Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment (original) (raw)
Abstract
Background and Aims
Polyploidy is a major component of plant evolution. The citrus gene pool is essentially diploid but tetraploid plants are frequently encountered in seedlings of diploid apomictic genotypes. The main objectives of the present study were to establish the origin of these tetraploid plants and to ascertain the importance of genotypic and environmental factors on tetraploid formation.
Methods
Tetraploid seedlings from 30 diploid apomictic genotypes were selected by flow cytometry and genotyped with 24 single sequence repeat (SSR) markers to analyse their genetic origin. Embryo rescue was used to grow all embryos contained in polyembryonic seeds of ‘Tardivo di Ciaculli’ mandarin, followed by characterization of the plantlets obtained by flow cytometry and SSR markers to accurately establish the rate of tetraploidization events and their potential tissue location. Inter-annual variations in tetraploid seedling rates were analysed for seven genotypes. Variation in tetraploid plantlet rates was analysed between different seedlings of the same genotype (‘Carrizo’ citrange; Citrus sinensis × Poncirus trifoliata) from seeds collected in different tropical, subtropical and Mediterranean countries.
Key Results
Tetraploid plants were obtained for all the studied diploid genotypes, except for four mandarins. All tetraploid plants were identical to their diploid maternal line for SSR markers and were not cytochimeric. Significant genotypic and environmental effects were observed, as well as negative correlation between mean temperature during the flowering period and tetraploidy seedling rates. The higher frequencies (20 %) of tetraploids were observed for citranges cultivated in the Mediterranean area.
Conclusions
Tetraploidization by chromosome doubling of nucellar cells are frequent events in apomictic citrus, and are affected by both genotypic and environmental factors. Colder conditions in marginal climatic areas appear to favour the expression of tetraploidization. Tetraploid genotypes arising from chromosome doubling of apomictic citrus are extensively being used as parents in breeding programmes to develop seedless triploid cultivars and have potential direct use as new rootstocks.
Keywords: Citrus, polyembryony, apomixis, tetraploid, SSR markers, flow cytometry, chromosome doubling
INTRODUCTION
Polyploidy is a major component of eukaryote evolution and particularly in angiosperms (Grant, 1981; Soltis and Soltis, 1993; Wendel and Doyle, 2005). Many plant species result from autopolyploidization or allopolyploidization events and polyploidization should be considered as the most common sympatric speciation mechanism (Otto and Whitton, 2000). According to Thompson and Lumaret (1992), the dynamics of polyploid plants is based on three processes: the origin (polyploidization events), the establishment and the persistence of new polyploids. The mechanisms leading to polyploidy were variously discussed during the 1970s, and for a long time chromosome doubling was considered the major cause. Most authors (e.g. Stebbins, 1971) considered that meiotic restitution played only a minor role in the evolution of polyploid complexes. However, Harlan and De Wet (1975) argued that spontaneous chromosome doubling must be relatively rare in nature while polyploidization arising from 2_n_ gametes seems quite common. They claimed that, ‘Almost all polyploids arise by way of unreduced gametes; other mechanisms occur, but are negligible.’ These conclusions are now endorsed by numerous plant evolutionists (e.g. Bretagnolle and Thompson, 1995; Ramsey and Schemske, 1998, 2002).
Diploidy is the general rule in Citrus and related genera of Aurantioideae, with a basic chromosome number x = 9 (Krug, 1943). However, some higher euploid genotypes have been found in the citrus germplasm. The most common euploid variations are triploid and tetraploids (Lee, 1988). Longley (1925) was the first to formally identify a tetraploid wild form: the ‘Hong Kong’ kumquat (Fortunella hindsii Swing.). Triploid ‘Tahiti’ lime (Citrus latifolia Tan.), tetraploid strains of Poncirus trifoliata (L.) Raf., allotetraploid Clausena excavata Burm. F., tetraploid Clausena harmandiana Pierre (Guill) and hexaploid Glycosmis pentaphylla Retz. (Corrêa) are other examples of the few natural polyploids found in the germplasm of Aurantioideae (Ollitrault et al., 2008).
Despite the scarcity of polyploid genotypes in citrus germplasm banks, it appears that polyploidization events are relatively frequent when seedling populations are analysed. The reported frequency of female 2_n_ gametes ranges from rates below 1 % to over 20 % (Soost, 1987; Iwamasa et al., 1988), probably due to the abortion of the second meiotic division in the megaspore (Esen et al., 1979). It appears that most of the spontaneous triploids arising from diploid parents are found in small and abnormal seeds (Esen and Soost, 1971, 1973; Geraci et al., 1975; Aleza et al., 2010_a_), which are unlikely to germinate without implementation of an embryo rescue procedure. Therefore, 2_n_ gametes cannot have played an important role in citrus evolution.
