The Genomic Tree as Revealed from Whole Proteome Comparisons (original) (raw)
Abstract
The availability of a number of complete cellular genome sequences allows the development of organisms’ classification, taking into account their genome content, the loss or acquisition of genes, and overall gene similarities as signatures of common ancestry. On the basis of correspondence analysis and hierarchical classification methods, a methodological framework is introduced here for the classification of the available 20 completely sequenced genomes and partial information for Schizosaccharomyces pombe, Homo sapiens, and Mus musculus. The outcome of such an analysis leads to a classification of genomes that we call a genomic tree. Although these trees are phenograms, they carry with them strong phylogenetic signatures and are remarkably similar to 16S-like rRNA-based phylogenies. Our results suggest that duplication and deletion events that took place through evolutionary time were globally similar in related organisms. The genomic trees presented here place the Archaea in the proximity of the Bacteria when the whole gene content of each organism is considered, and when ancestral gene duplications are eliminated. Genomic trees represent an additional approach for the understanding of evolution at the genomic level and may contribute to the proper assessment of the evolutionary relationships between extant species.
The determination of complete genome sequences from ≥20 organisms offers an unprecedented opportunity for the study of evolutionary problems in molecular biology and at a highly integrated level. One of the first problems to address in such a context concerns the derivation of the universal tree of life, which should reflect the global evolutionary relationships of whole organisms, and not only single-gene phylogenies. The universal tree of life was based on the 16S-like rRNA genes (Woese 1987; Woese et al. 1990) and led to the proposal of the three primary kingdoms or domains (Eukarya, Bacteria, and Archaea). However, this proposal has been criticized on different grounds (Gupta 1998; Mayr 1998). Although other molecular phylogenies have confirmed this analysis, many genes (particularly those encoding metabolic enzymes) give different topologies or even fail to support the three-domain classification of living organisms (Cavalier-Smith 1989; Forterre et al. 1992; Brown and Doolittle 1997; Doolittle 1998; Gupta 1998). Within the three-domain classification itself, a recurrent question concerns the controversial proximity of Archaea to either Eukarya or Bacteria (Brinkmann and Philippe 1999). Archaeal organisms appear to be close to Eukarya when the protein synthesis machinery (transcription and translation) is considered but close to Bacteria if metabolic genes are compared (Doolittle and Logsdon 1998).
Such unsettling differences not only reflect classical problems in phylogenetic reconstruction due to horizontal transfer (which may have been more intense during early cellular evolution, Woese 1998), unequal rates of nucleotide substitution, and gene displacement but also underline the fact that trees depict the evolutionary distances between genes and not between organisms or entire genomes.
Previous attempts to analyze the macrostructure of genomes for phylogenetic reconstruction have been based on a number of well-known techniques such as DNA hybridization studies and restriction enzyme fragment analyses (Li 1997). As in the case of gene-based phylogenies, such approaches are ultimately dependent on the degree of sequence divergence. On the contrary, analysis of comparative gene order provides quantitative models of genome evolution that become independent from the degree of sequence divergence once orthologs have been defined (Sankoff et al. 1992; Boore and Brown 1998). Likewise, a more integrative view of genome evolution is feasible with the shared gene trees proposed recently by Snel et al. (1999). Here we present a different but complementary approach, not on the basis of evolutionary descent but on a hierarchical classification of genomes involving their gene content and overall similarity. We call the resulting phenograms the genomic tree.
