Segmental Duplications: Organization and Impact Within the Current Human Genome Project Assembly (original) (raw)
Abstract
Segmental duplications play fundamental roles in both genomic disease and gene evolution. To understand their organization within the human genome, we have developed the computational tools and methods necessary to detect identity between long stretches of genomic sequence despite the presence of high copy repeats and large insertion-deletions. Here we present our analysis of the most recent genome assembly (January 2001) in which we focus on the global organization of these segments and the role they play in the whole-genome assembly process. Initially, we considered only large recent duplication events that fell well-below levels of draft sequencing error (alignments 90%–98% similar and ≥1 kb in length). Duplications (90%–98%; ≥1 kb) comprise 3.6% of all human sequence. These duplications show clustering and up to 10-fold enrichment within pericentromeric and subtelomeric regions. In terms of assembly, duplicated sequences were found to be over-represented in unordered and unassigned contigs indicating that duplicated sequences are difficult to assign to their proper position. To assess coverage of these regions within the genome, we selected BACs containing interchromosomal duplications and characterized their duplication pattern by FISH. Only 47% (106/224) of chromosomes positive by FISH had a corresponding chromosomal position by BLAST comparison. We present data that indicate that this is attributable to misassembly, misassignment, and/or decreased sequencing coverage within duplicated regions. Surprisingly, if we consider putative duplications >98% identity, we identify 10.6% (286 Mb) of the current assembly as paralogous. The majority of these alignments, we believe, represent unmerged overlaps within unique regions. Taken together the above data indicate that segmental duplications represent a significant impediment to accurate human genome assembly, requiring the development of specialized techniques to finish these exceptional regions of the genome. The identification and characterization of these highly duplicated regions represents an important step in the complete sequencing of a human reference genome.
A main goal of the Human Genome Project (HGP) is to provide the complete and accurate reference sequence of the euchromatic portions of all human chromosomes (Collins et al. 1998). It has been argued that this endeavor differs from previously sequenced invertebrate models not only in terms of scale but also in terms of repetitive complexity (Green 1997; Eichler 1998). Repetitive complexity leads to misassignment and misassembly of sequence. It has been suggested that segmental duplications may be particularly problematic in this regard because of their inconspicuousness, large size, and high degree of sequence similarity. The inability to identify such duplications, let alone differentiate their true position from paralogous positions, may confound sequence assembly, resulting in merging of distinct loci into the same sequence (Eichler 1998).
Segmental duplications are duplicated blocks of genomic DNA typically ranging in size from 1–200 kb (IHGSC 2001). They often contain sequence features such as high-copy repeats and gene sequences with intron–exon structure. Thus, being composed of apparently normal genomic DNA, segmental duplications cannot be detected a priori; rather, most segmental duplications have to date been discovered based on experimental analyses. Over the past decade a large number of both intra- and interchromosomal segmental duplications have been observed (Wong et al. 1990; Tomlinson et al. 1994; Eichler et al. 1997; Mazzarella and Schlessinger 1997; Regnier et al. 1997; Zimonjic et al. 1997; Eichler 1998; Trask et al. 1998a; Jackson et al. 1999; Ji et al. 1999). These data suggest numerous interchromosomal exchanges during recent hominoid evolution with apparent biases into and between pericentromeric and subtelomeric regions (Eichler et al. 1997, 1999; Monfouilloux et al. 1998; Trask et al. 1998a; Jackson et al. 1999; Horvath et al. 2000a). To date, however, no systematic analysis of the genome has been performed to quantify this bias. Another unanticipated finding has been the important role segmental duplications play in disease (for review, see Ji et al. 2000; Mazzarella and Schlessinger 1998). Aberrant homologous recombination between highly similar paralogs appears to be a major mechanism for many genomic disorders such as velocardiofacial/DiGeorge, Smith-Magenis, and Prader-Willi/Angelman syndromes (Chen et al. 1997; Amos-Landgraf et al. 1999; Christian et al. 1999; Edelmann et al. 1999; Shaikh et al. 2000).
A major step toward developing a final reference sequence has been the completion of the draft-sequencing phase of the HGP and its subsequent assembly. The assembly has occurred in three main steps: (1) Sequenced clones are placed into fingerprint contigs generated from the entire RPCI-11 BAC library; (2) fingerprint contigs are assigned and positioned to chromosomes using all available genetic and STS markers; and (3) the sequence within each contig is assembled by Jim Kent's Gigassembler (IHGMC 2001; IHGSC 2001). This landmark achievement has given us the ability to examine segmental duplications in a genome-wide and systematic manner. We reported an unprecedented amount (3.6%) of sequence was involved in recent segmental duplications with identity between 90%–98%. Additionally we provided examples of pericentromeric and subtelomeric regions that appear to be composed almost entirely of duplicated sequence (IHGSC 2001). However, further characterization of highly duplicated regions has yet to be accomplished.
In this article, we present our methodology for the analysis of such duplications and an in-depth analysis of segmental duplications in the current working draft assembly (January 2001, oo23 assembly), paying particular attention to the quality of assignment and assembly for the duplication-rich clones and regions. Because of the estimated error rates of sequence and the potential for misassembly in the draft assembly, we consider two categories of duplications: segments with >98% nucleotide identity, and segments with 90%–98% identity. For the first time, we quantify the genome-wide enrichment of duplicated sequence in both pericentromeric and subtelomeric regions. In addition, we examine more specifically the impact of these segments on the current assembly. We find duplicated sequences are enriched in sequence contigs that have not been mapped within the current assembly. We also find that clones containing duplications are often assigned to a chromosome inconsistent with FISH and only ∼50% of the chromosomes with FISH signals from these clones have a corresponding sequence similarity by BLAST analysis. This underrepresentation may be attributable to many factors: misassignment, merging, or reduced coverage in these paralogous regions. Taken together, the clustering of duplications combined with the difficulty in positioning and assembling them, suggests that large tracts of segmental duplications, particularly those located at pericentromeres, will be refractory to currently employed assembly methods. Specialized methods will be necessary to correctly integrate these regions into the reference human genome sequence. We propose that the determination of whether an observed overlap is allelic or paralogous will facilitate the final assembly of the human genome, helping to eliminate many gaps both within paralogous as well as unique sequence regions.
RESULTS
Detection of Segmental Duplications (January 2001 oo23 Assembly)
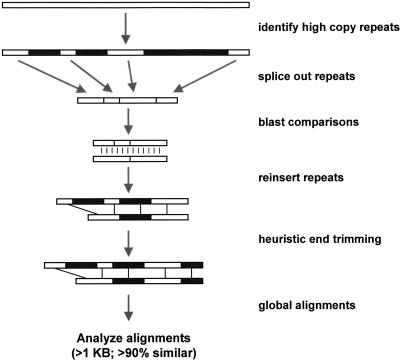
There are two major obstacles to in silico detection of large segmental duplications: (1) They may be composed of common high-copy repeats such as _Alu_s and LINEs; and (2) they may contain large insertion-deletions that hamper the characterization of contiguous segments. To overcome these obstacles we developed a method that we call “fuguization” (see Methods; Fig. 1). This refers to the compact genome of the puffer fish (Fugu rubripes), a genome largely devoid of high-copy repeats (Brenner et al. 1993). The central aspect of our method is to generate a compact version of the human genome sequence by first removing all RepeatMasked high-copy repeats from the sequence, which leaves putatively unique genomic DNA. Fuguization offers two main advantages: It yields faster BLAST searches because of the overall reduction in sequence content (∼50%) and repetitive complexity, and it easily traverses high-copy repeats because of their absence) generating larger contiguous alignments. This enhances our ability to detect duplications riddled with high-copy repeats that would otherwise be missed. It also increases the power to define the true junction boundaries of the duplication event. Additional heuristics were implemented to further refine the junction sequences, to traverse large gaps, and to assess various mapping properties (see Methods).