In citrus germplasm, apomictic (nucellar polyembryony) and non-apomictic genotypes are found (Frost and Soost, 1968). In non-apomictic citrus genotypes, cases of sexual polyembryony have been reported, probably originating by cleavage of the zygotic embryo or from two or more functional embryo sacs in a single ovule (Frost, 1926; Bacchi, 1943; Aleza et al., 2010_b_). The majority of citrus genotypes are apomictic, with the exception of all citron (C. medica L.), pummelo [C. grandis (L.) Osb.] and clementine (C. clementina Hort. ex Tan.) genotypes and some mandarin hybrids. Tetraploidization seems to occur frequently in apomictic citrus genotypes. Lapin (1937) found tetraploid seedlings among eight citrus species (rate ranging from less than 1 to 5·6 %) and Poncirus (4 %). Cameron and Frost (1968) estimated that 2·5 % of nucellar progenies from a broad range of citrus genotypes grown in California were tetraploid, whereas Barrett and Hutchison (1978) found less than 1 % of tetraploids in seedlings of Citrus, Poncirus and intergeneric hybrids (Citrus × Poncirus) obtained from seeds coming from California and Florida (0·75 and 0·90 %, respectively). In these pioneering studies, ploidy variation was estimated by observation of morphological traits and some tetraploids were confirmed by chromosome counts. The reliability of the estimated rates of tetraploidy is thus questionable. Chromosome doubling in somatic tissues was observed by Raghuvanshi (1962); however, very few tetraploid budsports have been identified as a consequence of unfavourable competition between diploid and tetraploid cells in the meristem (Iwamasa et al., 1988). The formation of fully developed tetraploid seeds from diploid female × tetraploid male hybridization has also been reported (Tachikawa et al., 1961; Cameron and Soost, 1969; Esen and Soost, 1972). Esen and Soost (1972) suggested that they originated from unreduced gametes fertilized by diploid pollen. However, it appears that most of the natural tetraploid lines of citrus arise from apomictic genotypes. Frost and Soost (1968) and Kobayashi et al. (1981) proposed that chromosome doubling in nucellar tissue might be the general mechanism underlying tetraploidization in apomictic citrus genotypes (Cameron and Frost, 1968). However, this hypothesis was not formally demonstrated for a large range of genetic diversity. Today, there is renewed interest in citrus tetraploid lines as parents for seedless triploid breeding programmes (for a bibliographic review see Ollitrault et al., 2008) and for rootstock breeding (Saleh et al., 2008). Several research groups are working to expand the tetraploid gene pool by somatic hybridization (Grosser et al., 2000, 2010) or tetraploid seedling selection (our group). Within this context, the origin and genetic structure of tetraploid seedlings must be clearly identified for their rational use in citrus breeding programmes.
The aim of the present work was to answer three basic questions. (1) What is the genetic origin of the tetraploid plantlets produced by diploid apomictic citrus? (2) What is the probable tissue associated with tetraploidization events and to what extent does the rate of tetraploid seedling occurrence reflect the frequency of these events? (3) How do genotypes and environmental conditions affect tetraploidization?
MATERIALS AND METHODS
Plant material
Genetic origin of tetraploid seedlings in Citrus species
Seeds of 30 apomictic genotypes representing 26 cultivars of three Citrus species and interspecific hybrids (Table 1) obtained by open pollination were collected from mature fruits of trees from two germplasm banks (IVIA Germplasm Bank, Valencia, Spain, and INRA/CIRAD Germplasm Bank, Corsica, France). The taxonomic classification of Swingle and Reece (1967) is used in this paper. Immediately after extraction, seeds were planted in trays of 54–96 alveoli, one seed per alveolus, containing a substrate of a mixture of six parts of peat moss (black/blond, 1 : 1) and one of perlite, in temperature-controlled greenhouses (18–27 °C).
Table 1.