RESULTS
Construction of Genomic Trees by Comparisons of All Predicted ORF Products for Completely Sequenced Genomes
In this work, we aim to derive the genomic tree from all of the available completely sequenced genomes through whole proteome comparisons, taking into account the predicted gene product content of each organism and their similarity. The construction of such a tree requires an appropriate methodological approach. The full set of predicted gene products of a completely sequenced organism is compared with itself and with that of every other organism considered. The possible similarity of a given open reading frame (ORF) product to any other is determined by appropriately defined statistical limits (see Methods). Comparison of organism j with organism i determines the proportion of ORFs in organism j that have at least one similar ORF in organism i (Tij). We call this proportion the “weight” of the common ancestry of j with respect to i (see Methods). The validation of the statistical limits used in such comparisons is discussed in Methods. The overall pairwise comparison of n organisms leads to a nxn matrix of Tijs. The appropriate method to handle such data matrices as a whole is correspondence analysis (Benzecri 1973; Greenacre 1984). The rationale of this method is to derive an orthogonal system of axes, called factors and denoted F1, F2,... F_n_−1 (a maximum of n − 1 such axes can be determined), which pass through the barycenter of the observations and correspond to a decreasing order of the amount of information each factor represents. Each organism is represented by its coordinates in this system. Thus, distances between organisms can be calculated, and their subsequent classification according to their neighborhood leads to a hierarchical tree, or the genomic tree. Such a tree is a graphical representation of the relationship between sets of organisms, which includes indirectly genome sizes, levels of internal redundancy due to ancestral duplications, and overall gene loss or acquisition events. This tree is independent of functional identity. Instead, it is based on the sole presence or absence of genes of common ancestry, as defined by comparison with all other genomes.
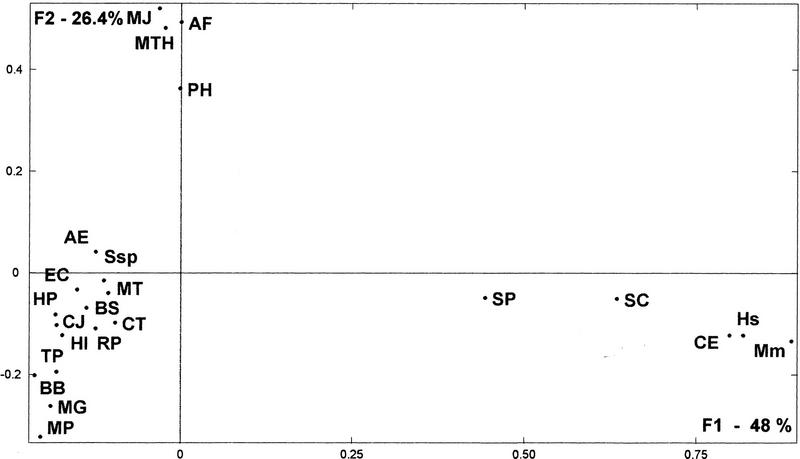
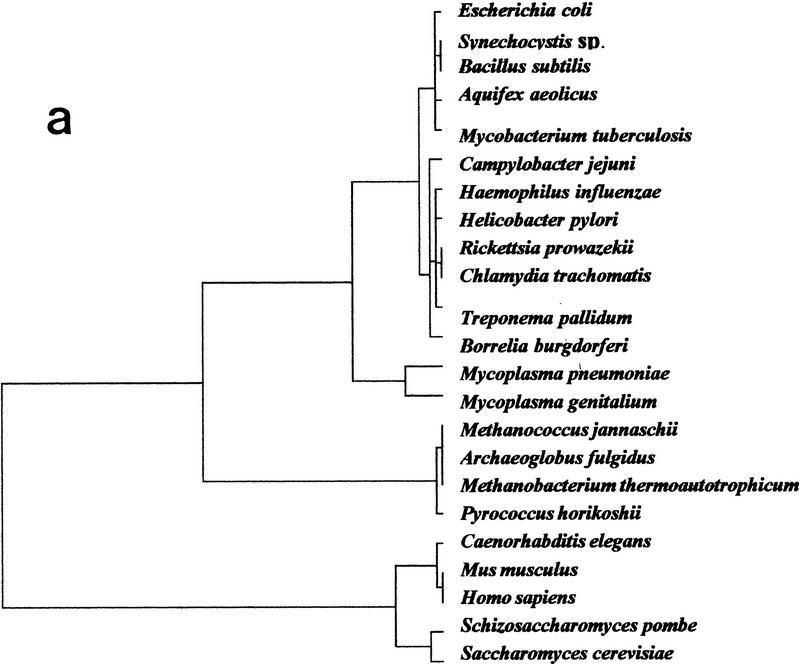
This method was applied to the data set, obtained from the comparison of the 20 completely sequenced organisms, plus the data available from human, mouse, and Schizosaccharomyces pombe (see Table 1). The results of our analysis are presented in Figure 1, in which organisms are represented on the best factorial space (i.e., the first and second factors). The distances between the surveyed organisms were calculated from their factorial coordinates and used to construct the genomic tree shown in Figure 2a. Four well-defined groups of organisms with similar profiles appear on this tree: (1) An archaeal cluster formed by Methanococcus jannaschii; Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, and Pyrococcus horikoshii; (2) a (eu)bacterial group formed by Escherichia coli, Synechocystis sp., Bacillus subtilis, Aquifex aeolicus, Mycobacterium tuberculosis, Campylobacter jejuni, Haemophilus influenzae, Helicobacter pylori, Rickettsia prowazekii, Chlamydia trachomatis, Treponema pallidum, and Borrelia burgdorferi; (3) a mycoplasma cluster (Mycoplasma pneumoniae and Mycoplasma genitalium) that groups with the Bacteria cluster; and (4) the eukaryotic group (Caenorhabditis elegans, Mus musculus, Homo sapiens, S. pombe, and Saccharomyces cerevisiae).
