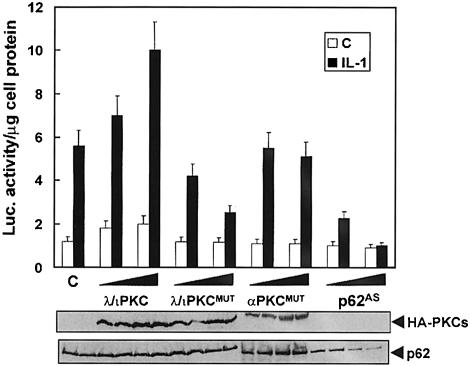
We initially determined whether dominant-negative mutants of these PKCs affected the activation of a κB-dependent reporter in IL–1-activated cells. Thus, HepG2 cells were transfected with a κB-dependent luciferase system along with either a control vector or two concentrations of expression plasmids for dominant-negative mutants of λ/ιPKC or ζPKC or the conventional PKC isoform, αPKC. The expression of the dominant-negative mutants of λ/ιPKC (Figure 1) or ζPKC (not shown), but not of αPKC (Figure 1), severely impaired NF–κB activation by IL–1. In contrast, the expression of wild-type λ/ιPKC favored the activation of NF–κB by IL–1 (Figure 1). We have previously shown that p62 is required for the activation of NF–κB by TNFα (Sanz et al., 1999). Therefore, in the next experiment we addressed whether this is also true for IL–1. HepG2 cells were transfected with two concentrations of a p62 antisense construct, and the ability of IL–1 to activate NF–κB was determined as above. Of potentially great interest, the depletion of p62 levels inhibited the activation of NF–κB by IL–1 (Figure 1).
Fig. 1. Role of λ/ιPKC and p62 in NF–κB activation by IL–1. Subconfluent cultures of HepG2 cells were transfected with 1 ng of the κB-luciferase reporter gene plasmid along with 1 or 2 μg of HA–tagged versions of wild-type or a dominant-negative mutant of λ/ιPKC, an HA–tagged dominant-negative mutant of αPKC, or the antisense p62 plasmid (p62AS), and enough empty vector to give 2.5 μg of total DNA. After 24 h, cells were stimulated or not with IL–1β (1 ng/ml for 4 h) and extracts were prepared and the levels of luciferase activity were determined as described in Materials and methods. Results are the mean ± SD of three independent experiments with incubations in duplicate. The panels below show representative control blots of the expression of the HA–tagged constructs and p62 levels from one of the experiments.
The aPKCs and p62 are required for the activation of NF–κB by TRAF6, IRAK and MyD88
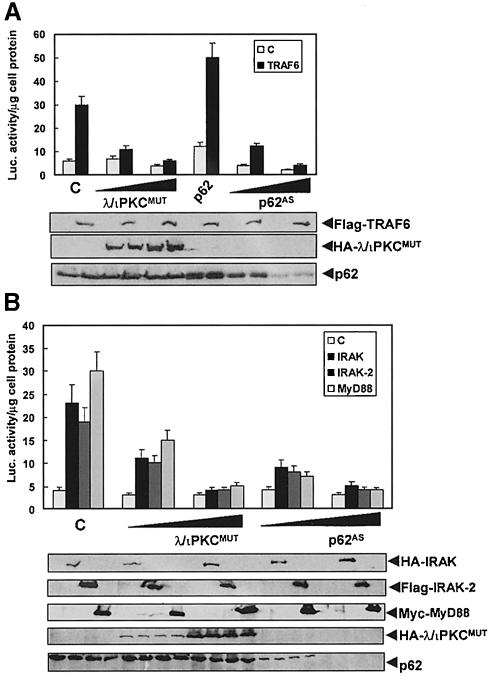
To address whether the aPKCs and/or p62 are required for TRAF6 actions, 293 cells were transfected with the κB-luciferase reporter along with either a plasmid control or a TRAF6 expression vector together with a p62 expression plasmid or increasing concentrations of λ/ιPKC or ζPKC dominant-negative mutants, or the p62 antisense construct. The results from these experiments demonstrate that the expression of dominant-negative λ/ιPKC (Figure 2A) or ζPKC (not shown), or the depletion of p62 (Figure 2A), impairs NF–κB activation by TRAF6 in a way similar to that reported for RIP (Sanz et al., 1999). Also, the overexpression of p62 favored the activation of NF–κB by TRAF6 (Figure 2A). IRAK is an important component upstream of TRAF6 in IL–1 signaling (see above). Therefore, it was of interest to determine whether the aPKCs and p62 are critical for IRAK signal transduction. To address this possibility, 293 cells were transfected with the κB-luciferase reporter along with either a plasmid control or expression vectors for IRAK or IRAK2 together with increasing concentrations of the λ/ιPKC dominant-negative mutant (Figure 2B) or the p62 antisense construct (Figure 2B). Interestingly, the expression of the λ/ιPKC mutant or the depletion of p62 severely abrogated NF–κB activation by the IRAKs, consistent with the notion that p62 and the aPKCs are targets of the IRAK–TRAF6 signaling pathway. Note that the ability of MyD88 to activate NF–κB is likewise inhibited by transfection of the dominant-negative mutant of λ/ιPKC or by depletion of p62 (Figure 2B).
Fig. 2. Role of λ/ιPKC and p62 in NF–κB activation by TRAF6, IRAK and MyD88. Subconfluent cultures of 293 cells were transfected with 1 ng of the κB-luciferase reporter gene plasmid along with 0.1 μg of Flag-TRAF6 (A), HA–IRAK, Flag-IRAK2 or Myc-MyD88 (B) with or without 1 or 2 μg of HA–λ/ιPKC dominant-negative mutant (HA–λ/ιPKC_MUT_) (A and B), 2 μg of an expression vector for HA–p62 (A), or the antisense p62 plasmid (p62_AS_) (A and B), and enough empty vector to give 2.5 μg of total DNA. After 24 h, cell extracts were prepared and the levels of luciferase activity were determined. Results are the mean ± SD of three independent experiments with incubations in duplicate. The panels below show representative control blots of the expression of the differently tagged constructs and p62 levels from one of the experiments.
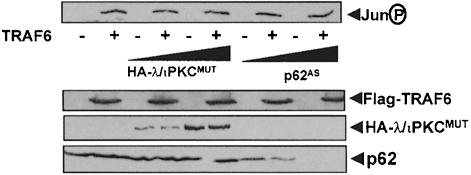
TRAF6 is not only involved in NF–κB but is also important for JNK activation in response to IL–1. In order to determine whether p62 and the aPKCs play any role in the JNK branch of the TRAF6 signaling pathway, 293 cells were transfected with the TRAF6 expression vector along with a plasmid control or expression vectors for the λ/ιPKC dominant-negative mutant or the p62 antisense construct. Afterwards, the activity of the JNK pathway was determined in cell extracts by immunoblotting with an anti-phospho-Jun antibody. Results in Figure 3 show that neither the λ/ιPKC mutant nor the depletion of p62 significantly inhibited the activation of JNK by TRAF6.
Fig. 3. Lack of a role for p62 and λ/ιPKC in TRAF6-induced JNK activation. Subconfluent cultures of 293 cells were transfected with 3 μg of Flag-TRAF6 with or without 6 or 10 μg of HA–λ/ιPKC dominant-negative mutant (HA–λ/ιPKC_MUT_) or the antisense p62 plasmid (p62_AS_), and enough empty vector to give 20 μg of total DNA. After 24 h, cell extracts were prepared and phospho-Jun was determined by immunoblot analysis with an anti-phospho-Jun antibody. Extracts were also immunoblotted with antibodies for the Flag and HA epitopes, as well as for p62. This is a representative experiment of another two with similar results.