Tetraploid rates in seedlings of 30 genotypes of the Citrus genus
| Species | Genotype | Accession no. | Origin | No. of seedlings | No. of 4_x_ seedlings | SP. 4_x_* | Percentage 4_x_ seedlings |
|---|---|---|---|---|---|---|---|
| C. sinensis | ‘Boukhobza’ | SRA-569 | Cirad, Corsica | 290 | 2 | 2 | 0·8 |
| ‘Tarocco Rosso’ | SRA-574 | Cirad, Corsica | 300 | 1 | 1 | ||
| ‘Cara Cara’ | SRA-666 | Cirad, Corsica | 210 | 1 | 1 | ||
| ‘Sanguinelli’ | SRA-243 | Cirad, Corsica | 150 | 1 | 1 | ||
| ‘Sanguinelli’ | IVIA-34 | Ivia, Moncada | 63 | 2 | 2 | ||
| ‘Moro’ | SRA-301 | Cirad, Corsica | 300 | 1 | 1 | ||
| ‘Shamouti’ | SRA-299 | Cirad, Corsica | 350 | 2 | 2 | ||
| ‘Parson Brown’ | SRA-144 | Cirad, Corsica | 150 | 1 | 1 | ||
| ‘Pineapple’ | SRA-42 | Cirad, Corsica | 300 | 6 | 5 | ||
| C. paradisi | ‘Star Ruby’ | SRA-293 | Cirad, Corsica | 150 | 2 | 2 | 2·8 |
| ‘Star Ruby’ | IVIA-197 | IVIA, Moncada | 61 | 1 | 1 | ||
| ‘Duncan’ | IVIA-274 | IVIA, Moncada | 78 | 5 | 5 | ||
| C. reticulata | ‘Tardivo di Ciaculli’ | IVIA-186 | IVIA, Moncada | 73 | 7 | 6 | 3·0 |
| ‘Anana’ | IVIA-390 | IVIA, Moncada | 70 | 4 | 4 | ||
| ‘Kara’ | IVIA-218 | IVIA, Moncada | 100 | 2 | 2 | ||
| ‘Salteñita’ | IVIA-361 | IVIA, Moncada | 84 | 0 | 0 | ||
| ‘Fremont’ | SRA-147 | Cirad, Corsica | 150 | 0 | 0 | ||
| ‘Beauty’ | SRA-261 | Cirad, Corsica | 150 | 0 | 0 | ||
| ‘Simeto’ | IVIA-413 | IVIA, Moncada | 73 | 0 | 0 | ||
| ‘Kinnow’ | SRA-276 | Cirad, Corsica | 150 | 2 | 2 | ||
| ‘Kinnow’ | IVIA-33 | IVIA, Moncada | 92 | 13 | 13 | ||
| C. reticulata × C. sinensis | ‘Murcott tangor’ | SRA-601 | Cirad, Corsica | 150 | 2 | 2 | 3·5 |
| ‘Murcott tangor’ | IVIA-196 | IVIA, Moncada | 179 | 10 | 8 | ||
| ‘Nadorcott tangor’ | IVIA-641 | IVIA, Moncada | 77 | 2 | 2 | ||
| ‘Ortanique tangor’ | IVIA-276 | IVIA, Moncada | 105 | 4 | 4 | ||
| C. reticulata × C. paradisi | ‘Mapo tangelo’ | IVIA-190 | IVIA, Moncada | 38 | 2 | 2 | 2·5 |
| ‘Minneola tangelo’ | IVIA-84 | IVIA, Moncada | 166 | 3 | 3 | ||
| Complex interspecific hybrids | ‘Page’ | IVIA-79 | IVIA, Moncada | 156 | 1 | 1 | 1·0 |
| ‘Sunburst’ | IVIA-200 | IVIA, Moncada | 112 | 2 | 2 | ||
| ‘Fairchild’ | IVIA-83 | IVIA, Moncada | 115 | 1 | 1 | ||
| Total | 4442 | 80 | 76 | 1·80 |
Accurate evaluation of tetraploidization events and their location
Seeds of ‘Tardivo di Ciaculli’ mandarin, obtained by open pollination, were collected from mature fruits of trees of the IVIA Germplasm Bank. Embryos were isolated from seeds under aseptic conditions with the aid of a stereoscopic microscope and were cultivated on Petri dishes containing Murashige and Skoog (1962) culture medium with 50 g L−1 sucrose, 500 mg L−1 malt extract supplemented with vitamins (100 mg L−1 _myo_-inositol, 1 mg L−1 pyridoxine hydrochloride, 1 mg L−1 nicotinic acid, 0·2 mg L−1 thiamine hydrochloride, 4 mg L−1 glycine) and 8 g L−1 Bacto agar (MS). After germination plantlets were transferred to 25 × 150-mm test tubes with MS culture medium without malt extract. Cultures were maintained at 24 ± 1 °C, 60 % humidity and 16 h daily exposure to 40 µE m−2 s−1 illumination.
Inter-annual and genotypic effect
Seeds of seven apomictic genotypes including one sour orange (C. aurantium L.), two mandarins, one Poncirus and three citranges obtained by open pollination were collected from mature fruits of trees from the INRA/CIRAD Germplasm Bank in three consecutive years (Table 2). Seedlings were grown as described above.