Table 1.
Completely Sequenced Organisms and Other Fragmentary Data Considered in this Analysis
| Organism | Domaina | Codeb | ORFsc | Partitionsd |
|---|---|---|---|---|
| H. influenzae | B | HI | 1713 | 1377 |
| M. genitalium | B | MG | 468 | 361 |
| Synechocystis sp. | B | Ssp | 3168 | 2002 |
| M. pneumoniae | B | MP | 677 | 424 |
| H. pylori | B | HP | 1577 | 1226 |
| E. coli | B | EC | 4290 | 2473 |
| B. subtilis | B | BS | 4100 | 2573 |
| B. burgdorferi | B | BB | 850 | 696 |
| A. aeolicus | B | AE | 1522 | 1157 |
| M. tuberculosis | B | MT | 3924 | 2329 |
| T. pallidum | B | TP | 1031 | 852 |
| C. trachomatis | B | CT | 877 | 718 |
| C. jejuni | B | CJ | 1731 | 1323 |
| R. prowazekii | B | RP | 837 | 653 |
| M. jannaschii | A | MJ | 1735 | 1180 |
| M. thermoautotrophicum | A | MTH | 1871 | 1227 |
| A. fulgidus | A | AF | 2437 | 1423 |
| P. horikoshii OT3 | A | PH | 2061 | 1373 |
| S. cerevisiae | E | SC | 6182 | 4437 |
| C. elegans | E | CE | 19,099 | 7558 |
| S. pombee | E | SP | 3579 | 2248 |
| H. sapiens | E | Hs | ||
| M. musculuse | E | Mm |
Figure 1.
Factorial representation of the weight of ancestral duplication and common ancestry in each genome, obtained by the multidimensional correspondence analysis method. First and second factorial axes (F1 and F2) represent, respectively, 48% and 26.4% of total information included in the ancestry weight matrix resulting from predicted gene product comparisons (see Methods). Dots represent the distribution of the surveyed organisms (abbreviations are as in Table 1).
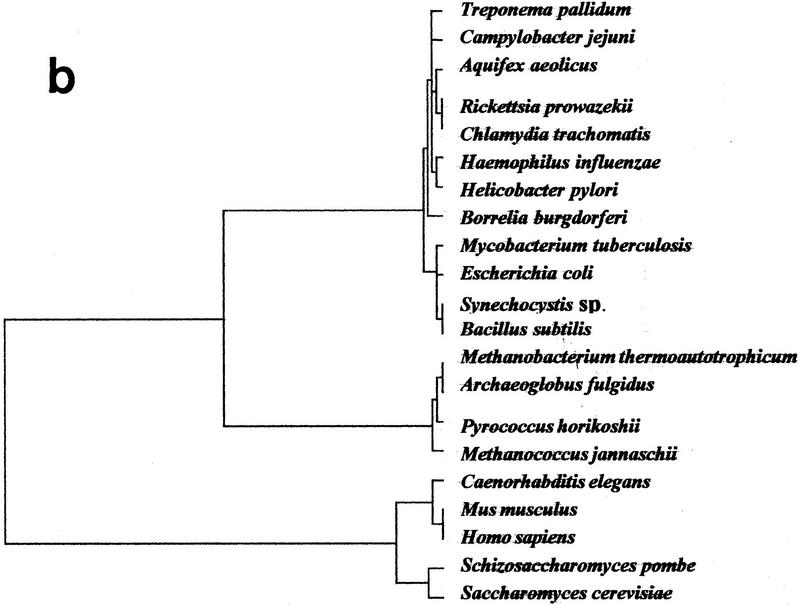
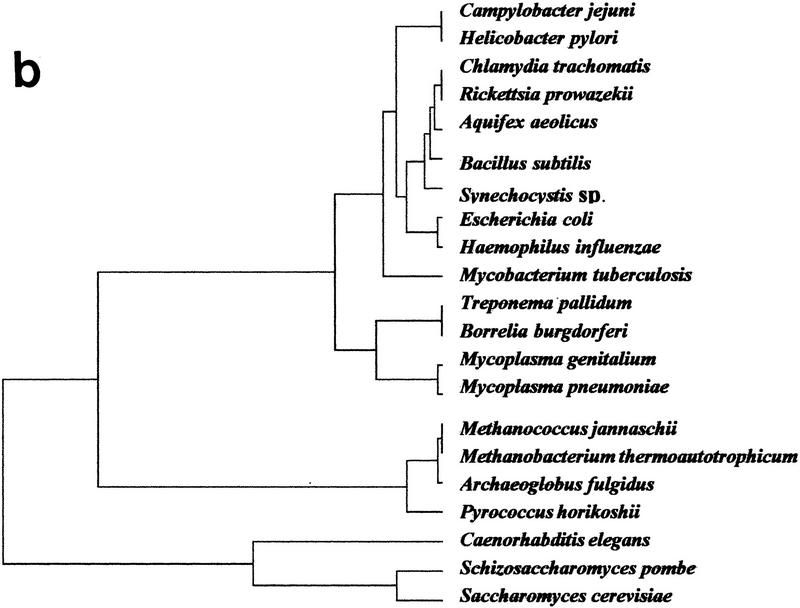
Figure 2.
Genomic tree. (a) This tree is obtained by a hierarchical classification of the organisms on the basis of their neighborhood distances. Distances between all pairs of organisms are calculated in the factorial space obtained by correspondence analysis. Horizontal lines between nodes are proportional to their similarity. (b) Same tree excluding data from M. genitalium and M. pneumoniae.
As indicated in Figure 2, the different species are not distributed at random in our trees, but their overall clustering follows the three-domain distribution, whose general topology is remarkably similar to unrooted 16S-like rRNA-based and gene-shared phylogenies (Woese 1987; Woese et al. 1990; Snel et al. 1999). Note, however, that although this tree is the outcome of a hierarchical classification, it carries a strong phylogenetic signature and can thus be considered a genomic tree of considerable assistance in understanding the evolutionary relationships between genomes.