Figure 1.
Detection Method. The method combines DNA sequence analysis software and a suite of Perl scripts that are optimized for the detection of large highly similar duplications. Briefly, the genome assembly (2.6 Gb) is broken into tractable 400-kb segments. For each segment, common repeats (blue) are identified with RepeatMasker. Repetitive sequence is then removed (“fuguized”) leaving putatively unique DNA. All fuguized pieces are then compared by BLAST. Repeats internal to an individual 400-kb segments are detected with BLASTZ. Relaxed affine gap parameters are used allowing gaps up to 1 kb in size to be traversed. Fuguized pairwise alignments (>0.87 similarity and >500 aligned bp) have their common repeats reinserted and then the alignment ends undergo heuristic trimming allowing for refinement of alignment end points which may lie within common repetitive sequence. The program ALIGN generates optimal global alignments from which final alignment statistics are calculated. Global alignments >1000 bases aligned and >90% identity were selected in this analysis.
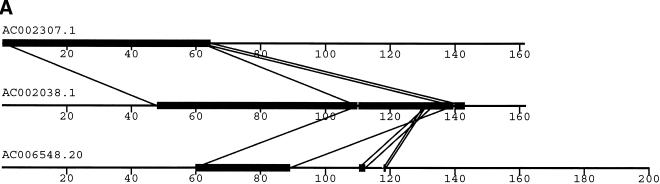
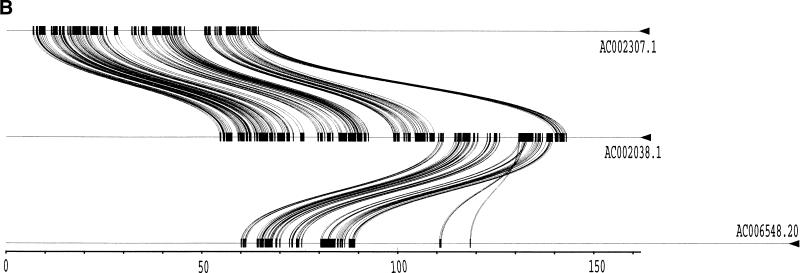
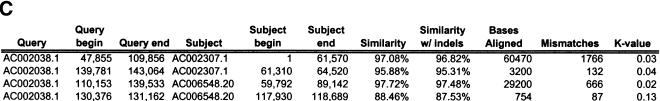
To validate our method, we selected a set of human sequences that contained known duplications with experimentally verified junctions (Eichler et al. 1996, 1997; Horvath et al. 2000a,b). The training set consisted of sequence alignments that ranged from 88%–99% nucleotide identity and contained insertion-deletions as large as 1250 nucleotides. Examination of the 24 alignments returned by our method found that 41 of the 46 alignment end positions were in complete agreement with those determined previously. The five cases that disagreed with previous alignments had ambiguous ends in which the differing end positions were equally valid choices (data not shown). An example is shown for duplications between three pericentromeric clones (Fig. 2). Our method (Fig. 2a) compares favorably to miropeats (Parsons 1995) analysis (Fig. 2b) in that the same duplications are detected, indicating no loss of sensitivity. In contrast, our method allows for the traversal of high-copy repeats and large-insertion deletions. This yields fewer, larger alignments, which allows for more accurate determination of the boundaries between unique and duplicated sequence. An example of a large insertion-deletion is shown in the partial view of a global alignment (Fig. 2d). A sample of the statistics generated for each global pairwise alignment by the program align_scorer (J.A. Bailey, unpubl.) is shown in Figure 2c.
Figure 2.
Example of pericentromeric duplication using fuguization method. (A) A graphical view of the output for our method as displayed in the program PARASIGHT (J.A. Bailey, unpubl.). Compared to miropeats (B; Parsons 1995), all of the positions of similarity have been captured as continuous large alignments (C). An example of a large insertion-deletion in an alignment (D) demonstrates the ability of fuguization to traverse such regions returning larger more meaningful alignments. Lower thresholds (>500 aligned bases; >90% identity) were used for this test case compared to our genome analysis.
Segmental Duplication Content of the Human Genome (January 2001 oo23 Assembly)
As part of the IHSC, we searched for the presence of duplicated sequences (July 2000 oo15 assembly). An unexpected large fraction of the human genome sequence 16.3% (442/2711 Mb) was found to be duplicated by this analysis. Because the majority of these duplications were >98% identical, we suspected that a significant proportion of these might have represented allelic overlaps missed during the assembly of working draft sequence. To help eliminate this artifact, algorithmic improvements in Jim Kent's Gigassembler and a more refined analysis of FPC contigs were implemented in the next major release of public assembly, based in part on our initial analysis (J. Kent, pers. comm..).
We analyzed the current 2692 Mb HGP assembly (January 2001 oo23 assembly) with our method, detecting a total of 48,651 alignments of ≥90% identity and ≥1 kb in size (Fig. 3). Supplement 1, available on-line at http://www.genome.org, contains a detailed breakdown of sequence coverage in terms of chromosome and sequence similarity. Overall, 13.2% (355/2692 MB) of the current assembly was identified as putative segmental duplications. Compared with the oo15 assembly, only a small fraction (<20%) of the highly similar alignments (>98% identity) have now been successfully merged, decreasing from 12.9% in oo15 to 10.6% in oo23. Analyses of other assembly versions, from May 2000 to the most current (oo23), have consistently shown large amounts of these highly similar “duplications” (10%–15% of assembled sequence). The 90%–98% identity compartment (Fig. 3a) has changed only slightly (3.64% in oo15 versus 3.62% in oo23). Within this compartment interchromosomal duplications comprise 1.77% (47.7 Mb) and intrachromosomal duplications comprise 2.29% (97.5Mb) of the overall sequence (on-line Supplement 1). (Note: There is overlap between categories because a given stretch of sequence may be involved in both inter- and intrachromosomal alignments as well as alignments of different percent identity.)