TRAF6 interacts with p62 in vivo
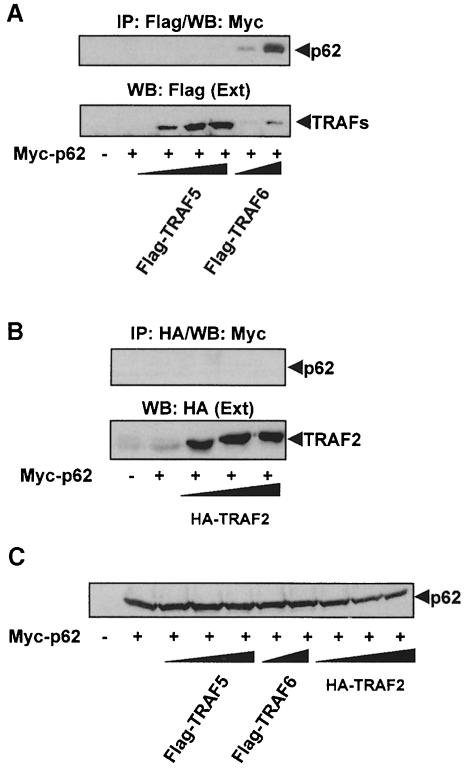
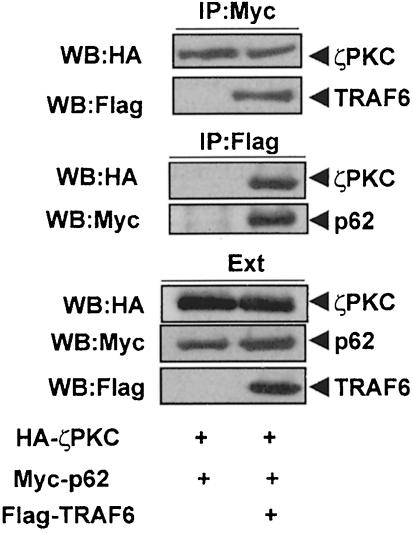
Because p62 binds to RIP in a TNFα-inducible manner (Sanz et al., 1999), and TRAF6 is a critical intermediary during NF–κB activation by IL–1 (Cao et al., 1996b; Kelliher et al., 1998; Lomaga et al., 1999), we wanted to determine in the next series of experiments whether p62 can interact with TRAF6. To address this possibility, 293 cells were transfected with a Myc-tagged version of p62 along with increasing concentrations of Flag-tagged TRAF6 or TRAF5, or an HA–tagged TRAF2 construct. These three are the only TRAFs involved in NF–κB activation (Arch et al., 1998). Cell extracts were immunoprecipitated with anti-Flag or anti-HA antibody and the immunoprecipitates were analyzed by immunoblotting with an anti-Myc antibody. Results in Figure 4A and B demonstrate that p62 co-immunoprecipitates with TRAF6 but not with TRAF5 or TRAF2. Since these were overexpression experiments the interaction of p62 with TRAF6 was ligand independent. Note that neither TRAF5 nor TRAF2 interacted with p62 even in the presence of IL–1 or TNFα (not shown). In addition, the experiments in Figure 5 show that the presence of TRAF6 does not affect the ability of p62 to interact with ζPKC and that the three proteins can be co-immunoprecipitated in a ternary complex.
Fig. 4. Interaction in vivo of p62 with TRAF6 but not with TRAF2 or TRAF5. Subconfluent cultures of 293 cells in 100–mm-diameter plates were transfected with 4 μg of the Myc-p62 expression plasmid either alone or in combination with 15 or 20 μg of Flag-TRAF6, 2.5, 5 or 10 μg of Flag-TRAF5 (A) or 2, 4 or 6 μg of HA–TRAF2 (B) and enough pCDNA3 plasmid to give 25 μg of total DNA. After transfection (24 h), cell extracts were immunoprecipitated with anti–Flag (A) or anti-HA (B) antibodies, after which the immunoprecipitates were analyzed by immunoblotting with an anti-Myc antibody. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the corresponding anti-tag antibodies. The gel shown in (C) demonstrates that the amount of Myc-p62 expressed was comparable in all the transfection points. Essentially identical results were obtained in another three independent experiments.
Fig. 5. ζPKC forms a ternary complex with p62 and TRAF6. Subconfluent cultures of 293 cells in 100–mm-diameter plates were transfected with 4 μg of Myc-p62 along with 5 μg of HA–ζPKC expression plasmid either alone or in combination with 20 μg of Flag-TRAF6 and enough pCDNA3 plasmid to give 30 μg of total DNA. After transfection (24 h), cell extracts were immunoprecipitated with anti-Myc or anti-Flag antibodies, after which the immunoprecipitates were analyzed by immunoblotting with anti-Flag, anti-Myc or anti-HA antibodies. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the corresponding anti-tag antibodies. Essentially identical results were obtained in another two independent experiments.
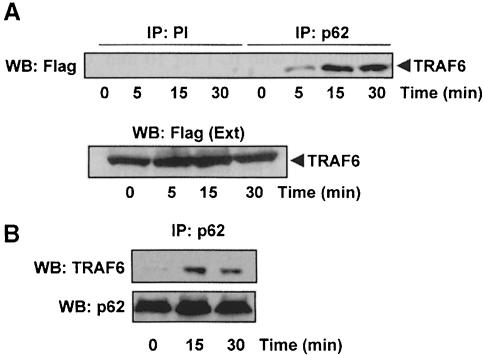
Collectively these results indicate that p62 specifically interacts with TRAF6 in vivo to an extent comparable to that of RIP (Sanz et al., 1999). To demonstrate this interaction in a more physiological setting, we generated HepG2 cells expressing a Flag-tagged version of TRAF6 to levels that were just 1.5- to 2–fold higher than the endogenous protein. These cells were either untreated or stimulated with IL–1 for different times, after which extracts were prepared and endogenous p62 was immunoprecipitated with a polyclonal affinity-purified anti-p62 antibody. As a control, cell extracts were also immunoprecipitated with a pre-immune serum. Interestingly, in the absence of IL–1 there is little or no association of endogenous p62 with TRAF6 (Figure 6A). However, the addition of IL–1 promoted a rapid and reproducible association of both proteins (Figure 6A). Next we analyzed the interaction of both endogenous molecules. Cell extracts from HepG2 cells treated or not with IL–1 for different times were incubated with the anti-p62 antibody to immunoprecipitate the endogenous p62. Afterwards, these immunoprecipitates were analyzed by immunoblotting with an anti-TRAF6 antibody. Results in Figure 6B demonstrate that there is little or no association of endogenous p62 and TRAF6 in unstimulated cells but that this interaction becomes evident upon IL–1 stimulation. These results, together with those demonstrating the critical role of p62 in IL–1-induced activation of NF–κB, strongly suggest that the TRAF6–p62 interaction is functionally relevant.
Fig. 6. The interaction of endogenous p62 with TRAF6 is IL–1 dependent. (A) Subconfluent cultures of HepG2 cells in 100–mm-diameter plates were transfected with 25 μg of an expression vector for Flag-TRAF6. Twenty-four hours post-transfection, cells were either untreated or stimulated with IL–1 (1 ng/ml) for different times. Afterwards, cell extracts were immunoprecipitated with a pre-immune or an anti-p62 polyclonal antibody, and the immunoprecipitates were analyzed by immunoblotting with a monoclonal anti-Flag antibody. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the anti-Flag antibody. Essentially identical results were obtained in another three independent experiments. (B) Subconfluent cultures of HepG2 cells in 100–mm-diameter plates stimulated or not with IL–1 (1 ng/ml) for different times. Afterwards, cell extracts were immunoprecipitated with the polyclonal anti-p62 antibody, and the immunoprecipitates were analyzed by immuno- blotting with the monoclonal anti-TRAF6 and anti-p62 antibodies. Essentially identical results were obtained in another two independent experiments.