Table 2.
Variation in tetraploid rates in seedlings of seven genotypes from the INRA/CIRAD Germplasm Bank (Corsica) over three years (the number of seedling analysed is given in parentheses)
| Species | Genotype | Accession | Year 2002 | Year 2003 | Year 2004 | P |
|---|---|---|---|---|---|---|
| C. aurantium | ‘Begaradier’ | SRA-952 | 3·1 % (1002) | 1·9 % (268) | – | <0·1 |
| C. reticulata | ‘Willow leaf’ | SRA-133 | 3·9 % (1207) | 2·5 % (1613) | 0·4 % (719) | <0·001 |
| C. reticulata | ‘Cleopatra’ | SRA-948 | 4·1 % (761) | 1·2 % (759) | 0·2 % (526) | <0·001 |
| P. trifoliata | ‘Pomeroy’ | SRA-1074 | – | 5·4 % (1600) | 1·0 % (487) | <0·001 |
| C. sinensis × P. trifoliata | ‘Carrizo’ | SRA-796 | 17·2 % (853) | 8·1 % (908) | 3·5 % (373) | <0·001 |
| C. sinensis × P. trifoliata | ‘Troyer’ | SRA-981 | 20·9 % (250) | 6·9 % (348) | – | <0·001 |
| C. sinensis × P. trifoliata | ‘C 35’ | SRA-731 | – | 2·6 % (349) | 0·3 % (726) | <0·001 |
| Homogeneity χ2 test | P < 0·001 | P < 0·001 | P < 0·001 |
Impact of geographical origin of seeds on tetraploid seedling rates for the same genotype
To complete the analysis of environmental effects, we used seeds of ‘Carrizo’ citrange from several countries worldwide (South Africa: Eastern Cape; Brazil: Bahia and San Paulo states; Uruguay: San José; USA: California and Florida; France: Corsica; Spain: Valencia; Table 3). For this experiment, tetraploid rates were evaluated in three samples of 80 seeds from each geographical origin, except for the South African origin, for which three samples of 100 seeds were used. Seedlings were grown as described above. Meteorological data were collected from the different sites during the period covering floral induction and the flowering period (August–November for the Southern Hemisphere and February–May for the Northern Hemisphere). Mean temperature, mean minimum temperature and mean maximum temperature during the flowering period are given in Table 3.
Table 3.
Origin of the seeds used for the analysis of environmental influence and mean temperatures during the blooming period on tetraploidization
| Origin | Latitude, longitude, altitude | Mean temp. (°C) | Minimum mean temp. (°C) | Maximum mean temp. (°C) |
|---|---|---|---|---|
| Station de Recherches Agronomiques de Corse – INRA-CIRAD, San Giuliano (Corsica) – France. | 46°98′N, 7°98′E, 55 m | 13·9 | 8·2 | 17·4 |
| Willits & Newcomb Inc., Arvin (CA) – USA. | 35°12′N, 118°47′W, 152·4 m | 14·6 | 7·8 | 18·8 |
| Instituto Valenciano de Investigaciones Agrarias, Moncada (Valencia) – Spain. | 39°34′50″N, 00°24′17″W, 55 m | 15·0 | 7·6 | 19·2 |
| Programa Nacional de Certificación de Cítricos – MGAP, Punta del Espinillo (San José) – Uruguay. | 34°47′3″S, 56°15′7″W, 48·8 m | 15·8 | 10·5 | 19·3 |
| Outspan Foundation Block, Uitenhage (Eastern Cape) – South Africa. | 33°76′67″S, 25°31′67″E, 110 m | 16·9 | 8·9 | 23·3 |
| Thompson Citrus Nursery, Winter Haven; Polk County (FL) – USA. | 27°59′N, 81°31′W, 15 m | 21·9 | 14·9 | 25·7 |
| Centro de Citricultura Sylvio Moreira – IAC, Cordeirópolis (San Paulo) – Brazil. | 22°32′S, 47°27′W, 639 m | 21·9 | 15·2 | 27·7 |
| Centro Nacional de Pesquisa de Mandioca e Fruticultura Tropical – EMBRAPA, Cruz das Almas (Bahia) – Brazil. | 12°40′19″S, 39°06′22″W, 220 m | 23·8 | 19·6 | 28·7 |
Ploidy level evaluation by flow cytometry
Ploidy level was determined by flow cytometry according to the methodology described by Aleza et al. (2009). Each sample comprised a small piece of leaf of the analysed plant (approx. 0·5 mm2) with a similar piece from a diploid control plant. For each genotype, one of the seedlings identified as tetraploid was subjected to detailed analysis. Ten samples of four mixed leaves harvested from the entire canopy of the tetraploid plants were studied without a diploid control to check their possible cytochimeric nature.
