MEME-ChIP: motif analysis of large DNA datasets (original) (raw)
Abstract
Motivation: Advances in high-throughput sequencing have resulted in rapid growth in large, high-quality datasets including those arising from transcription factor (TF) ChIP-seq experiments. While there are many existing tools for discovering TF binding site motifs in such datasets, most web-based tools cannot directly process such large datasets.
Results: The MEME-ChIP web service is designed to analyze ChIP-seq ‘peak regions’—short genomic regions surrounding declared ChIP-seq ‘peaks’. Given a set of genomic regions, it performs (i) ab initio motif discovery, (ii) motif enrichment analysis, (iii) motif visualization, (iv) binding affinity analysis and (v) motif identification. It runs two complementary motif discovery algorithms on the input data—MEME and DREME—and uses the motifs they discover in subsequent visualization, binding affinity and identification steps. MEME-ChIP also performs motif enrichment analysis using the AME algorithm, which can detect very low levels of enrichment of binding sites for TFs with known DNA-binding motifs. Importantly, unlike with the MEME web service, there is no restriction on the size or number of uploaded sequences, allowing very large ChIP-seq datasets to be analyzed. The analyses performed by MEME-ChIP provide the user with a varied view of the binding and regulatory activity of the ChIP-ed TF, as well as the possible involvement of other DNA-binding TFs.
Availability: MEME-ChIP is available as part of the MEME Suite at http://meme.nbcr.net.
Contact: t.bailey@uq.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The genomic regions identified as bound by a transcription factor (TF) in a chromatin immunoprecipitation followed by sequencing (ChIP-seq) experiment are a rich source of information about transcriptional regulation. These regions are defined by mapping the sequence tags to the genome, which identifies ‘peaks’ of (direct or indirect) binding by the ChIP-ed factor typically to a resolution of about 100 bp. This high resolution is of obvious utility for identifying which genes a TF regulates, but the genomic regions surrounding the peaks are typically highly enriched for binding sites of the ChIP-ed TF and other TFs. Hence, these regions can be mined computationally to understand the roles, interactions and functions of the ChIP-ed TF and its regulatory partners.
We describe here a web service called MEME-ChIP that automatically performs five types of analysis on ChIP-seq regions. (i) Ab initio motif discovery identifies novel sequence patterns (motifs) in the ChIP-seq regions that may be due to TF binding sites. (ii) Motif enrichment analysis looks for enrichment of known TF DNA-binding motifs in the data. (iii) Motif visualization displays the relative locations and binding strengths of TF binding sites in the input regions. (iv) Motif binding strength analysis computes an estimate of the total DNA-binding affinity of each input region for the TF corresponding to each discovered motif. (v) Motif identification compares the ab initio motifs to known TF DNA-binding motifs. The output of MEME-ChIP is thus a multifaceted view of the identities, prevalence, DNA-binding patterns and potential interactions of the ChIP-ed TF and its regulatory partners.
Ab initio motifs discovered in ChIP-seq data give an unbiased view of the in vivo DNA-binding propensities of TFs binding alone or in protein complexes. MEME-ChIP employs two motif discovery algorithms with complementary characteristics. The MEME (Bailey et al., 2006) algorithm uses expectation maximization (EM) to discover probabilistic models of DNA-binding by single TFs or TF complexes. MEME motifs can provide accurate thermodynamic models of TF binding. MEME is complemented by DREME (Bailey, 2011), which uses a simpler, non-probabilistic model (regular expressions) to describe the short binding motifs characteristic of single eukaryotic TFs. DREME is often able to detect very short motifs that are not found by MEME. MEME-ChIP also attempts to identify the motifs found by MEME and DREME by comparing them to a database of known TF motifs using the TOMTOM (Gupta et al., 2007) algorithm. Motif discovery thus identifies novel binding motifs and TFs that are regulatory partners of the ChIP-ed TF.
Motif enrichment analysis can identify additional regulatory motifs whose enrichment in the ChIP-seq regions is too slight to be detected by ab initio motif discovery. It achieves higher sensitivity by limiting the search for motifs to a set of previously known TF DNA-binding motifs. MEME-ChIP uses the AME (McLeay and Bailey, 2010) algorithm for motif enrichment analysis.
For motif visualization and binding strength analysis, MEME-ChIP utilizes the MAST (Bailey and Gribskov, 1998) and AMA (Buske et al., 2010) algorithms, respectively. MAST uses a threshold-based approach to identify a putative set of non-overlapping binding sites in the ChIP-seq regions for all the motifs discovered by MEME (or DREME). This allows associations among the locations of the different motifs (TF binding sites) to be seen by eye. The AMA algorithm computes a thermodynamic estimate of the average binding affinity of the TF (as described by the motif) for the each ChIP-seq sequence region.