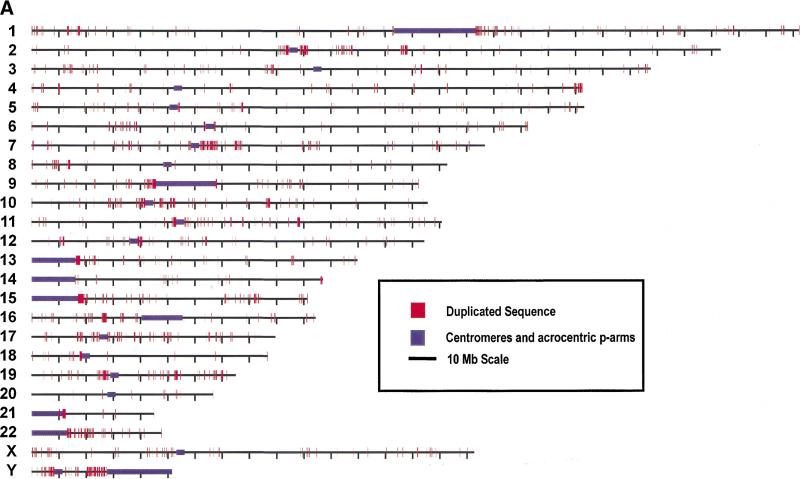
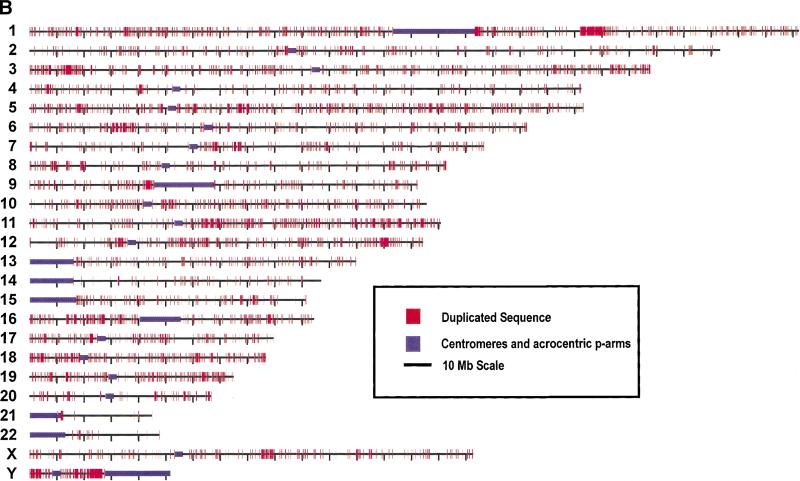
Figure 3.
Genome-wide view of segmental duplications. The positions of alignments are depicted in red for each of the 24 chromosomes. Panels separate alignments on the basis of similarity: (A) 90%–98% identity and (B) 98%–100% identity. Purple bars depict centromeric gaps as well as the p-arms of acrocentric chromosomes (13, 14, 15, 21, and 22).Because of scale constraints, only alignments >5 kb are visible. Views were generated with the program PARASIGHT (J.A. Bailey, unpubl.), a graphical pairwise alignment viewer.
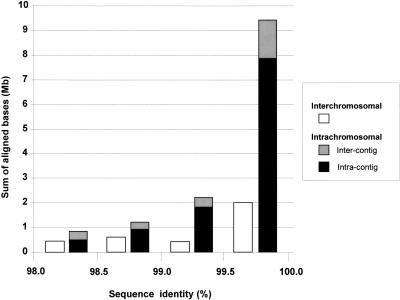
For the highly similar alignments (>98% identity; Fig. 3b), the amount of duplicated sequence is fivefold higher than expected, based on estimates generated from assemblies using only finished sequence (10.6% oo23 versus 2% expected). A more detailed breakdown of highly similar alignments is presented in Figure 4, in which both interchromosomal and intrachromosomal duplications are considered. Intrachromosomal duplications are further divided into two subgroups: duplications that occur within a sequence contig, and those that occur between two different sequence contigs (intracontig and intercontig, respectively). As can be seen in Figure 4, the overwhelming majority (69%) of such alignments are near allelic levels of similarity (99.5%–100% identity) and are located (74%) within the same contig. Taking into account estimated draft sequencing error rates (∼1 error/1000 bases) and potential difficulties owing to assembly misjoins (phrap misassemblies within working draft clones), this overabundance of highly similar intracontig duplications may be caused by missed true overlaps that have not been joined.
Figure 4.
Distribution of highly homologous duplications (>98% identity). A histogram showing the sum of aligned bases for different bins of percent identity. Colors denote interchromosomal alignments (red) and intrachromosomal alignments, which may further subdivided into intercontig (light blue) or intracontig (dark blue) “duplications.”
Segmental Duplications are Difficult to Integrate into the Assembly
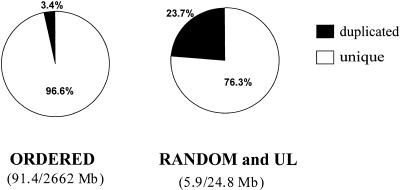
In the oo23 genome assembly, “ordered contigs” are contigs that have been assigned to a chromosome as well as to a unique map location within the chromosome sequence assembly. Two classes of sequence contigs have incomplete positions: unlocated (UL) contigs that lack chromosome assignment, and random contigs that have a chromosome assignment but lack an ordered position within that chromosome. To assess whether BACs containing duplications have been particularly problematic in assembly and chromosomal assignment, we analyzed the distribution of duplicated segments (90%–98% sequence identity, ≥1 kb) within the “random” bin and compared it to the distribution of ordered sequence contigs. The random and UL contigs account for a total of 24.8 Mb, which is the sequence equivalent of a small chromosome. The percent of the random and UL sequence that is duplicated is 23.7% (5.9/24.8 Mb) compared to 3.4% (91.4/2662 Mb) for ordered contigs (Fig. 5). This is a 6.6-fold enrichment compared to the genome average of 3.6%, demonstrating that duplicated sequences are less likely than unique sequences to be assigned a complete genomic position. When duplicated segments showing >98% sequence identity were considered, no significant difference in distribution was observed.
Figure 5.
Integration of segmental duplications into assembly. The two pie charts divide the assembly contigs into ordered contigs and unordered (random and unlocated) contigs. Random contigs have chromosomal assignment but no specific position in the chromosome. Unlocated contigs have no chromosome position. Duplicated sequence represents 3% and 25% of the sequence in the ordered and unordered bins, respectively.
Segmental Duplications are Enriched within Pericentromeric and Subtelomeric Regions
Our previous analyses have shown clusters of duplications in the pericentromeric regions of finished chromosomes 21 and 22 (IHGSC 2001). In addition, several groups have found large tracts of duplications associated with pericentromeric and subtelomeric repetitive marker sequences (Amann et al. 1996; Trask et al. 1998b; Eichler et al. 1999; Horvath et al. 2000b). With the advent of a working draft human reference sequence, we had the opportunity, for the first time, to quantitatively test for these biases in distribution. Because of the limitations of the current assembly, particularly with respect to duplicated regions in the vicinity of heterochromatin, two different approaches were undertaken to measure pericentromeric and telomeric biases: an assembly-based and a repeat-based approach.
For the assembly-based approach, we mapped all duplicated sequence between 90%–98% identity (Fig. 3a) and calculated the number of duplicated bases found in close proximity to predicted centromeric and telomeric locations. It is readily apparent that there are certain regions with megabases of sequence involved in segmental duplications. Associations (≥500 kb) with centromeres (purple) are seen in pericentromeric regions 1q, 2p, 2q, 5q, 7p, 7q, 9p, 9q, 10p, 10q, 11p, 13p, 15q, 17p, 17q, 18p, 19p, 21q, and 22q—roughly one-half (19/43) of the pericentromeres targeted by the HGP. To quantify this view, we defined the pericentromere as the most proximal 2 Mb from the centromere, which encompassed a total of 86 Mb around the 43 sequenced pericentromeres. These pericentromeric regions showed an enrichment of 3.7-fold for all alignments, containing 12% of all duplicated bases although comprising only 3.2% of all genomic sequence. Interchromosomal duplications were enriched 4.5-fold and intrachromosomal duplications were enriched 3.1-fold. For subtelomeric regions, the most proximal 500 kb from each chromosome end was analyzed. These regions showed an enrichment of 1.7-fold for all alignments. Interestingly, only interchromosomal alignments showed a clear bias (2.7-fold) whereas intrachromosomal alignments appear somewhat reduced (0.76-fold enrichment). Thus, based on the predicted location of telomeres and pericentromeric regions within HGP assembly, pericentromeric and subtelomeric regions are enriched for interchromosomal duplications, but only pericentromeric regions showed enrichment for intrachromosomal duplications.