In the approach discussed here, genome size, levels of ancestral gene redundancy due to duplications, and overall loss or acquisition of genes all contribute indirectly to the position of a given organism in the factorial space. An obvious concern is that the inclusion of small genomes, such as those of M. genitalium or M. pneumoniae, which may have undergone massive gene losses, may drastically alter the genomic tree by the limitation imposed on the proportions of genes of common ancestry in other genomes. To test this possibility, we eliminated the two Mycoplasma genomes from the data set and recomputed a novel tree (Fig. 2b). As shown in Figure 2b, whereas the exclusion of the mycoplasma produces no major changes in the overall tree topology, it affects the internal branching of the Bacteria, displacing A. aeolicus from a cluster that includes E. coli, B. subtilis, Synechocystis sp., and M. tuberculosis, to another branch with R. prowazekii and C. trachomatis. These changes probably are due to the small branch lengths among the inner nodes of the Bacteria. Removal of the two mycoplasma genomes affects slightly the Archaea, in which P. horikoshii is displaced by M. janaschii.
Because the positions of B. burgdorferi (850 genes), C. trachomatis (877 genes), R. prowazekii (837 genes), and T. pallidum (1031 genes) in the genomic tree are within the major bacterial branch, and not with the M. genitalium–M. pneumoniae cluster (468 and 677 genes, respectively), genome size is not overemphasized in the genomic tree. The fact that the two mycoplasma genomes form a deep branch within the Bacteria that is far removed from their close relative B. subtilis (as shown, e.g., by their 16S rRNA phylogeny) demonstrates that more realistic assessments of the different contributions of the variables defining the position of a given organism in the genomic tree are still required. We have tested the impact of the insertion of additional sequences on the topology of the tree by simulating the inclusion of artificial genomes with different degrees of ancestral conservation with the actual genomes (0, 20, 100) and found essentially the same results (data not shown).