MEME-ChIP complements other web-based ChIP-seq motif analysis tools. Like MEME-ChIP, the peak-motifs algorithm (Thomas-Chollier et al., 2008) performs several analyses including a plot of the positional distribution of each motif. Trawler (Ettwiller et al., 2007) performs motif discovery and (optionally) analyzes the conservation of predicted motif sites. Peak-motifs and Trawler both perform word-based motif discovery. MEME-ChIP does as well (via DREME) and complements this with MEME, a non-word-based approach. Unlike MEME-ChIP, peak-motifs and Trawler do not perform motif enrichment analysis or motif binding strength analysis.
2 IMPLEMENTATION
The MEME-ChIP web service simplifies the analysis of ChIP-seq data by executing a computational pipeline on a set of genomic regions uploaded by the user. The uploaded regions should be FASTA-formatted sequences of at least 100 bp in length, each centered on a ChIP-seq tag peak. Prior to motif discovery and motif enrichment analysis, MEME-ChIP centers and trims each sequence to 100 bp; the full-length sequences are used in the subsequent motif visualization step. All trimmed sequences are input to the DREME motif discovery algorithm, whereas, due to computational complexity, a maximum of 600 sequences (randomly selected from the input) are input to the MEME algorithm. MEME and DREME output novel motifs as position-specific probability matrices along with a wealth of other information about the motifs discovered. MEME-ChIP runs the AME motif enrichment algorithm on all of the trimmed sequences. AME computes and outputs the statistical enrichment in the sequences of matches to each motif in the JASPAR CORE database (Portales-Casamar et al., 2010) of TF motifs. AMA computes and outputs the average binding affinity score for each motif MEME finds and for each input sequence. MEME-ChIP uses the MAST algorithm to visualize the locations of (putative) matches to each of the MEME and DREME motifs in the untrimmed input sequences. It also compares each of the MEME and DREME motifs to each of the motifs in the JASPAR CORE database to identify possible TFs binding to each motif.
3 EXAMPLE
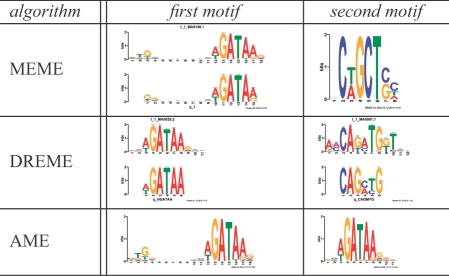
To demonstrate the functionality of MEME-ChIP, we use it to analyze the ChIP-seq peak regions reported by Kassouf et al. (2010) for SCL (also called Tal1), a key regulator of erythropoeisis. (Complete results are available at http://meme.nbcr.net/meme/doc/examples/memechip_example_output_files.) The two ab initio motif discovery algorithms (MEME and DREME) and motif enrichment analysis algorithm (AME) all identify a known SCL binding motif. In the case of MEME and AME, the most significant motif found is a composite motif believed to represent binding of a protein complex involving SCL and GATA-1, another transcription factor that plays a central role in erythropoeisis (Fig. 1, column 1, rows 1 and 3). The value of running two types of motif discovery algorithms is illustrated by the fact that although DREME does not discover this composite motif, it finds a better match to the canonical SCL binding motif (Fig. 1, column 2, row 2) than MEME does. Interestingly, DREME reports that the SCL motif is less significant in this ChIP-seq dataset than the canonical GATA-1 motif is (Fig. 1, column 1, row 2), suggesting that SCL binds more frequently in complex with GATA-1 than alone. This is supported by the fact that the motif enrichment analysis by AME also reports that the canonical SCL motif is less enriched than both the SCL-GATA-1 motif and the GATA-1 motif (data not shown). In all, AME reports that the SCL ChIP-seq regions are enriched for 15 known vertebrate motifs. DREME reports nine significant motifs, six of which match known vertebrate TF motifs, and three of which are novel. MEME finds three significant motifs, one of which is novel. Such novel motifs are possible candidates for further study, such as by using Gene Ontology enrichment analysis (Buske et al., 2010) to predict their transcriptional roles. Additional details on the implementation and use of MEME-ChIP are given in the Supplementary materials.
Fig. 1.
Two most significant motifs found by the MEME, DREME and AME algorithms in the SCL ChIP-seq data. For MEME and DREME motifs, the motif Logo (bottom) is shown aligned with the most similar JASPAR motif Logo (top) if the similarity is significant (E ≤ 0.05).
Funding: ARC Centre of Excellence in Bioinformatics (to P.M.); National Institutes of Health grant (R0-1 RR021692-05 to T.L.B.).
Conflict of Interest: none declared.
Supplementary Material
Supplementary Data
REFERENCES
- Bailey TL. Bioinformatics. DREME: Motif discovery in transcription factor ChIP-seq data. (in press) accepted for publication April 15, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bailey TL, et al. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, et al. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics. 2010;26:860–866. doi: 10.1093/bioinformatics/btq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwiller L, et al. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat. Methods. 2007;4:563–565. doi: 10.1038/nmeth1061. [DOI] [PubMed] [Google Scholar]
- Gupta S, et al. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, et al. Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;8:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeay RC, Bailey TL. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, et al. Jaspar 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data