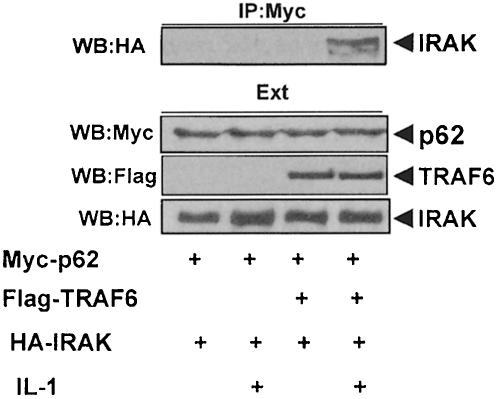
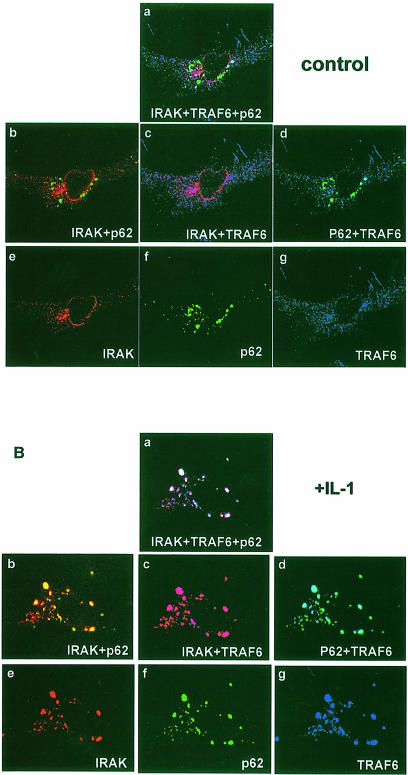
IRAK is a critical component in IL–1 signaling upstream of TRAF6 (see above). Therefore, it was of interest to investigate whether p62 could form a complex with IRAK through TRAF6. Thus, HepG2 cells were transfected with the expression vectors for HA–IRAK and Myc-p62 either with or without Flag-TRAF6, after which cells were either untreated or stimulated with IL–1. Cell extracts were immunoprecipitated with anti-Myc antibody to immunoprecipitate p62, and the immunoprecipitates were analyzed with anti-HA antibody to detect the associated IRAK. Results in Figure 7 demonstrate that IRAK co-immunoprecipitates with p62 only in stimulated cells that have been transfected with TRAF6, indicating that TRAF6 actually forms a bridge between IRAK and p62. In order to obtain other evidence of the existence of a p62–TRAF6–IRAK complex, we transfected HepG2 cells with Flag-TRAF6 and HA–IRAK, along with a green fluorescent protein (GFP)–p62 construct, after which cells were either untreated or stimulated with IL–1 for 10 min. Cells were then analyzed by triple confocal laser scanning microscopy with a rabbit polyclonal anti-HA antibody and an anti-rabbit Alexa 594 antibody (red fluorescence) to detect IRAK, and a monoclonal anti-Flag antibody and an anti-mouse Cy5 antibody (blue fluorescence) to detect TRAF6. Green fluorescence indicated the expression of GFP–p62. The expression of GFP–p62 gives a characteristic punctuate cytosolic staining (Figure 8A and B, panels a, b, d and f; Sanchez et al., 1998). According to the data shown in Figure 8A, in unstimulated HepG2 cells there was a partial colocalization of TRAF6 and IRAK detected as a pink signal in panels a and c. However, there was little or no colocalization of TRAF6 and p62 (Figure 8A, panel d) probably due to the fact that the expression levels of both proteins are lower in these cells than in 293 cells, in which the interaction between both proteins is easily detected in the absence of any stimulus. The observation that there is at least a partial colocalization of IRAK with TRAF6 in unstimulated HepG2 cells (Figure 8A, panels a and c) may indicate that the affinity of their interaction may be stronger than that of TRAF6 with p62. Note that there is no colocalization at all of IRAK with p62 (Figure 8A, panels a and b). However, when HepG2 cells were exposed to IL–1, there was a dramatic relocalization of the IRAK–TRAF6 complex to the p62 signal. Thus, the data in Figure 8B show a very good colocalization of IRAK and TRAF6 with p62, detected as a yellow signal in panel b, pink in panel c and light blue in panel d. In panel a the signal is white, indicative of the colocalization of the three molecules.
Fig. 7. TRAF6 links p62 to IRAK. Subconfluent cultures of HepG2 cells in 100–mm-diameter plates were transfected with 4 μg of the Myc-p62 expression plasmid either alone or in combination with 4 μg of HA–IRAK with or without 15 μg of Flag-TRAF6 and enough pCDNA3 plasmid to give 25 μg of total DNA. After transfection (24 h), cells were either untreated or stimulated with IL–1 for 10 min. Cell extracts were immunoprecipitated with an anti-Myc antibody, after which the immunoprecipitates were analyzed by immunoblotting with an anti-HA antibody. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the corresponding anti-tag antibodies. Essentially identical results were obtained in another two independent experiments.
Fig. 8. Colocalization of p62 with TRAF6 and IRAK in IL–1-stimulated cells. HepG2 cells were transfected with Flag-TRAF6 and HA–IRAK along with a GFP–p62 construct, after which cells were either untreated (A) or stimulated with IL–1 for 10 min (B). Cells were then analyzed by triple confocal laser scanning microscopy with a rabbit polyclonal anti-HA antibody and an anti-rabbit Alexa 594 antibody (red fluorescence) to detect IRAK, and a monoclonal anti-Flag antibody and an anti-mouse Cy5 antibody (blue fluorescence) to detect TRAF6. Green fluorescence indicates the expression of GFP–p62. Essentially identical results were obtained in another two independent experiments.
Mapping the regions of p62 responsible for the interaction with TRAF6
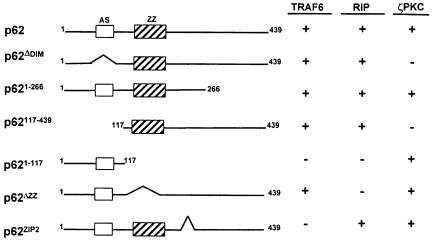
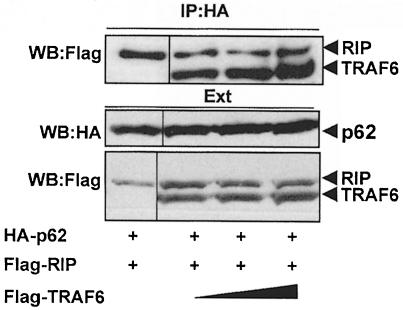
In order to determine which region of p62 is required for the interaction with TRAF6, differently tagged p62 deletion mutants (Figure 9) were transfected into 293 cells along with Flag-tagged TRAF6, and their association was determined by immunoprecipitation assays as above. As reported previously, the region encompassing amino acids 1–117 of p62 is necessary and sufficient to account for the interaction with ζPKC but not with RIP or TRAF6 (Figure 9; Sanz et al., 1999). This region contains a novel acidic motif (amino acids 69–81) homologous to a sequence of the yeast protein CDC24. Interestingly, the simple deletion of that motif is sufficient to abolish the interaction of p62 with ζPKC with no effect on the binding of RIP or TRAF6. Similarly to what has been reported for RIP (Sanz et al., 1999), the expression of the p62 amino acids 1–266 or 117–439 is sufficient to account for TRAF6 interaction (Figure 9), whereas deletion of amino acids 117–439 completely abolishes it (Figure 9). These results indicate that the region encompassing amino acids 117–266 is required for the interaction of p62 with TRAF6. Note that this is the same region that is responsible for the binding of p62 to RIP in the TNFα signaling cascade (Sanz et al., 1999). Interestingly, the expression of this fragment that is dominant-negative for the activation of NF–κB by RIP (Sanz et al., 1999) also inhibits the TRAF6 effects on this parameter (not shown). This region includes the ZZ domain, a novel atypical zinc-finger motif of unknown function (Ponting et al., 1996). To determine whether this ZZ region is important for the interaction of p62 with RIP or TRAF6, it was deleted to generate the p62ΔZZ mutant. The potential interaction of an HA–tagged version of this construct with Flag-tagged TRAF6 or RIP was assessed as above. Interestingly, the deletion of the ZZ region completely abrogated the interaction of p62 with RIP but not with TRAF6 (Figure 9). This is the first evidence for a role of a ZZ motif as a protein–protein interaction module. We next sought to determine which region of p62 could account for the interaction with TRAF6. In this regard, the existence of an isoform of p62, termed ZIP2, which is specifically expressed in brain, has recently been reported (Gong et al., 1999). This isoform is generated by alternative splicing and lacks amino acids 225–251. The results of the experiments summarized in Figure 9 demonstrate that the deletion of these 27 amino acids severely impairs the association of TRAF6 but not of RIP to p62. Together these results indicate that p62 can anchor both RIP and TRAF6 simultaneously and that they will not compete for the same binding site in p62. This conclusion is consistent with the results in Figure 10 showing that the expression of increasing amounts of TRAF6 does not affect RIP binding to p62.