Construction of Genomic Trees by Comparison of the Minimized Sets of Predicted ORF Products from Completely Sequenced Genomes
In the approach presented here, gene families are represented by their respective weights, and this corresponds to the whole genome picture. This approach can be complemented by highlighting the functional classification of genes. Because the organisms included here have genomes with different sizes and may exhibit important variations in the degree of gene acquisition and loss, it is important to construct a tree derived from genomes reduced to their minimal content by eliminating ancestral gene duplications that are still recognizable. Thus, in a second alternative approach, we have reduced each organism to its minimum genomic content by eliminating ancestral gene duplications and derived a second genomic tree. This was achieved with the following approximation: Each organism is represented by its partitions (i.e., genes with common ancestry; see Methods). Accordingly, instead of considering ancestry weight as the outcome of gene similarity, we considered it as the result of partition similarity. This neutralizes the variable rates of gene acquisition and losses because now each given gene family is represented by only one member (i.e., the corresponding partition). The resulting data set was analyzed following the previous methodology for the construction of a hierarchical tree, which represents the constituent set of genes in the organisms considered. The conservation rate of organism j in organism i is defined by Pij, which is obtained by dividing the number of distinct partitions of j including members having at least one significant match in i, by the total number of distinct partitions in j. Thus, for instance, 11.4% of yeast partitions share ancestral conservation with B. subtilis, 9.6% with H. influenzae, and so on.
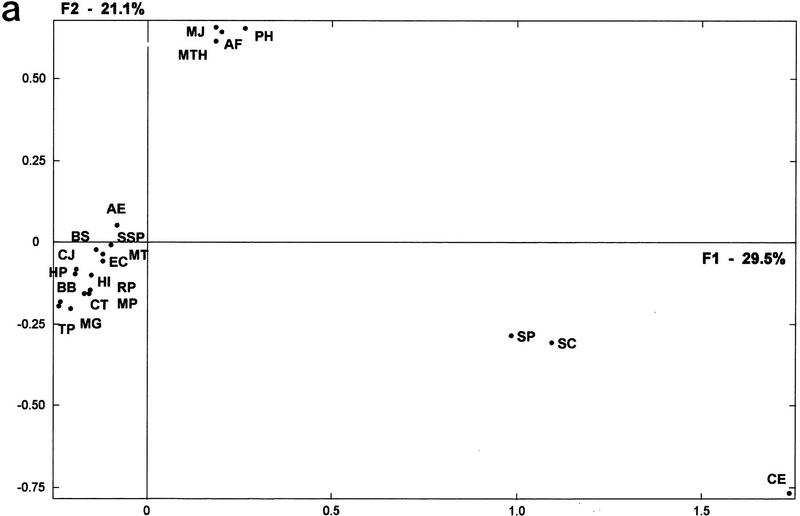
Figure 3a shows the organisms’ distribution on the first factorial space, and Figure 3b shows the genomic tree obtained by the hierarchical classification of their genomes by their distances as calculated in the whole factorial space. This new genomic tree represents the synthesis of the minimal content relationships in the considered organisms, and can be mapped onto the small subunit (SSU) rRNA phylogeny discussed by Woese (1987) and Woese et al. (1990). This result bears upon the current debates on the major divisions in the living world (Gupta 1998; Mayr 1998).
Figure 3.
(a) Factorial representation of the constituent ancestry in each genome. First and second axes (F1 and F2) represent, respectively, 29.5% and 21.1% of the total information included in the ancestry weight matrix resulting from the organism’s partitions (see Methods). Organismal distribution on this factorial plane is very similar to that in Fig. 1. Human and mouse, for which no accurate ancestral gene duplications can be presently calculated, were not considered in this analysis (abbreviations are as in Table 1). (b) Genomic tree for the considered organisms (minus human and mouse) as obtained from the whole factorial space resulting from the corresponding analysis (see Fig. 2a for methods of analysis).