As we have already shown, duplicated sequences are prone to be misassigned; thus, a method that depends solely on map position in the assembly might fail to detect some sequences. Therefore, to examine clustering and enrichment in a manner free from assembly location, we used repetitive sequence markers to identify putative pericentromeric and subtelomeric sequence (Table 1; see Methods). To identify pericentromeric regions, five markers were considered: alpha satellite, beta satellite, CER satellite, gamma satellite, CAGGG repeat, and duplicon 4 (Willard 1990; Eichler et al. 1999; Horvath et al. 2000a). To identify subtelomeric sequence contigs we utilized telomeric associated repeat (TAR) and the classic TTAGGG telomere repeat. We found that the repeat-based analysis showed a higher enrichment compared to the assembly-based method. For all repeat-identified subtelomeric and pericentromeric sequences (PeriSubALL), one-quarter of all duplicated sequence fell within this 4% of the genome, representing a 6.5-fold enrichment. Interchromosomal duplications showed a greater association with repetitive marker sequence (ninefold enrichment) when compared to intrachromosomal duplications (4.9-fold). Unfortunately, the separation of subtelomeric and pericentromeric compartments using a repeat-based strategy is confounded by sequence overlap between the two compartments, which is consistent with observations that telomere-associated sequences such as TAR are also occasionally identified within pericentromeric sequence (Eichler 1999). Given this caveat, the repeat-based subtelomeric compartment shows an 8.3-fold enrichment for duplicated sequences with enrichments for both interchromosomal (11.7-fold) and intrachromosomal (5.9-fold) duplications.
Table 1.
Duplication Content of Pericentromeric and Subtelomeric Compartments
| Method | Sequence | All pairwise | Interchromosomal pairwise | Intrachromosomal pairwise | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kb | % genome | Kb | % pair | fold* | Kb | % pair | fold* | Kb | % pair | fold* | |
| Assembly-Based** | |||||||||||
| Peri2Mb | 86,000 | 3.2% | 11,548 | 11.8% | 3.7 | 7,348 | 14.5% | 4.5 | 6,341 | 10.0% | 3.1 |
| Sub500kb | 21,000 | 0.8% | 1,290 | 1.3% | 1.7 | 1,062 | 2.1% | 2.7 | 376 | 0.6% | 0.8 |
| Repeat-Based** | |||||||||||
| Perialpha | 58,561 | 2.2% | 9,999 | 10.3% | 4.7 | 7,565 | 14.9% | 6.9 | 5,106 | 8.1% | 3.7 |
| PeriαβCERγ | 76,143 | 2.8% | 16,245 | 16.7% | 5.9 | 11,689 | 23.0% | 8.1 | 8,618 | 13.6% | 4.8 |
| Periduplicons | 23,425 | 0.9% | 9,995 | 10.2% | 11.8 | 8,824 | 17.4% | 20.0 | 3,556 | 5.6% | 6.5 |
| PeriALL | 92,090 | 3.4% | 22,769 | 23.3% | 6.8 | 17,350 | 34.2% | 10.0 | 10,863 | 17.2% | 5.0 |
| Sub1kb | 23,535 | 0.9% | 7,038 | 7.2% | 8.3 | 5,179 | 10.2% | 11.7 | 3,261 | 5.2% | 5.9 |
| PeriSubALL | 106,678 | 4.0% | 25,157 | 25.8% | 6.5 | 18,763 | 37.0% | 9.3 | 12,211 | 19.3% | 4.9 |
| TOTAL ASSEMBLY | 2,692,604 | — | 97,514 | — | — | 50,752 | — | — | 63,160 | — | — |
For pericentromeric regions, several different marker subcategories were considered independently as well as combined (Table 1). When all five markers were analyzed (PeriSubALL), we found that >23% of all duplicated bases were associated with pericentromeric repeats (representing a 6.8-fold enrichment). Interchromosomal duplications show the strongest association, in which more than one-third of all duplicated bases (34.2%) are located near such repeats. Within smaller pericentromeric subcategories (Perialpha, PeriαβCERγ, and Periduplicons), interchromosomal enrichment varies considerably from 6.9-fold to 20-fold. The most enriched pericentromeric subcompartment (Periduplicons) consists of two recently characterized interspersed pericentromeric repeats that were originally identified in close proximity to duplicated genomic segments (Eichler et al. 1996; Horvath et al. 2000b). It is not surprising that virtually none of these elements exist in the absence of a nearby duplicated segment. However, even if the classical marker of centromeric DNA (alpha satellite) is solely considered, a strong interchromosomal duplication bias is evident (6.9-fold, Table 1).
Segmental Duplications are Underrepresented and/or Misassigned
To assess the potential role highly homologous duplicated sequences play in the assembly of the human genome sequence, we selected 37 RPCI-11 BAC clones containing interchromosomal duplications by standard metaphase FISH analysis. Each clone had been sequenced as part of the HGP; its clone identity had been verified, was not chimeric in organization (see Methods), and had been predicted by in silico analysis to harbor several interchromosomal duplications (see Methods). Observed FISH signals can be used as a standard with which to compare the completeness and accuracy of the assembly in terms the assignment of interchromosomal duplications. First, we used similarity searches to simulate the potential location of multi-site signals within the current assembly. We set low-stringency search criteria for a FISH equivalent hit, as a BLAST result with sequence alignment ≥90% identity and ≥5000 unique bases within a 400-kb segment (Table 2). By these parameters, we would expect that many significant alignments of ∼90% similarity and 5000 bases would be false positives (as they are small diverged sequences that would not generate a strong FISH signal within the context of a whole BAC hybridization). Such a low threshold, however, should minimize false negatives, chromosomes in the assembly that contain sequence (undetected by BLAST) that were positive by FISH. FISH analysis of our 37 sequenced BAC clones identified a total of 224 interchromosomal signals of which 47% (106/224) lacked a corresponding BLAST hit within the current genome assembly. There are two likely causes for this absence: The sequence is missing from the working draft, or the sequence is not assigned to its proper chromosome. Because the exact equivalence of BLAST sequence identity and length compared to whole-BAC FISH has not been precisely quantified, we generated a series of BLAST versus FISH simulations using various thresholds for a positive BLAST hit (Table 3). Even after lowering the threshold to 90% and 2500 unique bases, 42% of the FISH positive chromosomes remain undetected by BLAST. If we combine our results with a larger subset of characterized multi-site clones (Cheung et al. 2001), similar results are obtained with 49% (278/569) of paralogous chromosomes undetected by in silico analysis of the working draft sequence at 90% and 5000 bp (Supplement 2, available on-line at http://www.genome.org).
Table 2.