Fig. 9. Schematic representation of the various constructs used for the mapping of the p62–TRAF6 and p62–RIP interaction domains. Constructs were prepared as described in Materials and methods. AS, acidic sequence and ζPKC-interacting region; ZZ, zinc-finger domain.
Fig. 10. Simultaneous interaction of p62 with TRAF6 and RIP. Subconfluent cultures of 293 cells in 100–mm-diameter plates were transfected with 4 μg of HA–p62 expression plasmid along with 5 μg of Flag-RIP either alone or in combination with 5, 10 or 15 μg of Flag-TRAF6 and enough pCDNA3 plasmid to give 25 μg of total DNA. After transfection (24 h), cell extracts were immunoprecipitated with anti-HA antibody, after which the immunoprecipitates were analyzed by immunoblotting with anti-Flag antibody. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the corresponding anti-tag antibodies. Essentially identical results were obtained in another three independent experiments.
Interaction of p62 with the C–terminal domain of TRAF6
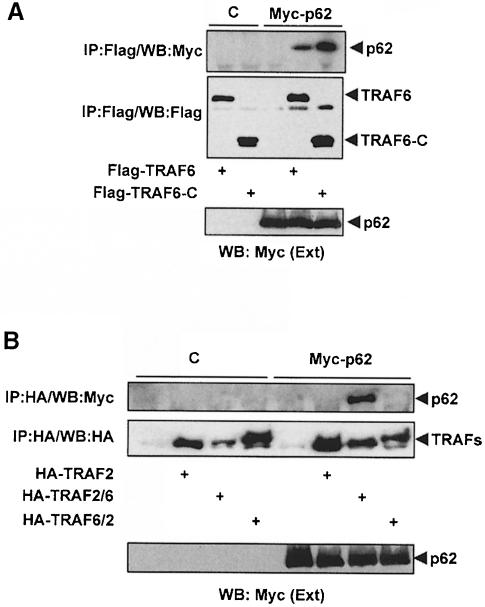
All TRAFs, except TRAF1, have an N–terminal region consisting of a ring and several zinc-fingers, and a conserved C–terminal module (TRAF domain) containing an N–proximal TRAF–N coiled-coil sequence and a C–proximal TRAF–C domain (Arch et al., 1998). In order to determine which sequences in the TRAF6 molecule are responsible for the interaction with p62, 293 cells were transfected with a control vector or an expression plasmid for Myc-p62 along with a Flag-tagged construct consisting of the TRAF domain (including the N- and C–proximal regions). For comparison, cells were also transfected with the Flag-tagged full-length TRAF6 expression vector. The immunoprecipitation experiments shown in Figure 11A demonstrate that p62 binds the TRAF domain of TRAF6 to an extent comparable to that of the full-length molecule. The TRAF domain is the most conserved region in all the TRAFs. However, TRAF6 shares only a 29% identity in this sequence, which makes it the most divergent (Cao et al., 1996b; Ishida et al., 1996; Arch et al., 1998). This may explain why p62 is capable of binding selectively to TRAF6 and not to TRAF2 or TRAF5.
Fig. 11. Interaction of p62 with the TRAF domain of TRAF6. Subconfluent cultures of 293 cells were transfected with expression plasmids for Flag-tagged wild-type TRAF6 (15 μg) or a mutant in which the N–terminal domain of TRAF6 was deleted (20 μg; TRAF6–C) (A), or with HA–TRAF2 (7.5 μg) or 25 μg of the HA–tagged chimeric constructs TRAF2/6 or TRAF6/2 (B) along with 4 μg of a control vector or a Myc-p62 expression plasmid. Cell extracts were immunoprecipitated with anti-Flag (A) or anti-HA (B) antibodies and the immunoprecipitates were analyzed by immunoblotting with anti-Myc (A and B), anti-Flag (A) or anti-HA (B) antibodies. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the anti-Myc antibody. Essentially identical results were obtained in another three independent experiments.
The TRAF domain serves as a receptor docking and oligomerization region as well as the recruitment module for signaling molecules in the NF–κB pathway such as RIP2 (McCarthy et al., 1998) or ECSIT (Kopp et al., 1999), which like p62 are selective for TRAF6. On the other hand, deletion of the N–terminal region of TRAF6 containing the ring and zinc-fingers not only suppresses the ability of TRAF6 to activate NF–κB (Cao et al., 1996b; Arch et al., 1998; Baud et al., 1999) but also generates a dominant-negative molecule. This indicates that the N–terminal region must be intact for TRAF6 to be able to signal and that mutants lacking that domain may keep the signaling molecules in an inactive complex unable to activate downstream components of the pathway. To determine whether p62 may also bind the N–terminal region of TRAF6, 293 cells were transfected with a control vector or an expression plasmid for an HA–tagged mutant of TRAF6 in which the TRAF domain was deleted (TRAF6ΔC), along with a control vector or a Myc-p62 expression plasmid. Cell extracts were immunoprecipitated with an anti-HA antibody and the immunoprecipitates were analyzed by immunoblotting with an anti-Myc antibody. Results from these experiments demonstrate that there is not a detectable association of p62 with the N–terminal portion of TRAF6 (not shown). This indicates that p62, like ECSIT (Kopp et al., 1999), interacts with the TRAF domain of TRAF6. However, Karin and co-workers have recently shown that the oligomerization of TRAF6 and TRAF2 is a required event for interactions in the N–terminal domain to occur (Baud et al., 1999). To address the possibility that oligomerization could trigger the ability of the N–terminal domain of TRAF6 to interact with p62, we took advantage of the fact that p62 does not bind TRAF2, and that the C–terminal region of TRAF2, like that of TRAF6, is capable of forming oligomers upon overexpression (Arch et al., 1998). Thus, we generated a chimeric molecule (TRAF6/2) in which the N–terminal domain of TRAF6 was fused to the C–terminal region of TRAF2. As a control, we generated the complementary construct (TRAF2/6) by fusing the N–terminal portion of TRAF2 to the TRAF domain of TRAF6. HA–tagged versions of these two constructs and of wild-type TRAF2, as a negative control, were transfected into 293 cells along with the Myc-p62 plasmid. Cell extracts were immunoprecipitated with the anti-HA antibody and the immunoprecipitates were analyzed by immunoblotting with an anti-Myc antibody. Results in Figure 11B show that p62 coprecipitated with TRAF2/6 but not with TRAF6/2. As expected, the TRAF2 control did not bind p62 (Figure 11B). These results are consistent with the data in Figure 11A, demonstrating that p62 binds to the TRAF domain of TRAF6 and indicating that the presence of the N–terminal region of TRAF2 does not affect this interaction. The lack of interaction of p62 with the TRAF6/2 mutant indicates that the N–terminal domain of TRAF cannot interact, even in its oligomerized form, with p62.