DISCUSSION
Genomic and Gene Trees
On the basis of correspondence analysis and a hierarchical classification of gene content and overall gene similarities as ancestry weight, we have developed a new approach for genomic analysis that allows the construction of genomic trees that carry a strong phylogenic signature and whose overall topology strongly resembles the SSU rRNA-based evolutionary trees (Woese et al. 1990). Although our approach provides an excellent equivalent to the 16S-like rRNA-based branching orders of Archaea and the Eukarya (Fig. 2a), it leads to a different branching order within the Bacteria domain. This is particularly true of Gram-positive bacteria, from which the two mycoplasma are widely separated, forming a branch distant from both B. subtilis and M. tuberculosis. The latter species group into a non-natural cluster together with E. coli, Synechocystis sp., and A. aeolicus. It is of interest that A. aeolicus, whose exact phylogenetic position has been debated (Deckert et al. 1998), is firmly located in our tree within the Bacteria, as in the case of rRNA phylogenies (Burggrof et al. 1992; Pitulle et al. 1994; Reysenbach et al. 1996). However, it does not branch off early and instead clusters with M. tuberculosis.
In contrast to the rooted universal phylogenies that pair Archaea with the eukaryotic branch (Gogarten et al. 1989; Iwabe et al. 1989; Brown and Doolittle 1995), our methodology places the two prokaryotic kingdoms closer to each other than any one of them is to Eukarya (Fig. 2a). This is in accordance with the reconstruction of the universal tree, which eliminates artifacts due to long branch attraction and places Archaea as a sister group of Bacteria (Brinkmann and Philippe 1999).
Because the genomic tree shown in Figure 2a is based solely on the ancestral duplication and conservation proportions, its coherence at a gross level with the small subunit rRNA tree suggests that the average duplication and loss events that have taken place through evolutionary time are statistically similar in related organisms. That is, the weight of ancestry contributes to define the overall properties of a genome and groups it in a way that is strongly reminiscent of rRNA-based phylogenies over extended periods of evolutionary time.
The strong similarity between our genomic trees, which embody sequence divergence, gene losses, and acquisitions, with the 16S-like RNA phylogenies, which are based solely on sequence divergence, raises the issue of their consistency with phylogenetic trees constructed from other genes common to all the surveyed organisms. To analyze this issue, 75 partitions of universal genes were determined [i.e., each partition of structural orthologous genes (see Methods) includes at least one member from each of the organisms considered]. Phylogenetic trees were constructed by the neighbor-joining method with a bootstrap value of 1000, by use of the Clustal W program (Thompson et al. 1994). These 75 trees are scarcely consistent with each other and with the rooted universal tree. The resulting gene trees can be divided roughly as follows: (1) 36% are consistent with the rooted universal tree (i.e., that branch archaeal genes with eukaryal ones) and include, among others, those of genes encoding ribosomal proteins, as well as those involved in DNA metabolism and a few metabolic pathways; (2) in 21%, archaeal genes branch with bacterial ones, and this group includes, among others, genes involved in electron transport, gluconeogenesis/glycolysis, and RNA processing; and (3) 43% are a mixture of the previous topologies, that is, some genes branch with eukaryal genes, whereas others branch with bacterial genes, and eukaryal sometimes branch with bacterial genes (F. Tekaia, A. Lazcano, and B. Dujon, in prep.). These results show the phylogenetic distortion effects on gene trees, and emphasize the conflict between species and gene trees. In light of these distortions, the last genomic tree shown in Figure 3b may be considered as the average tree of all orthologous genes.
Future genome sequences will allow further refinement of the genomic trees presented here, and critical comparison with the sequence-based (Woese 1987) and shared-gene (Snel et al. 1999) trees will lead to a proper assessment of the value of our results. The trees presented here are less likely to suffer from the pitfalls of traditional methods such as variable changes in sequences and reliability of sequence alignments (Gupta 1998), because our approach is insensitive to such problems. However, our methodology is not intended to substitute for evolutionary inference on the basis of sequence comparisons but, rather, to provide a snapshot of the molecular evolution whether the large variations of genome sizes between organisms, the level of internal redundancy in each genome, and the losses or acquisitions of genes during evolution are considered or not. The observed differences between the topology of the genomic trees very likely are due to the different weights of gene families and their ancestry. The proximity between the Archaea and the Bacteria observed in the two genomic trees has to be confirmed once more completely sequenced eukaryal and archaeal genomes are available. Nevertheless, the statistical analysis of the degree of ancestral duplication and evolutionary conservation discussed here may help in the development of novel approaches to the management and understanding of large volumes of genomic data. Thus, our results represent an additional approach for the understanding of evolution at the genomic level and may contribute to the proper assessment of the evolutionary relationships between extant species.