Chromosome counts
For each genotype we performed a chromosome count of the tetraploid plant selected for inclusion in the IVIA or CIRAD/INRA Germplasm Banks. Tips and small leaves were collected and incubated in a solution of 0·04 % 8-hydroxyquinoline (4 h at room temperature and 4 h at 4 °C). They were then incubated in an ethanol/acetic acid solution (3 : 1) for 48 h and stored at 4 °C in a 70 % ethanol solution. The preparations were then treated for 20 min in 5 m HCl and washed with distilled water. Finally, the tissue was deposited on microscope slides, stained with a drop of DAPI (4–6-diamine-2-phenylindol) and squashed. Observations were carried out with UV light using an E800 eclipse Nikon microscope.
Molecular marker characterization
Genetic analysis was performed with 26 single sequence repeat (SSR) markers: TAA 1, TAA 15 and CAC 15 published by Kijas et al. (1997), Ci01C07, Ci02B07, Ci07C07, Ci07C09 and mCrCIR07E12 (Froelicher et al., 2008), mCrCIR02D04b (Kamiri et al., 2011), mCrCIR01C06, mCrCIR02D09, mCrCIR02F12, mCrCIR02G02, mCrCIR03C08, mCrCIR03G05, mCrCIR04H06, mCrCIR05A05 and mCrCIR07D06 (Cuenca et al., 2011) and eight new SSR markers (described in Table 4). These markers display a broad distribution in the clementine genetic map (Ollitrault et al., 2011).
Table 4.
SSR markers used for the genetic analysis
| SSR marker | Sequence forward 5′ → 3′ | Sequence reverse 5′ → 3′ | Linkage group* | Position (cM) | SSR motif | Reference |
|---|---|---|---|---|---|---|
| TAA 15 | GAAAGGGTTACTTGACCAGGC | CTTCCCAGCTGCACAAGC | 1 | 119·734 | TAA | Kijas et al. (1997) |
| mCrCIR02D09 | AATGATGAGGGTAAAGATG | ACCCATCACAAAACAGA | 2 | 13·378 | GA | Cuenca et al. (2011) |
| mCrCIR04H06 | GGACATAGTGAGAAGTTGG | CAAAGTGGTGAAACCTG | 2 | 27·906 | GA | Cuenca et al. (2011) |
| CAC 15 | TAAATCTCCACTCTGCAAAAGC | GATAGGAAGCGTCGTAGACCC | 2 | 52·561 | CAC | Kijas et al. (1997) |
| mCrCIR03C08 | CAGAGACAGCCAAGAGA | GCTTCTTACATTCCTCAAA | 2 | 98·689 | GA | Cuenca et al. (2011) |
| mCrCIR05A05 | ATACCTGTGAGCGTGAG | CCTCTTCCCTTCCATT | 2 | 153·129 | GA | Cuenca et al. (2011) |
| Ci01C07 | GTCACTCACTCTCGCTCTTG | TTGCTAGCTGCTTTAACTTT | 2 | 153·604 | TC | Froelicher et al. (2008) |
| mCrCIR07D06 | CCTTTTCACAGTTTGCTAT | TCAATTCCTCTAGTGTGTGT | 4 | 16·354 | TG GA | Cuenca et al. (2011) |
| mCrCIR03G05 | CCACACAGGCAGACA | CCTTGGAGGAGCTTTAC | 4 | 75·074 | GA | Cuenca et al. (2011) |
| mCrCIR02D04b | CTCTCTTTCCCCATTAGA | AGCAAACCCCACAAC | 4 | 85·835 | GA | Kamiri et al. (2011) |
| mCrCIR03D12a | GCCATAAGCCCTTTCT | CCCACAACCATCACC | 4 | 90·063 | TGGA | New published marker |
| Mest 15 | TTATTACGAAGCGGAGGTGG | GCCTCGCATTCTCTTGACTC | 5 | 16·214 | GAG | New published marker |
| Mest 88 | GCCTGTTTGCTTTCTCTTTCTC | ATGAGAGCCAAGAGCACGAT | 5 | 57·056 | TC | New published marker |
| mCrCIR07E12 | TGTAGTCAAAAGCATCAC | TCTATGATTCCTGACTTTA | 5 | 95·430 | GAAAGA | Froelicher et al. (2008) |
| Mest 56 | AGTCCGCCTTTGCTTTTTCT | GGTGCAAAAGAGAGCGAGAG | 5 | 104·503 | CA | New published marker |
| Mest 123 | GGGATGGACTCCCAGTGTTA | AAGAAAGATTTGCTGGCAGAG | 6 | 26·839 | TC | New published marker |
| mCrCIR02F12 | GGCCATTTCTCTGATG | TAACTGAGGGATTGGTTT | 6 | 60·841 | GA | Cuenca et al. (2011) |
| mCrCIR01C06 | GGACCACAACAAAGACAG | TGGAGACACAAAGAAGAA | 6 | 88·819 | GA | Cuenca et al. (2011) |
| Mest 132 | TTATTTCCTTTGACGGTGGG | TTCTTTGGAGCCGAACAACT | 6 | 91·877 | GA | New published marker |
| TAA 1 | GACAACATCAACAACAGCAAGAGC | AAGAAGAAGAGCCCCCATTAGC | 6 | 93·386 | TAA | Kijas et al. (1997) |
| Ci07C07 | TATCCAGTTTGTAAATGAG | TGATATTTGATTAGTTTGG | 7 | 98·019 | TG | Froelicher et al. (2008) |
| Ci02B07 | CAGCTCAACATGAAAGG | TTGGAGAACAGGATGG | 9 | 0 | GA | Froelicher et al. (2008) |
| mCrCIR02G02 | CAATAAGAAAACGCAGG | TGGTAGAGAAACAGAGGTG | No map | – | GA | Cuenca et al. (2011) |