BLAST vs. FISH for BACs with Interchromosomal Duplications: Comparison of Chromosome Assignment
| Accession no. | RPCl-11 clone | oo23 assignment (chr/contig) | Assignment consistency with FISH* | BLAST chr # | BLAST chr (>=90% identity >=5000 unique bases)** | FISH chr # | FISH chr (Standard Metaphase Spread) | FISH chr positive by BLAST** |
|---|---|---|---|---|---|---|---|---|
| AC006379 | 456n16 | 7/ctg15064 | I | 1 | 7 | 2 | 16,19 | 0 |
| AC007276 | 226o1 | 7/ctg15064 | C | 2 | 7,12 | 3 | 7,9,12 | 2 |
| AC008166 | 155j2 | 2/ctg15424 | C | 1 | 2 | 3 | 2,3,10 | 1 |
| AC009954 | 226l21 | 6/ctg16045 | l | 10 | 1,4,6,7,8,10,13,16,20,Y | 3 | 8,17,18 | 1 |
| AC010098 | 400j9 | 2/ctg14798 | C | 13 | 1,2,4,5,7,10,13,15,16, 17,21,22,U,L,Y | 10 | 1,2,7,9,10,14,15,16,17,22 | 8 |
| AC011244 | 497h16 | 5/ctg12770 | C | 4 | 5,6,20,22 | 2 | 5,6 | 2 |
| AC011881 | 17l12 | UL/ctg14414 | UL | 5 | 2,5,6,10,15,UL | 9 | 1,3,4,9,13,14,15,21,22 | 1 |
| AC012661 | 413e6 | 3/ctg14246 | C | 10 | NA,UL,Y,3,4,5,8,9,11, 15,16,17 | 18 | 1,3,4,7,8,9,10,11,12,13,14, 15,16,19,20,21,22,Y | 8 |
| AC013633 | 12p21 | 22/ctg25248 | C | 4 | 2,4,9,22,UL | 6 | 9,13,14,15,21,22 | 2 |
| AC015973 | 366c6 | 1/ctg14824 | C | 3 | 1,15,UL,Y | 3 | 1,8,Y | 2 |
| AC016745 | 480c16 | 2/ctg16335 | C | 3 | 2,6,9 | 3 | 2,8,9 | 2 |
| AC018687 | 462g22 | 4/ctg16 | C | 1 | 4 | 2 | 4,14 | 1 |
| AC018692 | 555k2 | 4/ctg25230 | C | 3 | 1,4,9 | 5 | 4,9,13,14,21 | 2 |
| AC018963 | 452l16 | 4/ctg25230 | l | 8 | 1,2,4,5,15,16,19,22,UL | 3 | 2,15,16 | 3 |
| AC020590 | 150n22 | 3/ctg17028 | C | 6 | 2,3,4,9,17,UL,Y | 12 | 3,4,9,13,14,15,16,18,20, 21,22,Y | 4 |
| AC020715 | 446k10 | 19/ctg25122 | C | 1 | 19 | 2 | 5,19 | 1 |
| AC022030 | 358b14 | 18/ctg25448 | C | 2 | 16,18 | 2 | 16,18 | 2 |
| AC022192 | 23b24 | 2/ctg100 | l | 8 | 2,5,9,10,13,15,18,21,UL | 9 | 3,4,9,13,14,15,21,22,Y | 4 |
| AC022702 | 33b1 | 4/ctg15540 | C | 8 | 1,4,6,7,9,10,13,19 | 9 | 1,3,4,7,9,10,11,16,19 | 6 |
| AC023099 | 264m14 | 16/ctg16860 | C | 8 | 2,5,10,15,16,19,22,UL,Y | 3 | 2,15,16 | 3 |
| AC024117 | 327i12 | 10/ctg13284 | C | 1 | 10 | 2 | 2,10 | 1 |
| AC024119 | 542j23 | 1/ctg17190 | C | 2 | 1,2 | 3 | 1,3,19 | 1 |
| AC024345 | 384k6 | 4/ctg15540 | C | 9 | 1,4,6,7,8,10,13,20,Y | 15 | 1,2,3,4,5,6,7,8,9,10,11,16, 19,20,Y | 8 |
| AC024410 | 125l9 | 5/ctg12474 | l | 1 | 5 | 5 | 13,14,15,21,22 | 0 |
| AC025222 | 297j4 | 9/ctg25280 | l | 5 | 1,4,9,12,15 | 2 | 1,11 | 1 |
| AC026205 | 61i9 | 3/ctg25118 | C | 1 | 3 | 2 | 3,4 | 1 |
| AC026495 | 509a17 | 15/ctg25305 | C | 6 | 1,2,4,15,16,22,UL | 10 | 1,2,9,10,13,14,15,16,17,22 | 5 |
| AL078621 | 395l14 | 2/ctg16335 | C | 11 | 2,6,7,8,9,15,16,19,21, 22,X | 16 | 1,2,3,6,7,8,9,10,11,12,15, 16,17,19,20,22 | 9 |
| AL132658 | 59j12 | 10/ctg13655 | l | 2 | 1,10 | 2 | 1,8 | 1 |
| AL133173 | 453n3 | 4/ctg3364 | l | 8 | 1,2,4,11,15,16,22,UL,X | 7 | 2,9,10,14,15,16,22 | 4 |
| AL133216 | 291l22 | 10/ctg13655 | C | 11 | 1,4,6,7,8,9,10,13,19, 20,Y | 13 | 1,3,4,5,6,7,9,10,11,16,19, 20,Y | 9 |
| AL137070 | 251o17 | 2/ctg14048 | C | 6 | 2,9,13,14,18,21 | 2 | 2,9 | 2 |
| AL138715 | 77p19 | 13/ctg14226 | C | 10 | 2,5,7,10,13,15,16,17,21, 22,UL | 7 | 2,13,14,15,18,21,22 | 5 |
| AL138998 | 119m6 | 4/ctg25230 | C | 1 | 4 | 4 | 4,13,14,21 | 1 |
| AL158811 | 139o21 | 22/ctg13153 | C | 7 | 1,2,5,7,13,21,22,UL | 8 | 1,2,7,13,14,15,21,22 | 6 |
| AL161418 | 149i22 | 22/ctg13153 | C | 7 | 1,2,5,7,13,21,22,UL | 5 | 13,14,15,21,22 | 3 |
| AL354817 | 85c8 | UL/ctg15409 | UL | 12 | 2,3,4,5,7,8,9,10,13,18, 19,21,UL | 12 | 1,2,3,4,9,13,14,15,20,21, 22,Y | 6 |
| Total | 37 | 201 | 224 | 118 |
Table 3.