METHODS
The rationale for the construction of genomic trees is based on the systematic comparison of the predicted translation products of all surveyed organisms (data taken from original publications; see Table 1) as a means to determine the presence or absence of genes of common ancestry with an internally calculated threshold of significance. For each organism included in the analysis, every gene product is successively used as a query sequence against all the gene products of the same organism, and against all the gene products of each of the other organisms considered; the former is used to define partitions of genes, and the latter to measure the weight of common ancestry (see below).
Definition of a Partition
A set of ORF products in a given organism defines a partition if, and only if, the following three properties are verified: (1) Each member of the set has at least one highly significant match with one other member of the set; (2) no member of the set has highly significant matches with members not included in the set; and (3) the set cannot be partitioned into subsets verifying (1) and (2) (i.e., the set is minimal).
Note that an ORF product that has no significant match in its own organism fulfills these properties and therefore is considered as a single member partition. Thus, a partition includes all ORF products with common ancestry in a given organism (the number of distinct partitions are shown in Table 1). Note that such construction of partitions is sometimes referred to as the single-linkage clustering method.
Definition of the Weight of Common Ancestry
The weight of common ancestry is the proportion of gene products that share a common ancestry with the gene products of other organisms, with all species being examined serially. The presence of homolog(s) defines the degree of ancestral duplication within each genome and of conservation between genomes.
Organism-Specific Comparisons
Comparisons of each query sequence with the complete proteome databases of the same and every other organism were performed with BLASTP (Altschul et al. 1990), version 1.4.8, using the pam250 substitution matrix, which favors large segment pairs and, hence, detects distantly related ORFs and the seg (Wootton and Federhen 1993) filter to eliminate compositionally biased regions in the query sequence. The M. musculus (Mmuniq) and human (Hsuniq) databases (see Table 1) serve solely as targets for comparison by queries of other genomes, with the TBLASTN (Altschul et al. 1990) program.
Because the genomes analyzed here exhibit important differences in size and complexity, we first determined a limit of significance of the BLASTP probability scores for each of the genomes considered (Tekaia and Dujon 1999). This was achieved by use of sets of random sequences, equivalent in number to the number of ORFs of each genome, and generated with sizes and amino acid compositions equal to the average size and composition of the actual proteome of each organism. Each of these random sequences was compared against the entire database of the cognate organism, and the best probability scores were recorded as for actual sequences. For each organism, the highest BLASTP probability score leaving <5% of pseudosignificant matches was considered as the limit of significance when that organism is used as target. Probability score limits were set at 10−9 for S. cerevisiae, 10−5 for B. subtilis, B. burgdorferi, M. tuberculosis, M. jannaschii, and C. elegans, 10−3 for S. pombe, T. pallidum, P. horikoshii, and R. prowazekii, 10−2 for C. trachomatis, and 10−4 for all other genomes.
Ancestry Weight Matrix using ORF Products
The data table T resulting from the pairwise comparisons of the organisms considered here can be found in http://www-alt.pasteur.fr/∼tekaia/dupcons.html. In this table, Tij is the proportion of ORF products from organism j having a common ancestry with one or several ORF product(s) of organism i (note that Tij is normalized because it is divided by the total number of ORFs in j). Tjj is the proportion of ORF products in organism j having a common ancestry with one or several other ORF product(s) of the same organism. In this study, i = 1, 23 and j = 1, 21, the difference between i and j correspond to the sequences from man (Hsuniq) and mouse (Mmuniq), which serve solely as targets for comparisons not as queries. As an example, in the S. cerevisiae genome, 16.7% of the ORFs share ancestral conservation with the B. subtilis genome, 12.7% with the H. influenzae genome, and so on. We refer to such proportions as the weight of common ancestry in the S. cerevisiae genome when compared with B. subtilis, H. influenzae, and others. Because of variable genome sizes and internal redundancy, the matrix is not symmetrical, for example, the weight of common ancestry in the B. subtilis genome when compared with S. cerevisiae is 15.5%.