| mCrCIR02G12 | AAACCGAAATACAAGAGTG | TCCACAAACAATACAACG | No map | – | GA | New published marker |
| Ci07C09 | GACCCTGCCTCCAAAGTATC | GTGGCTGTTGAGGGGTTG | No map | – | GA | Froelicher et al. (2008) |
| Mest 192 | CGCGGATCATCTAGCATACA | CTTGGCACCATCAACACATC | No map | – | AT | New published marker |
Extraction of genomic DNA was performed according to Dellaporta and Hicks (1983). PCR products were separated by vertical denaturalized electrophoresis (acrylamide, 6 % bis-acrylamide, 7 m urea) 0·5× TBE buffer (Tris, boric acid and 0·5 m EDTA, pH 8) in DCodeTM Biorad® buckets according to the methodology described by Froelicher et al. (2008). Silver staining was performed according to Benbouzas et al. (2006). The number of heterozygous loci analysed (n) was recorded for each genotype. The recombination rate (r) for each pair of adjacent SSR markers in a single linkage group was evaluated from their genetic distance (Kosambi map function) in the clementine genetic map (Ollitrault et al., 2011). The probability that a zygotic plant (arising from selfing) would not be identified by the analysed marker was estimated per linkage group based on P n = P n_−1(1 – 2_r n (1–r n)), where _P_1 = 0·5 and r n is the recombination rate between the _n_–1 and n heterozygous loci in the ordered linkage group. The global probability was calculated by multiplying the probability per each linkage group. As four of the markers used are not mapped and not taken into account, this probability is an overestimation of the risk to consider a zygotic plant as apomictic.
To synthesize the genotype data, a cluster analysis was done with the Darwin v.5·0·155 software (Perrier et al., 2003; Perrier and Jacquemoud-Collet, 2006) according to the weighted neighbour-joining method, using the simple-matching dissimilarity index, where d ij is the dissimilarity between units i and j, L is the number of loci, π is the level of ploidy and m l is the number of matching alleles for locus l:
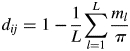
Statistical analysis
Chi square tests of homogeneity were used to evaluate the significance of annual and genotypic factors in the genotypic and inter-annual variation of the frequency of tetraploid seedlings.
Analysis of variance was used to study the impact of geographical origin of ‘Carrizo’ citrange on tetraploidy rates. The Duncan test was used for comparisons between means.
RESULTS
Identification of tetraploid plants from seedling populations of several apomictic citrus genotypes and analysis of their genetic origin
Seedlings of 30 apomictic diploid genotypes of the Citrus genus were analysed to investigate the genetic origin of tetraploid seedlings. We found 80 tetraploid plants (2_n_ = 4_x_ = 36) arising from 76 seeds from the 4442 seedlings analysed (Table 1). Tetraploid plants were identified for all genotypes studied, except for ‘Beauty’, ‘Fremont’, ‘Salteñita’ and ‘Simeto’ mandarins. Overall, the frequency of tetraploid plants was 1·80 %. The lowest frequency was found for sweet oranges (0·80 %) and tangelos (1·04 %) and the highest for tangors (3·52 %); rates between 2·5 and 3 % were observed for the other species. Intraspecific variability has been observed, with rates varying from 0 to 9 % between the aforementioned mandarins and ‘Tardivo di Ciaculli’ mandarin, for example. Tetraploid seedlings were obtained from fruits producing a majority of diploid seedlings. For 15 genotypes, a more detailed analysis of the ploidy level of plantlets arising from seeds producing at least one tetraploid seedling was performed. Twenty-eight seeds produced only one tetraploid plant. When more than one plant per seed was obtained, in most cases (25/28) diploid plantlets were found associated with the tetraploid ones (Table 5). In total, only three seeds among 4442 produced two tetraploid plants (Table 1).