Interaction of ζPKC with the N–terminal effector domain of TRAF6
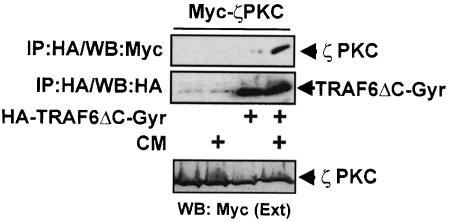
Altogether these results suggest that there is a striking parallel between p62 and ECSIT. Thus, p62 interacts with the aPKCs (Sanchez et al., 1998) that have been implicated in the activation of IKKβ (Lallena et al., 1999). ECSIT interacts with MEKK–1 (Kopp et al., 1999), a kinase that like the aPKCs has been reported to activate the IKK system (Lee et al., 1998; Baud et al., 1999). Interestingly, both p62 and ECSIT (Kopp et al., 1999) bind the TRAF domain of TRAF6. It should be noted, however, that MEKK–1 also interacts with the N–terminal domain of TRAF6 in an oligomerization-dependent manner (Baud et al., 1999). These observations could be explained by a model whereby MEKK–1 is linked to TRAF6 through two different modes: to the TRAF domain through the interaction with ECSIT, and to the N–terminal effector region by direct interaction. We reasoned that a similar model could apply for the p62–aPKC system. Thus, the aPKCs could be brought to the TRAF6 signaling complex through the interaction of p62 with the C–terminal region and by direct interaction with the N–terminal domain, the latter being dimerization dependent like that of MEKK–1 (Baud et al., 1999). To address this possibility, we fused the N–terminal domain of TRAF6 (TRAF6ΔC) to the B subunit of the bacterial DNA gyrase (GyrB) to generate the construct HA–TRAF6ΔC-Gyr. The Streptomyces product coumermycin (CM) binds GyrB with a stoichiometry of 1:2, acting as a natural dimerizer of GyrB (Farrar et al., 1996). Upon cell incubation with CM, chimeras that contain GyrB undergo dimerization (Farrar et al., 1996). Therefore, 293 cells were then transfected with the HA–TRAF6ΔC-Gyr construct along with Myc-ζPKC. Cells were incubated or not with CM for 15 min, after which the HA–TRAF6ΔC-Gyr protein was immunoprecipitated and the association of Myc-ζPKC was determined by immunoblotting with the anti-Myc antibody. Results in Figure 12 demonstrate that ζPKC was unable to interact with the N–terminal domain of TRAF6 in its monomeric form but that incubation with CM dramatically triggers that interaction. The same result was obtained when the interaction with the kinase-inactive mutant of ζPKC was investigated (not shown). Note that p62 was unable to interact with the N–terminal domain of TRAF6 in this system (not shown).
Fig. 12. The N–terminal domain of TRAF6 interacts with ζPKC in a dimerization-dependent manner. Subconfluent cultures of 293 cells in 100–mm-diameter plates were transfected with 10 μg of either pCDNA3 or an expression plasmid for the HA–TRAF6ΔC-Gyr construct along with 7.5 μg of a Myc-ζPKC expression vector. After transfection (24 h), cells were stimulated or not with CM for 15 min, after which cell extracts were immunoprecipitated with an anti-HA antibody and the immunoprecipitates were analyzed with anti-Myc and anti-HA antibodies. An aliquot [one-tenth of the amount of extract (Ext) used for the immunoprecipitation] was loaded in the gels and analyzed by immunoblotting with the anti-Myc antibody. Essentially identical results were obtained in another three independent experiments.
Discussion
Previous results from this laboratory have demonstrated that the aPKC-interacting protein p62 binds to the intermediary domain of RIP in a TNFα-dependent manner (Sanz et al., 1999). This interaction is potentially of great relevance because it serves to link these kinases to NF–κB activation in the TNFα signal transduction pathway. In fact, the down-regulation of p62 with an antisense plasmid or its inhibition by transfection of dominant-negative mutants of this protein severely impairs the activation of NF–κB by RIP but not by TRAF2 (Sanz et al., 1999). The aPKCs have previously been implicated in IL–1 signaling (Rzymkiewicz et al., 1996; Herrera-Velit et al., 1997). Here we show that the interaction of p62 with TRAF6 may account for the involvement of the aPKCs in this pathway. Thus, p62 interacts with TRAF6 but not with TRAF2 or TRAF5, indicating that the binding of p62 to TRAF6 is specific. This is in marked contrast to other TRAF-interacting proteins like the kinase NIK, which interacts indiscriminately with all TRAFs (Song et al., 1997). In addition, the TRAF6–p62 interaction appears to be of functional relevance since transfection of an aPKC dominant-negative mutant or the down-regulation of p62 not only impaired the activation of NF–κB by IL–1 but also by TRAF6 and its upstream regulators IRAK and MyD88. Therefore, a picture emerges whereby p62 acts as an adapter connecting the aPKCs to different NF–κB pathways through the interaction with either RIP or TRAF6. We report here that while the ZZ zinc-finger domain of p62 is critical for the interaction with RIP, the interaction with TRAF6 is mediated by a region that is missing in the ZIP2 isoform of p62. This isoform is selectively expressed in brain and in differentiated neurons (Gong et al., 1999). The significance of this observation in the activation of NF–κB in neurons is intriguing, especially in light of the proposed role of TRAF6 in NGF signaling toward NF–κB activation (Khursigara et al., 1999). In addition, we show here for the first time that the ZZ ‘atypical’ zinc-finger serves as a protein–protein interaction module. However, this is not completely unexpected as, for example, we have previously demonstrated that the zinc-finger is a critical region where the aPKCs interact with proteins such as LIP (Diaz–Meco et al., 1996b) and Par–4 (Diaz–Meco et al., 1996a).
TRAF6 has been shown to bind at least two other proteins involved in IL–1 signaling pathways. The kinase TAK1 and its regulator TAB1 were found to associate with TRAF6 and to mediate the activation of the IKK complex through NIK-dependent (Ninomiya-Tsuji et al., 1999) or –independent (Sakurai et al., 1999) pathways. The other TRAF6-binding protein described is ECSIT, a molecule cloned by the two-hybrid system using full-length TRAF6 as a bait (Kopp et al., 1999). ECSIT interacts with the TRAF domain of TRAF6 but not with TRAF2 or TRAF5. Interestingly, ECSIT also interacts with MEKK–1, which has been implicated in the activation of NF–κB through the phosphorylation of the IKK complex (F.S.Lee et al., 1997, 1998; Baud et al., 1999). Therefore, there is a remarkable functional similarity between ECSIT and p62, since both bind the upstream activator TRAF6 and both interact with a downstream kinase proposed to act upstream of the IKK complex, MEKK–1 and aPKC, respectively.