Ancestry Weight Matrix Using Partitions
The data table P resulting from the pairwise comparisons of the organisms can also be found in http://www-alt.pasteur.fr/∼tekaia/dupconsparts.html. In this table, Pij is the proportion of distinct partitions in organism j having ancestry with at least one predicted gene product in organism i. Because each partition is unique in its organism, Pjj = 100.% (i.e., when comparing a given organism with itself, each partition is its unique match).
Structural Orthologous Genes
Two genes belonging to two distinct organisms are called structural orthologs, if and only if each shows the most similarity to the other when comparing it with its counterpart organism.
Partitions are obtained by applying the same definition (as in organisms) and by considering the whole set of orthologous genes obtained from the considered organisms.
Acknowledgments
We thank Stewart Cole for helpful discussion. We are indebted to Henri Buc, Edouard Yeramian, and the members of the Unité de Génétique Moléculaire des Levures for their encouragement and several useful discussions. Support from the Manlio Cantarini Foundation (Paris) and Universidad Autonoma de Mexico—Direction General de Asunto del Personal Academico (UNAM–DGAPA) project PAPIIT-IN213598 support to A.L. is gratefully acknowledged. B.D. is Professor of Molecular Genetics at University Pierre et Marie Curie and a member of the Institut Universitaire de France.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tekaia@pasteur.fr; FAX 33 1 40 61 34 56.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Benzecri J-P. L’analyse des données. Vol 2: L’analyse des correspondances. Paris, France: Dunod; 1973. [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD. Establishing a human transcript map. Nat Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- Brinkmann, H. and H. Philippe. 1999. Archaea sister-group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol. Biol. Evol. (In press). [DOI] [PubMed]
- Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Burggraf S, Olsen GJ, Stetter KO, Woese CR. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:353–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Molecular phylogeny. Archaebacteria and Archezoa. Nature. 1989;339:100–101. doi: 10.1038/339100a0. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE 3rd, Tekaia F, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. Microbial genomes opened up. Nature. 1998;392:339–342. doi: 10.1038/32789. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Logsdon JM., Jr Archaeal genomics: Do archaea have a mixed heritage? Curr Biol. 1998;8:R209–R211. doi: 10.1016/s0960-9822(98)70127-7. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J-F, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Forterre P, Benachenhou-Lahfa N, Confalonieri F, Duguet M, Elie C, Labedan B. The nature of the last universal ancestor and the root of the tree of life, still open questions. Biosystems. 1992;28:15–32. doi: 10.1016/0303-2647(92)90004-i. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Aert R, Agostini-Carbone ML, Ahmed A, Aigle M, Alberghina L, Albermann K, Albers M, Aldea M, Alexandraki D, et al. The Yeast Genome Directory. Nature(Suppl) 1997;387:5–105. [Google Scholar]
- Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, et al. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenacre M. Theory and application of correspondence analysis. London, UK: Academic Press; 1984. [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acid Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bologin A, Borchert S, et al. The complete genome sequence of the Grampositive bacterium Bacillus subtilis. Nature. 1997;390:249–259. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Li W-H. Molecular evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- Mayr E. Two empires or three? Proc Natl Acad Sci. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulle C, Yang Y, Marchiani M, Moore ER, Siefert JL, Aragno M, Junrtshuk P, Fox GE. Phylogenetic position of the genus Hydrogenobacter. Int J Syst Bact. 1994;44:620–626. doi: 10.1099/00207713-44-4-620. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Wickham GS, Pace NR. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D, Leduc G, Antoine N, Paquin B, Lang BF, Cedergren R. Gene order comparisons for phylogenetic inference: Evolution of the mitochondrial genome. Proc Natl Acad Sci. 1992;89:6575–6579. doi: 10.1073/pnas.89.14.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Tekaia, F. and B. Dujon. 1999. Pervasiveness of gene conservation and persistence of duplicates in cellular genomes. J. Mol. Evol. (In press). [DOI] [PubMed]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode Caenorhabditis elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The universal ancestor. Proc Natl Acad Sci. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 1993;17:149–163. [Google Scholar]