Table 5.
Number of seeds that germinated producing different combinations of tetraploid and diploid plantlets for 15 accessions of Citrus
| Variety | Accession | 1 × 4_x_ + 0 × 2_x_ | 1 × 4_x_ + 1 × 2_x_ | 1 × 4_x_ + 2 × 2_x_ | 1 × 4_x_ + 3 × 2_x_ | 1 × 4_x_ + 4 × 2_x_ | 2 × 4_x_ |
|---|---|---|---|---|---|---|---|
| ‘Anana’ | IVIA-390 | 1 | 3 | 0 | 0 | 0 | 0 |
| ‘Duncan’ | IVIA-274 | 1 | 4 | 0 | 0 | 0 | 0 |
| ‘Fairchild’ | IVIA-83 | 1 | 0 | 0 | 0 | 0 | 0 |
| ‘Kara’ | IVIA-218 | 2 | 0 | 0 | 0 | 0 | 0 |
| ‘Kinnow’ | IVIA-33 | 7 | 5 | 1 | 0 | 0 | 0 |
| ‘Mapo’ | IVIA-190 | 2 | 0 | 0 | 0 | 0 | 0 |
| ‘Minneola’ | IVIA-84 | 1 | 2 | 0 | 0 | 0 | 0 |
| ‘Murcott’ | IVIA-196 | 2 | 2 | 0 | 1 | 1 | 2 |
| ‘Nadorcott’ | IVIA-641 | 1 | 1 | 0 | 0 | 0 | 0 |
| ‘Ortanique’ | IVIA-276 | 2 | 1 | 1 | 0 | 0 | 0 |
| ‘Page’ | IVIA-79 | 1 | 0 | 0 | 0 | 0 | 0 |
| ‘Sanguinelli’ | IVIA-34 | 2 | 0 | 0 | 0 | 0 | 0 |
| ‘Star Ruby’ | IVIA-197 | 1 | 0 | 0 | 0 | 0 | 0 |
| ‘Sunburst’ | IVIA-200 | 1 | 1 | 0 | 0 | 0 | 0 |
| ‘T. Ciaculli’ | IVIA-186 | 3 | 2 | 0 | 0 | 0 | 1 |
| Total | 28 | 21 | 2 | 1 | 1 | 3 |
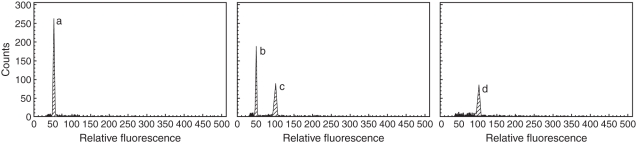
One tetraploid plant of each cultivar was thoroughly analysed using ten samples of four mixed leaves coming from different branches covering the entire canopy. All the samples analysed displayed a single peak corresponding to the tetraploid level. This result demonstrates that these plants were not chimeric for ploidy level (Fig. 1). The same plants were analysed for chromosome counts and all of them displayed 36 chromosomes (2_n_ = 4_x_ = 36).
Fig. 1.
Histograms of the control diploid plant of ‘Murcott’ tangor (a), the control diploid plant and tetraploid plant of ‘Murcott’ tangor (b,c) and tetraploid ‘Murcott’ tangor plant without diploid control plant (d).