The TRAF proteins display a very interesting structure. All except TRAF1 have an N–terminal region consisting of a ring and several zinc-fingers and a conserved C–terminal portion denoted the TRAF domain (Arch et al., 1998). This conserved region in TRAF6 is the most divergent among all the TRAFs, displaying only a 29% identity at the amino acid level. The low homology displayed by this TRAF domain may explain why ECSIT and p62 can interact selectively with TRAF6 and show no interaction at all with TRAF2 or TRAF5. In addition, the TRAF domain is responsible for the oligomerization of the molecule, an event that is critical for signaling (Nakano et al., 1998; Baud et al., 1999; Park et al., 1999; Ye et al., 1999). Interestingly, deletion mutants of the N–terminal sequence of TRAF6 and TRAF2 have a completely impaired capability to signal, and can even display a dominant-negative phenotype when overexpressed (Cao et al., 1996b; Hsu et al., 1996a,b; Arch et al., 1998). This suggests that the N–terminus of TRAF6 is an important domain for signaling. This notion has received recent support by results from Karin and co-workers (Baud et al., 1999). These investigators replaced the TRAF domain of TRAF2 and TRAF6 with repeats of the FKBP12 protein, and demonstrated that the oligomerization of the N–terminal effector domains of both TRAFs was sufficient to trigger their interaction with MEKK–1. This may indicate that the binding of adapters such as p62 or ECSIT to the TRAF domain may be necessary to bring the effector kinase, aPKC or MEKK–1, respectively, into close proximity with the N–terminal effector domain of TRAF6, which is required for effective signaling. This would explain why N-deletion mutants of TRAF6 act as dominant-negatives because they could keep p62 and other adapters such as ECSIT, and their associated kinases, in inactive complexes lacking the additional required interaction with the N–terminus. In this study we demonstrate that the N–terminal effector region of TRAF6 interacts with ζPKC in a dimerization-dependent manner, lending further support to that model.
Therefore, it seems that downstream of TRAF6 there is a bifurcation toward ECSIT–MEKK–1 and p62–aPKC. The interaction of ECSIT with MEKK–1 serves to promote its proteolytic cleavage and subsequent activation (Kopp et al., 1999). Our preliminary results (P.Sanchez and J.Moscat) suggest that although p62 does not modify the ability of the aPKCs to phosphorylate physiologically irrelevant substrates such as the myelin basic protein, recombinant p62 favors the activation of IKKβ by ζPKC in in vitro reconstitution experiments. In this regard, p62 would behave similarly to ECSIT. Further experiments are needed to determine whether the interaction of TRAF6 with p62 or ECSIT modulates the ability of both proteins to activate their respective target kinases.
Materials and methods
Reagents and cell culture
Recombinant human IL–1β was purchased from Calbiochem. The monoclonal 12CA5 anti-HA and anti-Flag antibodies were from Boehringer and Sigma, respectively. The rabbit anti-Myc and anti-HA epitope and the anti-phospho-Jun antibodies as well as the monoclonal anti-TRAF6 antibody were from Santa Cruz Biotechnologies, Inc. The rabbit affinity-purified anti-p62 has been described previously (Sanchez et al., 1998). HepG2 and 293 cells were obtained from the American Type Culture Collection (ATCC). Cultures of 293 cells were maintained in high glucose Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS), penicillin G (100 μg/ml) and streptomycin (100 μg/ml) (Flow). HepG2 cells were maintained in Eagle's minimum essential medium (MEM) supplemented with 0.1 mM non-essential amino acids, 1.0 mM sodium pyruvate and 10% FCS, penicillin G (100 μg/ml) and streptomycin (100 μg/ml) (Flow). Subconfluent cells were transfected by the calcium phosphate method (Clontech, Inc.).
Plasmids
pCDNA3-HA–λ/ιPKCMUT, pCDNA3-HA–λ/ιPKC, pcDNA3-HA–ζPKC, pcDNA3-HA–αPKCMUT, pCDNA3-Myc-ζPKC, pCDNA3-HA–p62, pCDNA3-Myc-p62, pRK5-p62AS, 3EconA-Luc, pRK5-Flag-RIP and pCDNA3-HA–TRAF2 have been described previously (Diaz–Meco et al., 1996a,b; Sanchez et al., 1998; Sanz et al., 1999). The GFP–p62 construct was generated by inserting the _Eco_RI digested full-length p62 cDNA into the pEGFP-C1 vector. The different domains of p62 or RIP were subcloned into pCDNA3-HA, pCDNA3-Flag or pCDNA3-Myc vectors. The pCR-Flag-TRAF6, pCR-Flag-TRAF6-C and pCR-Flag-TRAF5 plasmids were described previously (Akiba et al., 1998). The TRAF6ΔC construct encompassing amino acids 1–274 of TRAF6 was subcloned into the pCDNA3-HA vector. The TRAF2/6 chimera containing amino acids 1–272 of TRAF2 fused to amino acids 273–531 of TRAF6, and the TRAF6/2 chimera containing amino acids 1–273 of TRAF6 fused to amino acids 273–501 of TRAF2 were subcloned into pCDNA3-HA. pCDNA3-HA–TRAF6ΔC-Gyr was constructed by fusing amino acids 1–273 of TRAF6 to the B subunit of the bacterial DNA gyrase (GyrB) (Farrar et al., 1996) into pCDNA3-HA. pcDNA3-HA–IRAK was obtained from pcDNA3-IRAK generously provided by Dr Keith Ray (Glaxo Wellcome, UK). Plasmids Flag-IRAK2 and Myc-MyD88 were a gift from Dr David Goeddel (Tularik Inc.).
Immunoprecipitations
For immunoprecipitations of endogenous proteins, HepG2 cells were used. For co-immunoprecipitations, subconfluent 293 or HepG2 cells plated on 100–mm dishes were transfected with the expression plasmids indicated. After transfection (24 h), HepG2 cells were stimulated or not with 1 ng/ml IL–1. Cells were then harvested and lysed in buffer PD (40 mM Tris–HCl pH 8.0, 500 mM NaCl, 0.1% NP–40, 6 mM EDTA, 6 mM EGTA, 10 mM β–glycerophosphate, 10 mM NaF, 10 mM phenyl phosphate, 300 μM Na3VO4, 1 mM benzamidine, 2 M phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM dithiothreitol). Extracts were centrifuged twice at 15 000 g for 15 min and 1 mg of whole-cell lysate was diluted in PD buffer and incubated with 5–10 μg of the antibody indicated. This reaction mixture was incubated on ice for 2 h, and then 25 μl of protein A or G beads were added and the mixture was left to incubate with gentle rotation for an additional 1 h at 4°C. The immunoprecipitates were then washed five times with PD buffer. Samples were fractionated on 8% SDS–PAGE, transferred to nitrocellulose ECL membrane (Amersham) and subjected to Western blot analysis with the corresponding antibody. Proteins were detected with the ECL reagent (Amersham). The TRAF6–p62 interaction was also detected when the immunoprecipitations were performed in a more stringent buffer (0.1% NP–40, 0.7 M NaCl).
Confocal immunofluorescence microscopy
HepG2 cells were grown on glass coverslips to 80% confluence in MEM supplemented with FCS (10%), transfected with 5 μg each of HA–IRAK, Flag-TRAF6 and GFP–p62, after which they were either untreated or stimulated with 1 ng/ml IL–1β for 10 min. Cells were rapidly washed twice in ice-cold phosphate-buffered saline (PBS), and fixed in 4% formaldehyde for 15 min at room temperature. Cells were washed four times with PBS and permeabilized with 0.2% Triton X–100. Free aldehyde groups were quenched with 10 mM glycine, and the fixed cells were incubated with primary antibodies for 1 h at 37°C. The secondary antibodies used were anti-rabbit Alexa 594 (Molecular Probes) and anti-mouse Cy5 (Amersham). Glass coverslips were mounted on Mowiol and were examined with the Radiance 2000 Bio-Rad confocal system (Bio-Rad, Richmond, CA) mounted on a Zeiss Axiovert S100 TV microscope (Zeiss, Oberkochen, Germany).