BLAST vs. FISH for BACs with Interchromosomal Duplications: Multiple Thresholds for a Postive BLAST Signal
| Statistic | Minimum sum of bases for positive chromosome** | ||||||
|---|---|---|---|---|---|---|---|
| 2500 | 5000 | 10000 | 20000 | 40000 | |||
| Minimum percent identity for BLAST HSPs | 90% | BLAST chr number | 240 | 201 | 148 | 104 | 53 |
| % BLAST chr over FISH chr* | 107% | 90% | 66% | 46% | 24% | ||
| % FISH chr identified by BLAST | 58% | 53% | 44% | 34% | 17% | ||
| % BLAST chr confirmed by FISH | 0.47 | 0.51 | 0.59 | 0.65 | 0.68 | ||
| 94% | Sum of BLASTNUM | 216 | 183 | 140 | 92 | 52 | |
| % BLAST chr over FISH chr* | 96% | 82% | 63% | 41% | 23% | ||
| % FISH chr identified by BLAST | 54% | 50% | 43% | 30% | 17% | ||
| % BLAST chr confirmed by FISH | 49% | 53% | 60% | 65% | 69% | ||
| 96% | BLAST chr number | 166 | 152 | 118 | 81 | 47 | |
| % BLAST chr over FISH chr* | 74% | 68% | 53% | 36% | 21% | ||
| % FISH chr identified by BLAST | 46% | 43% | 38% | 26% | 16% | ||
| % BLAST chr confirmed by FISH | 54% | 55% | 63% | 64% | 68% | ||
| 98% | BLAST chr number | 119 | 105 | 78 | 53 | 36 | |
| % BLAST chr over FISH chr* | 53% | 47% | 35% | 24% | 16% | ||
| % FISH chr identified by BLAST | 36% | 33% | 25% | 19% | 13% | ||
| % BLAST chr confirmed by FISH | 62% | 65% | 69% | 75% | 75% | ||
| 99% | BLAST chr number | 70 | 57 | 46 | 39 | 32 | |
| % BLAST chr over FISH chr* | 31% | 25% | 21% | 17% | 14% | ||
| % FISH chr identified by BLAST | 22% | 19% | 16% | 14% | 12% | ||
| % BLAST chr confirmed by FISH | 63% | 68% | 72% | 77% | 81% |
A reciprocal analysis was also performed, in which BLAST criteria were set to include only large highly similar sequences (≥40,000 unique bases and ≥99% identity) that are almost certain to produce a FISH signal. If no FISH signal is seen, then the sequence has the wrong chromosomal assignment or there exists considerable heteromorphic variation in the distribution of these segments within the human population. However, for the 32 BLAST positive chromosomes that passed this strict threshold, 19% of them could not be confirmed by FISH, suggesting that these large highly similar sequences have been placed on the wrong chromosome. Not surprisingly, these highly similar hits are nearly equivalent to an analysis comparing FISH to the assembly position for each of the 37 clones. Of the 35 with chromosomal assignments, 21% are inconsistent with FISH localizations suggesting that they have been assigned to nonallelic and nonparalogous locations (Table 2).
DISCUSSION
Our results revealed several interesting features of segmental duplications—both biological and practical—that had not been characterized previously. This was the first genome-wide analysis quantifying pericentromeric and subtelomeric duplication biases. Because of limitations of the oo23 assembly, we pursued two independent methods to assess this effect. We first defined pericentromeres and subtelomeres solely on the basis of their position in the assembly. However, because our FISH analysis of both duplicated and heterochromatic (data not shown) clones often revealed incorrect chromosomal assignment, we sought to examine sequence based only on its association with centromeric and telomeric repetitive markers. Both analyses revealed a strong pericentromeric duplication bias with enrichment levels ranging from 4.7-fold (assembly-based approach) to 11.8-fold (repeat-based approach). Because the sequence markers used in this study localize almost exclusively to centromeric and/or subtelomeric regions (Willard 1990; Eichler 1999; Lee et al. 1999; Horvath et al. 2000b), we believe that the observed increase was due to the ascertainment of additional pericentromeric and subtelomeric sequence, rather than the inclusion of DNA from outside of these regions.
It is interesting to note that this bias does not appear to be uniformly distributed among all chromosomes. Associations (≥500 kb) between duplications and centromeres are observed for only one-half (19/43) of all possible pericentromeric regions (Fig. 3a: 1q, 2p, 2q, 5q, 7p, 7q, 9p, 9q, 10p, 10q, 11p, 13p, 15q, 17p, 17q, 18p, 19p, 21q, and 22q). There are two possible explanations: (1) The degree of sequence coverage within these regions is inadequate such that the apparent lack of duplication is attributable to the absence of representative sequence and/or misassignment. Although this may be true for some chromosomes, it is unlikely to be the case for chromosomes 6, 20, and the X chromosome where intensive mapping and sequencing efforts have included pericentromeric regions (Bentley et al. 2001; M. Schuler, unpubl.). (2) Alternatively, there are two models for the organization of sequence within the euchromatin-heterochromatin transition—chromosomes that show mosaic patterns of duplication and those that lack this architecture. Another noteworthy observation from our analysis is that the interchromosomal bias appears more pronounced within these regions than that seen for intrachromosomal duplications. It should be noted, however, that intrachromosomal events may be particularly underrepresented in the current assembly as a result of either, again, reduced sequence representation or misassembly of paralogous copies. This effect may be exacerbated if intrachromosomal duplications on average share greater sequence identity (IHGSC 2001). Despite this possible ascertainment bias, some intrachromosomal enrichment within pericentromeric regions could be observed by our assays. No intrachromosomal duplication effect, however, could be identified within assembled subtelomeric regions. Although final verification of the biological trends observed in our study awaits finished sequence, the available data support previous claims that within recent evolutionary time nonhomologous chromosomal exchanges have occurred preferentially within pericentromeres and subtelomeres. Pericentromeric, and to a lesser extent subtelomeric, chromosomal regions are among the most evolutionarily dynamic in the genome.
The other major finding of our paper is more practical in nature, addressing the effect that segmental duplications have in terms of placement and assembly of HGP working draft clones. First, we found that duplicated sequences were difficult to assign to their true genomic locations in the current assembly (oo23). Duplicated sequence was overrepresented among sequenced contigs that could not be mapped easily using traditional methods (23.7% of the 24.8 Mb of random and UL bins compared to ordered sequence where duplicated sequence was 3.4% of the total 2662 Mb). Second, we found evidence for a large fraction (20%) of gross misassignment—a chromosome assignment that could not be confirmed by FISH. This rate was much higher than that observed for single site BACs, for which there was a chromosomal discordance rate of 3.6% (Cheung et al. 2001). The likely explanation is that this increased discordance is caused by the BACs duplicative nature; however, it is difficult to reason why duplication would cause the assignment of a BAC to a nonparalogous location. Third, our analysis indicates that nearly half (47%) of duplicated loci cannot be identified within the current assembly, suggesting either underrepresentation or misassembly of paralogous sequence. Finally, an unusually large amount of highly similar alignments (>98% identity) were identified (10.6%). We suggest that most of these represent artifactual duplications created during the assembly of working draft sequence. It is likely that a significant fraction of these artifacts will be resolved on completion of the finished sequence. It should be emphasized that in contrast to the duplication-rich regions, analyses considering unique regions indicate that the current assembly is remarkably well assembled (Cheung et al. 2001; IHGSC 2001). These highly duplicated regions should be considered exceptional both in terms of assembly and potential biology. The computational tools and concomitant paralogy map of the human genome we have generated should facilitate final assembly of the human genome reference sequence by highlighting these regions for further study.