Reporter assays
For reporter gene assays, either HepG2 or 293 cells were seeded into six-well plates. Cells were transfected the following day by the calcium phosphate precipitation method with 1 ng of κB-luciferase reporter gene plasmid (3EconA-Luc), and various amounts of each expression construct. The total DNA transfected was kept constant by supplementation with the control vector pCDNA3. After 24 h, the HepG2 cell cultures were stimulated with IL–1β (1 ng/ml) for 4 h, extracts were prepared, and luciferase activity was determined as described previously (Diaz–Meco et al., 1996a).
Acknowledgments
Acknowledgements
We are indebted to Esther Garcia, Carmen Ibañez and Beatriz Ranera for technical assistance, to Carlos Sanchez for help with the confocal microscope, and Gonzalo Paris and Isabel Perez for help and enthusiasm. Research at the laboratory of J.M. is supported by grants SAF99-0053 from CICYT, PM96-0002-C02 from DGICYT and BIO4-CT97-2071 from the European Union, and by funds from Glaxo Wellcome Spain, and has benefited from an institutional grant from Fundación Ramón Areces to the CBM. The laboratory of H.N. is supported by a grant from the Core Research for Evolutional Science and Technology (CREST).
References
- Akiba H. et al. (1998)CD27, a member of the tumor necrosis factor receptor superfamily, activates NF–κB and stress-activated protein kinase/c-Jun N–terminal kinase via TRAF2, TRAF5 and NF–κB-inducing kinase. J. Biol. Chem., 273, 13353–13358. [DOI] [PubMed] [Google Scholar]
- Akimoto K. et al. (1996)EGF or PDGF receptors activate atypical PKCλ through phosphatidylinositol 3-kinase. EMBO J., 15, 788–798. [PMC free article] [PubMed] [Google Scholar]
- Arch R.H., Gedrich, R.W. and Thompson, C.B. (1998) Tumor necrosis factor receptor-associated factors (TRAFs)—a family of adapter proteins that regulates life and death. Genes Dev., 12, 2821–2830. [DOI] [PubMed] [Google Scholar]
- Baud V., Liu, Z.G., Bennett, B., Suzuki, N., Xia, Y. and Karin, M. (1999) Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino–terminal effector domain. Genes Dev., 13, 1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E., Diaz–Meco, M.T., Dominguez, I., Municio, M.M., Sanz, L., Lozano, J., Chapkin, R.S. and Moscat, J. (1993) Protein kinase C ζ isoform is critical for mitogenic signal transduction. Cell, 74, 555–563. [DOI] [PubMed] [Google Scholar]
- Berra E., Diaz–Meco, M.T., Lozano, J., Frutos, S., Municio, M.M., Sanchez, P., Sanz, L. and Moscat, J. (1995) Evidence for a role of MEK and MAPK during signal transduction by protein kinase C ζ. EMBO J., 14, 6157–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E., Municio, M.M., Sanz, L., Frutos, S., Diaz–Meco, M.T. and Moscat, J. (1997) Positioning atypical protein kinase C isoforms in the UV-induced apoptotic signaling cascade. Mol. Cell. Biol., 17, 4346–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G., Overvatn, A., Diaz–Meco, M.T., Moscat, J. and Johansen, T. (1995) Evidence for a bifurcation of the mitogenic signaling pathway activated by Ras and phosphatidylcholine-hydrolyzing phospholipase C. J. Biol. Chem., 270, 21299–21306. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G., Perander, M., Overvatn, A. and Johansen, T. (1997) Reversion of Ras- and phosphatidylcholine-hydrolyzing phospholipase C-mediated transformation of NIH 3T3 cells by a dominant interfering mutant of protein kinase C λ is accompanied by the loss of constitutive nuclear mitogen-activated protein kinase/extracellular signal-regulated kinase activity. J. Biol. Chem., 272, 11557–11565. [DOI] [PubMed] [Google Scholar]
- Cao Z., Henzel, W.J. and Gao, X. (1996a) IRAK: a kinase associated with the interleukin-1 receptor. Science, 271, 1128–1131. [DOI] [PubMed] [Google Scholar]
- Cao Z., Xiong, J., Takeuchi, M., Kurama, T. and Goeddel, D.V. (1996b) TRAF6 is a signal transducer for interleukin-1. Nature, 383, 443–446. [DOI] [PubMed] [Google Scholar]
- Colledge M. and Scott, J.D. (1999) AKAPs: from structure to function. Trends Cell Biol., 9, 216–221. [DOI] [PubMed] [Google Scholar]
- Diaz–Meco M.T. et al. (1993)A dominant-negative protein kinase C ζ subspecies blocks NF–κB activation. Mol. Cell. Biol., 13, 4770–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz–Meco M.T., Dominguez, I., Sanz, L., Dent, P., Lozano, J., Municio, M.M., Berra, E., Hay, R.T., Sturgill, T.W. and Moscat, J. (1994a) ζPKC induces phosphorylation and inactivation of IκBα in vitro. EMBO J., 13, 2842–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz–Meco M.T., Lozano, J., Municio, M.M., Berra, E., Frutos, S., Sanz, L. and Moscat, J. (1994b) Evidence for the in vitro and in vivo interaction of Ras with protein kinase C ζ. J. Biol. Chem., 269, 31706–31710. [PubMed] [Google Scholar]
- Diaz–Meco M.T., Municio, M.M., Frutos, S., Sanchez, P., Lozano, J., Sanz, L. and Moscat, J. (1996a) The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell, 86, 777–786. [DOI] [PubMed] [Google Scholar]
- Diaz–Meco M.T., Municio, M.M., Sanchez, P., Lozano, J. and Moscat, J. (1996b) λ-interacting protein, a novel protein that specifically interacts with the zinc finger domain of the atypical protein kinase C isotype λ/ι and stimulates its kinase activity in vitro and in vivo. Mol. Cell. Biol., 16, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez I., Diaz–Meco, M.T., Municio, M.M., Berra, E., Garcia de Herreros, A., Cornet, M.E., Sanz, L. and Moscat, J. (1992) Evidence for a role of protein kinase C ζ subspecies in maturation of Xenopus laevis oocytes. Mol. Cell. Biol., 12, 3776–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar M.A., Alberol, I. and Perlmutter, R.M. (1996) Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature, 383, 178–181. [DOI] [PubMed] [Google Scholar]
- Folgueira L., McElhinny, J.A., Bren, G.D., MacMorran, W.S., Diaz-Meco, M.T., Moscat, J. and Paya, C.V. (1996) Protein kinase C ζ mediates NF–κB activation in human immunodeficiency virus-infected monocytes. J. Virol., 70, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Xu, J., Bezanilla, M., van Huizen, R., Derin, R. and Li, M. (1999) Differential stimulation of PKC phosphorylation of potassium channels by ZIP1 and ZIP2. Science, 285, 1565–1569. [DOI] [PubMed] [Google Scholar]
- Herrera-Velit P., Knutson, K.L. and Reiner, N.E. (1997) Phos– phatidylinositol 3-kinase-dependent activation of protein kinase C ζ in bacterial lipopolysaccharide-treated human monocytes. J. Biol. Chem., 272, 16445–16452. [DOI] [PubMed] [Google Scholar]
- Hsu H., Huang, J., Shu, H.B., Baichwal, V. and Goeddel, D.V. (1996a) TNF–dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity, 4, 387–396. [DOI] [PubMed] [Google Scholar]
- Hsu H., Shu, H.B., Pan, M.G. and Goeddel, D.V. (1996b) TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell, 84, 299–308. [DOI] [PubMed] [Google Scholar]
- Ishida T. et al. (1996)Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino–terminal domain of the CD40 cytoplasmic region. J. Biol. Chem., 271, 28745–28748. [DOI] [PubMed] [Google Scholar]