Our findings point to duplication-rich pericentromeres as particularly problematic in terms of genome assembly. Pericentromeres often contain a megabase or more of wall-to-wall duplications, which provides no unique STSs to allow for clone assignment to a unique genomic position. In addition, these regions are often associated with satellite sequence that may confound efforts to map by fingerprinting. Thus, pericentromeres are the regions most intransigent to assembly as they confound the current overarching method for contig assignment based on BAC clone fingerprinting. The inability to assign and distinguish such paralogous sequences creates gaps in the current genome assembly that cannot be resolved by directing the closure of existing clones or by simply identifying a clone that bridges two existing contigs. Furthermore, not all duplicated sequence is restricted to pericentromeric and subtelomeric regions. Our analysis, in conjunction with previous reports, suggests that the euchromatic portions of human chromosomes are littered with highly homologous duplicated material (Dunham et al. 1999; Loftus et al. 1999; Hattori et al. 2000). Many of these regions are implicated in disease-causing recurrent chromosomal structural rearrangements. It is therefore essential that specialized techniques be developed to identify and assemble these exceptional regions of the human genome. Such strategies should become a priority in the final two years of the HGP.
METHODS
Detection of Segmental Duplications
Our detection method used a combination of published sequence analysis software and a suite of Perl programs to optimize the detection of large recent duplications (≥1 kb and ≥90% identity). Parallel batch processing was incorporated whenever possible to analyze gigabases of sequence in a timely fashion. The basic methodology involved identifying high-copy repeats, removing these repeats from the genomic sequence, searching all sequence for similarity, reinserting repeats into resulting pairwise alignments, trimming the ends of alignments, and the generation of global alignments with statistics (Fig. 1).
For the January 2001 oo23 assembly (2.6 Gb), large contigs were broken into tractable 400 kb segments. High-copy repeats identified by RepeatMasker (Smit and Green http://repeatmasker.genome.washington.edu, version 7/16/2000 with quick option) were spliced out of the sequence: “fuguization.” The resulting unique genomic DNA then underwent global BLAST similarity searches with reduced affine gap extension parameters, which allowed large gaps up to 1 kb to be traversed. NCBI's BLAST (Altschul et al. 1997) generated alignments between 400 kb segments (parameters: -G 180 –E 1 –q –80 –r 30 -z 3 × 10−9 –Y 3 × 10−9 –e 1e−10 –F F). A modified version of BLASTZ (W. Miller, unpubl.) that ignores self-identity compared each 400 kb piece to itself (parameters: B = 2 M = 30 I = −80 V = −80 O = 180 E = 1 W = 14 Y = 1400). The BLAST results were parsed for alignments with >1 kb of aligned bases and >88% identity. Each alignment was “defuguized” (the high-copy repeats were reinserted) and then alignment end trimming was done with the program blast_end_trim (J.A. Bailey, unpubl.). End trimming more precisely defined the alignment end positions, which may have been incorrect as a result of the relaxed gap parameters used or because the true end positions resided in a high copy repeat. Blast_end_trim is a heuristic program that attempted to extend the alignment (up to 2 kb) beyond the defined end position using global alignments generated by the program ALIGN (Myers and Miller 1988). When extension failed, the length of the attempted extension is recursively decreased until it converges on a given end position. After trimming, ALIGN was used to generate global alignments from which statistics were calculated using the program align_scorer (J.A. Bailey, unpubl.). Global alignments that equal or exceed the threshold of 1000 bases aligned and >90% identity (i.e., gaps excluded) were retained for further analysis. Generation of global alignments also acted as a safeguard against false positives from BLAST analysis.
Alignments ≥1 kb and ≥90% were considered in this analysis. The rationale for this decision was as follows: Size selection of ≥1 kb would potentially eliminate any uncharacterized transposons as sources of contamination; sequence similarity ≥90% would allow us to detect duplication events within the last 25 million years of primate evolution (neutral rates of nucleotide substitution). Below this threshold, detection of large-scale segmental duplication events becomes problematic because of extensive deletion, retroposition, and rearrangement of noncoding sequences. In cases of extremely large gaps (>1KB), alignments were fractured. Gaps were joined after the initial generation of BLAST alignments (although the sequence still lacked repeats) for gaps up to 5 kb and a deletion side of gap ±10 bp. Later, after the generation of final global alignments, larger gaps (up to 20 kb insertion side; minimum side of gap ±20 bp) were merged with the program alignment_joiner (J.A. Bailey, unpubl.). For oo23, the entire process of detection, from RepeatMasking through the generation of global alignments, takes roughly three weeks on a Linux computer cluster consisting of 32 600-MHZ Pentium processors. About one-half of this time is required for the initial identification of the high-copy repeats using RepeatMasker. For oo23 we utilized the RepeatMasker output that had already been generated for the assembly process using the −q option (J. Kent, unpubl.).
The training set consisted of 10 GenBank accessions: AC000382.1, AC002038.1, AC002041.1, AC002307.1, AC004222.1, AC004527.2, AC006359.3, U36341.1, U41302.1, and U52111.1. Large gaps (>1 kb) were not joined with alignment_joiner (J.A. Bailey, unpubl.), thus gaps were only traversed in the fuguization and trimming steps.
Measures of Duplicated Sequence
From the alignments, two main forms of statistics were generated. First, nonredundant bases involved in all duplications were calculated in terms of total bases duplicated and percentage of sequence duplicated with the program table_seqoverlap_combine (J.A. Bailey, unpubl.). The calculation was simply whether or not a base lies within a pairwise alignment. Alignments were broken down into various subsets based on categories such as chromosome location, contig type (ordered and unordered), similarity (90%–98% and >98 identity), and duplication type (inter- and intrachromosomal). For categories, such as similarity and duplication type, certain bases were involved in more than one subset, which resulted in the total numbers of bases involved in all alignments being less than the sum of the subsets. Second, the alignments themselves were broken down into categories (similarity, length, inter vs. intra, etc.) and the number alignments and the sum of aligned bases were calculated (Fig. 4). This measure is redundant because a base was counted each time it was involved in a pairwise.
Subtelomeric and Pericentromeric Localizations
To investigate possible enrichment in pericentromeric and subtelomeric regions, we first used the assembled chromosomes to define the pericentromere as the most centromeric 2 Mb and the subtelomere as the most telomeric 500 kb. The second method involved a repeat-based strategy whereby we assigned sequence, within 500 kb of clusters of known pericentromeric and subtelomeric repetitive markers, as putative pericentromeric and subtelomeric regions. Assembly contig boundaries were not crossed when defining sequence within 500 kb. Clusters were defined as a minimum amount of repetitive sequence within a 400-kb segment of sequence. If repeats did not pass this threshold in a 400-kb segment, they were not included. Minimum thresholds for clustering used for the various combinations of repeats were: 10 kb of alpha satellite for Perialpha; 10 kb of alpha, beta, CER, and/or gamma satellite for PeriabCERy; 1 kb of CAGGG and/or duplicon4 for Periduplicons; and 1 kb of TAR or TTAGGG for Sub1kb. PeriALL combined the sequence in Perialpha, PeriabCERy, and Periduplicons. PeriSubALL combined all of the identified repeat-based sequence. Once the putative sequence for a region had been defined, the region was assayed for duplicated bases using the program seqpos_intersection (J.A. Bailey, unpubl.). Enrichment was calculated as the fraction of the total genome duplicated bases in a region divided by the fraction of the genome that the region represented.