- Izumi Y., Hirose, T., Tamai, Y., Hirai, S., Nagashima, Y., Fujimoto, T., Tabuse, Y., Kemphues, K.J. and Ohno, S. (1998) An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol., 143, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher M.A., Grimm, S., Ishida, Y., Kuo, F., Stanger, B.Z. and Leder, P. (1998) The death domain kinase RIP mediates the TNF–induced NF–κB signal. Immunity, 8, 297–303. [DOI] [PubMed] [Google Scholar]
- Khursigara G., Orlinick, J.R. and Chao, M.V. (1999) Association of the p75 neurotrophin receptor with TRAF6. J. Biol. Chem., 274, 2597–2600. [DOI] [PubMed] [Google Scholar]
- Kopp E., Medzhitov, R., Carothers, J., Xiao, C., Douglas, I., Janeway, C.A. and Ghosh, S. (1999) ECSIT is an evolutionarily conserved intermediate in the Toll/IL–1 signal transduction pathway. Genes Dev., 13, 2059–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S., Nakagawa, N., Tokunaga, C., Tatematsu, K. and Tanizawa, K. (1999) Mammalian homologue of the Caenorhabditis elegans UNC–76 protein involved in axonal outgrowth is a protein kinase C ζ-interacting protein J. Cell Biol., 144, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena M.J., Diaz–Meco, M.T., Bren, G., Pay, C.V. and Moscat, J. (1999) Activation of IκB kinase β by protein kinase C isoforms. Mol. Cell. Biol., 19, 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F.S., Hagler, J., Chen, Z.J. and Maniatis, T. (1997) Activation of the IκB α kinase complex by MEKK1, a kinase of the JNK pathway. Cell, 88, 213–222. [DOI] [PubMed] [Google Scholar]
- Lee F.S., Peters, R.T., Dang, L.C. and Maniatis, T. (1998) MEKK1 activates both IκB kinase α and IκB kinase β. Proc. Natl Acad. Sci. USA, 95, 9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Reichlin, A., Santana, A., Sokol, K.A., Nussenzweig, M.C. and Choi, Y. (1997) TRAF2 is essential for JNK but not NF–κB activation and regulates lymphocyte proliferation and survival. Immunity, 7, 703–713. [DOI] [PubMed] [Google Scholar]
- Liao D.F., Monia, B., Dean, N. and Berk, B.C. (1997) Protein kinase C ζ mediates angiotensin II activation of ERK1/2 in vascular smooth muscle cells. J. Biol. Chem., 272, 6146–6150. [DOI] [PubMed] [Google Scholar]
- Lomaga M.A. et al. (1999)TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40 and LPS signaling. Genes Dev., 13, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.V., Ni, J. and Dixit, V.M. (1998) RIP2 is a novel NF–κB-activating and cell death-inducing kinase. J. Biol. Chem., 273, 16968–16975. [DOI] [PubMed] [Google Scholar]
- Mochly–Rosen D. (1995) Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science, 268, 247–251. [DOI] [PubMed] [Google Scholar]
- Mochly–Rosen D. and Gordon, A.S. (1998) Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J., 12, 35–42. [PubMed] [Google Scholar]
- Murray N.R. and Fields, A.P. (1997) Atypical protein kinase C ι protects human leukemia cells against drug-induced apoptosis. J. Biol. Chem., 272, 27521–27524. [DOI] [PubMed] [Google Scholar]
- Nakano H., Shindo, M., Sakon, S., Nishinaka, S., Mihara, M., Yagita, H. and Okumura, K. (1998) Differential regulation of IκB kinase α and β by two upstream kinases, NF–κB-inducing kinase and mitogen-activated protein kinase/ERK kinase kinase-1. Proc. Natl Acad. Sci. USA, 95, 3537–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z. and Matsumoto, K. (1999) The kinase TAK1 can activate the NIK-I κB as well as the MAP kinase cascade in the IL–1 signalling pathway. Nature, 398, 252–256. [DOI] [PubMed] [Google Scholar]
- Park Y.C., Burkitt, V., Villa, A.R., Tong, L. and Wu, H. (1999) Structural basis for self-association and receptor recognition of human TRAF2. Nature, 398, 533–538. [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Blake, D.J., Davies, K.E., Kendrick-Jones, J. and Winder, S.J. (1996) ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci., 21, 11–13. [PubMed] [Google Scholar]
- Puls A., Schmidt, S., Grawe, F. and Stabel, S. (1997) Interaction of protein kinase C ζ with ZIP, a novel protein kinase C-binding protein. Proc. Natl Acad. Sci. USA, 94, 6191–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzymkiewicz D.M., Tetsuka, T., Daphna-Iken, D., Srivastava, S. and Morrison, A.R. (1996) Interleukin-1β activates protein kinase C ζ in renal mesangial cells. Potential role in prostaglandin E2 up-regulation. J. Biol. Chem., 271, 17241–17246. [DOI] [PubMed] [Google Scholar]
- Sakurai H., Miyoshi, H., Toriumi, W. and Sugita, T. (1999) Functional interactions of transforming growth factor β-activated kinase 1 with IκB kinases to stimulate NF–κB activation. J. Biol. Chem., 274, 10641–10648. [DOI] [PubMed] [Google Scholar]
- Sanchez P., De Carcer, G., Sandoval, I.V., Moscat, J. and Diaz–Meco, M.T. (1998) Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol. Cell. Biol., 18, 3069–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Sanchez, P., Lallena, M.J., Diaz–Meco, M.T. and Moscat, J. (1999) The interaction of p62 with RIP links the atypical PKCs to NF–κB activation. EMBO J., 18, 3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonwasser D.C., Marais, R.M., Marshall, C.J. and Parker, P.J. (1998) Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel and atypical protein kinase C isotypes. Mol. Cell. Biol., 18, 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.Y., Regnier, C.H., Kirschning, C.J., Goeddel, D.V. and Rothe, M. (1997) Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-κB and c-jun N–terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc. Natl Acad. Sci. USA, 94, 9792–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E., Sontag, J.M. and Garcia, A. (1997) Protein phosphatase 2A is a critical regulator of protein kinase C ζ signaling targeted by SV40 small t to promote cell growth and NF–κB activation. EMBO J., 16, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.A., Allen, J.L., Tsen, M., Dubnicoff, T., Danao, J., Liao, X.C., Cao, Z. and Wasserman, S.A. (1999) Impaired cytokine signaling in mice lacking the IL–1 receptor-associated kinase. J. Immunol., 163, 978–984. [PubMed] [Google Scholar]
- Wang Y.M., Seibenhener, M.L., Vandenplas, M.L. and Wooten, M.W. (1999) Atypical PKC ζ is activated by ceramide, resulting in coactivation of NF–κB/JNK kinase and cell survival. J. Neurosci. Res., 55, 293–302. [DOI] [PubMed] [Google Scholar]
- Wooten M.W. (1999) Function for NF–κB in neuronal survival: regulation by atypical protein kinase C. J. Neurosci. Res., 58, 607–611. [DOI] [PubMed] [Google Scholar]
- Wooten M.W., Seibenhener, M.L., Zhou, G., Vandenplas, M.L. and Tan, T.H. (1999) Overexpression of atypical PKC in PC12 cells enhances NGF-responsiveness and survival through an NF–κB dependent pathway. Cell Death Differ., 6, 753–764. [DOI] [PubMed] [Google Scholar]
- Ye H., Park, Y.C., Kreishman, M., Kieff, E. and Wu, H. (1999) The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell, 4, 321–330. [DOI] [PubMed] [Google Scholar]
- Yeh W.C. et al. (1997)Early lethality, functional NF–κB activation and increased sensitivity to TNF–induced cell death in TRAF2-deficient mice. Immunity, 7, 715–725. [DOI] [PubMed] [Google Scholar]