Different thresholds and repetitive sequences were combined to generate different regional compartments. The ascertainment process for any given region was consistent. First, for each 400-kb segment, a segment was assayed for a minimum number of bases of relevant repeat. If so, these repeats were then used to define a region within 500 kb of any of these repeats in the larger fingerprint contig. (Contig boundaries were not crossed when defining these bases, but 400-kb segment boundaries were crossed.) The amount of duplicated bases that fell within any of these compartments was calculated using the program seqpos_intersection. Enrichment was defined as the fraction of total duplicated bases within a region over the fraction of the total assembled sequence that was contained in the region.
Clone Analysis
Based on database searches of GenBank (ver. 118, June 2000), we identified RPCI-11 BACs with potential duplications on the basis of sharing large overlaps with other clones (94%–98% identity; ≥10 kb aligned bases). These overlaps were detected in a global comparison of the human htgs and nt databases by BLAST. Representative RPCI-11 BACs from paralogous clusters were isolated and end-sequenced to confirm clone identity. Further, clones that showed no significant (<e−12) overlap with other RPCI-11 by fingerprinting (http://genome.wustl.edu/gsc/human/human_database.shthml) were excluded as possible chimeric clones. Eighty-three BACs consistent with their GenBank sequences were analyzed by standard metaphase FISH (Cheung et al. 2001). For our analysis of the oo23 assembly, we selected the 37 BACs that showed multichromosomal FISH localizations (as opposed to single site or multiple signals within single chromosome). As Cot-1 DNA was used to block repetitive signal in FISH, we used 400-kb segments, which lacked high-copy repeats, as our target genome database. We queried this database with the fuguized sequence of each of the 37 BACs. A FISH equivalent database match within a 400-kb segment was chosen to be >5000 aligned bases among HSPs with alignments ≥100 bases and ≥90% identity. If a BLAST-positive 400-kb segment had a chromosomal assignment, the chromosome was scored as BLAST-positive (Table 2). Because the correlation between FISH-positive and BLAST-positive sequences is not precisely known, we used a series of different thresholds for percent similarity and total aligned bases (Table 3).
Acknowledgments
We thank Anthony Popkie and Laurie Christ for technical assistance, and Dr. Ann Moormann and Devin Locke for helpful comments during the preparation of this manuscript. We also acknowledge Jim Kent, Dr. David Haussler, and Dr. Greg Schuler for providing access to sequence assemblages prior to publication. This work was supported by grants NIH GM58815, DOE ER62862–1013741–0005006, and a Basil O'Connor Scholar award (FY99–0519) to E.E.E.; it was also supported by NIH grant CA80295 to B.J.T. J.A.B. was supported in part by a Medical Sciences Training Program Grant. The financial support of the W.M Keck Foundation and a Howard Hughes Medical Institute grant to Case Western Reserve University, School of Medicine are also gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eee@po.cwru.edu; FAX (216) 368-3432.
Article published on-line before print: Genome Res., 10.1101/gr.187101.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.187101.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann J, Valentine M, Kidd VJ, Lahti JM. Localization of chi1-related helicase genes to human chromosome regions 12p11 and 12p13: Similarity between parts of these genes and conserved human telomeric-associated DNA. Genomics. 1996;32:260–265. doi: 10.1006/geno.1996.0113. [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Deloukas P, Dunham A, French L, Gregory SG, Humphray SJ, Mungall AJ, Ross MT, Carter NP, Dunham I, et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20, and X. Nature. 2001;409:942–943. doi: 10.1038/35057165. [DOI] [PubMed] [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Chen K, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault A, Lee C, Lupski J. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- Cheung VE, Nowak N, Jang W, Kirsch IR, Zhao S, Chen X-N, Furey TS, Kim U-J, Kuo W-L, Olivier M, et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. Human genome project: 1998–2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo- cardio-facial syndrome. Am J Hum Genet. 1999;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE. Masquerading repeats: Paralogous pitfalls of the Human Genome. Genome Res. 1998;8:758–762. doi: 10.1101/gr.8.8.758. [DOI] [PubMed] [Google Scholar]
- ————— Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet. 1996;5:899–912. doi: 10.1093/hmg/5.7.899. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum Mol Genet. 1997;6:991–1002. doi: 10.1093/hmg/6.7.991. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 1999;9:1048–1058. doi: 10.1101/gr.9.11.1048. [DOI] [PubMed] [Google Scholar]
- Green P. Against a whole-genome shotgun. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. The chromosome 21 mapping and sequencing consortium. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Horvath J, Schwartz S, Eichler E. The mosaic structure of a 2p11 pericentromeric segment: A strategy for characterizing complex regions of the human genome. Genome Res. 2000a;10:839–852. doi: 10.1101/gr.10.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath J, Viggiano L, Loftus B, Adams M, Rocchi M, Eichler E. Molecular structure and evolution of an alpha/non-alpha satellite junction at 16p11. Hum Mol Genet. 2000b;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- IHGMC (International Human Genome Mapping Consortium) A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- IHGSC (International Human Genome Sequencing Consortium) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications, and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Lee C, Stanyon R, Lin CC, Ferguson-Smith MA. Conservation of human gamma-X centromeric satellite DNA among primates with an autosomal localization in certain Old World monkeys. Chromosome Res. 1999;7:43–47. doi: 10.1023/a:1009223327818. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Kim UJ, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby ML, Barnstead M, Cronin L, et al. Genome duplications and other features in 12 Mbp of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- Mazzarella R, Schlessinger D. Duplication and distribution of repetitive elements and non-unique regions in the human genome. Gene. 1997;205:29–38. doi: 10.1016/s0378-1119(97)00477-0. [DOI] [PubMed] [Google Scholar]
- ————— Pathological consequences of sequence duplications in the human genome. Genome Res. 1998;8:1007–1021. doi: 10.1101/gr.8.10.1007. [DOI] [PubMed] [Google Scholar]
- Monfouilloux S, Avet-Loiseau H, Amarger V, Balazs I, Pourcel C, Vergnaud G. Recent human-specific spreading of a subtelomeric domain. Genomics. 1998;51:165–176. doi: 10.1006/geno.1998.5358. [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–7. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Parsons J. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Regnier V, Meddeb M, Lecointre G, Richard F, Duverger A, Nguyen VC, Dutrillaux B, Bernheim A, Danglot G. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Hum Mol Genet. 1997;6:9–16. doi: 10.1093/hmg/6.1.9. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Tomlinson IM, Cook GP, Carter NP, Elaswarapu R, Smith S, Walter G, Buluwela L, Rabbitts TH, Winter G. Human immunglobulin VH and D segments on chromosomes 15q11.2 and 16p11.2. Hum Mol Genet. 1994;3:853–860. doi: 10.1093/hmg/3.6.853. [DOI] [PubMed] [Google Scholar]
- Trask B, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998a;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen OT, Eichler EE, van den Engh G, Rouquier S, Shizuya H, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet. 1998b;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- Willard HF. Centromeres of mammalian chromosomes. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- Wong Z, Royle N, Jeffreys A. A novel human DNA polymorphism resulting from transfer of DNA from chromosome 6 to chromosome 16. Genomics. 1990;7:222–234. doi: 10.1016/0888-7543(90)90544-5. [DOI] [PubMed] [Google Scholar]
- Zimonjic D, Kelley M, Rubin J, Aaronson S, Popescu N. Fluorescence in situ hybridization analysis of keratinocyte growth factor gene amplification and dispersion in evolution of great apes and humans. Proc Natl Acad Sci. 1997;94:11461–11465. doi: 10.1073/pnas.94.21.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]