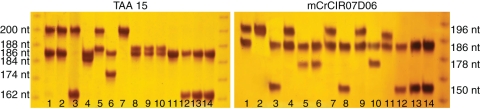
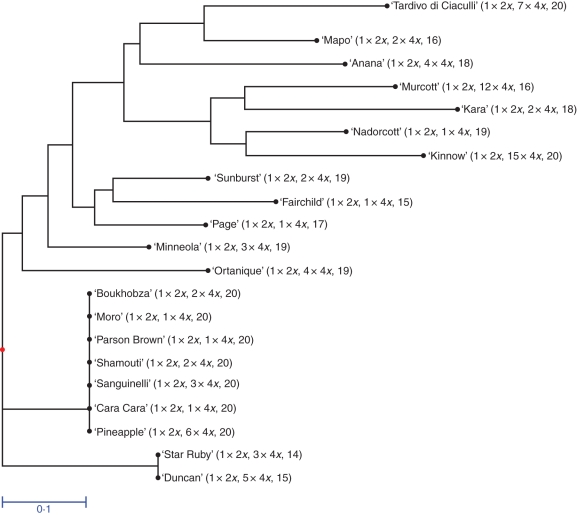
All the tetraploid plants and their parental diploid lines were characteriZed by using 24 SSR markers. These SSR markers showed considerable polymorphism among the parental lines and the observed segregation confirmed the monolocus status of the markers and thus the heterozygosity of the cultivars displaying two allelic bands. For example, all the cultivars analysed in Fig. 2 display heterozygotic profiles for the TAA15 and mCrCIR07D06 SSR markers. According to positions of the heterozygous markers for each parental genotype, we have estimated the probability of a diploid zygotic seedling from self-fertilization being identical to the mother plant. This is also the rate for a potential doubled diploid zygotic plant. Regardless, the probability of a zygotic plant originating from outcrossing being identical to the mother plant would be lower than it would from selfing. According to the diploid parental genotypes, between 14 and 20 of the analysed markers were heterozygous, corresponding to a probability of having zygotic seedlings identical to nucellar ones below 1·07 × 10−3 (see Supplementary Data Table S1, available online). All the tetraploid plants were found to be identical to their diploid parent for all the markers analysed, as shown in the neighbour-joining tree (Fig. 3 and Supplementary Data Table S1).
Fig. 2.
Allelic variability of TAA15 and mCrCIR07D06 SSR markers for some of the diploid parental lines. 1, ‘Nadorcott’; 2, ‘Murcott’; 3, ‘Ortanique’; 4, ‘Kara’; 5, ‘T. Ciaculli’; 6, ‘Anana’; 7, ‘Kinnow’; 8, ‘Fairchild’; 9,‘Sunburst’; 10, ‘Mapo’; 11, ‘Minneola’; 12, ‘Sanguinelli’; 13, ‘Duncan’; 14, ‘Star Ruby’.
Fig. 3.
Cluster analysis of tetraploid plants and their diploid parental lines based on 24 SSR markers: neighbour-joining analysis using simple-matching dissimilarity index. The numbers of diploid and tetraploid plants analysed are indicated, and the last number corresponds to the number of heterozygotic SSR markers used for genetic analysis.
Frequency of tetraploidization events and potential tissular origin in ‘Tardivo di Ciaculli’ mandarin
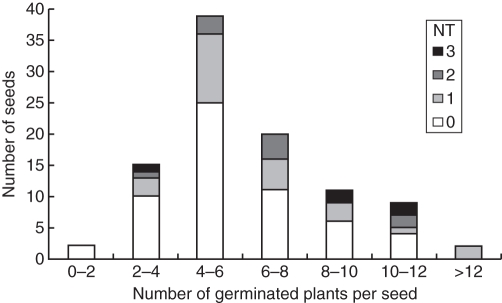
Under greenhouse conditions the rate of germination of apomictic embryos is low because many embryos are very small (Fig. 4). To estimate the relative number of diploid and tetraploid embryos in the same seed better, we conducted a systematic in vitro rescue of all the embryos contained in the apomictic seeds of ‘Tardivo di Ciaculli’ mandarin. This is a highly polyembryonic cultivar producing a high rate of tetraploid seedlings (Table 1). Among the 96 seeds studied, we found between two and 29 embryos per seed (mean 8·1). Among 777 cultivated embryos, 634 successfully germinated in vitro (6·6 plantlets per seed), of which 573 were diploids and 61 tetraploids (9·6 % tetraploids). For greenhouse seedlings of ‘Tardivo di Ciaculli’ mandarin, among 73 germinated seeds only 110 plantlets were recovered (1·5 plantlets per seed), of which 103 were diploids and seven were tetraploids (6·3 % tetraploids). These data indicate that the germination rate of both diploid and tetraploid embryos is much higher in vitro than in vivo, as expected. However, in vitro we found that 40 of 96 seeds contained at least one tetraploid embryo while only six of 73 seeds produced a tetraploid plant in the greenhouse. The number of successful tetraploidization events (producing tetraploid embryos) per seed is therefore at least more than four times higher than that estimated by analysing seedlings produced under greenhouse conditions. All seed containing tetraploid embryos also contained diploid embryos. Among the seeds containing tetraploid embryos, most contained a single tetraploid (25), while ten, four and one seed contained two, three and four tetraploid embryos, respectively (Fig. 5).
Fig. 4.
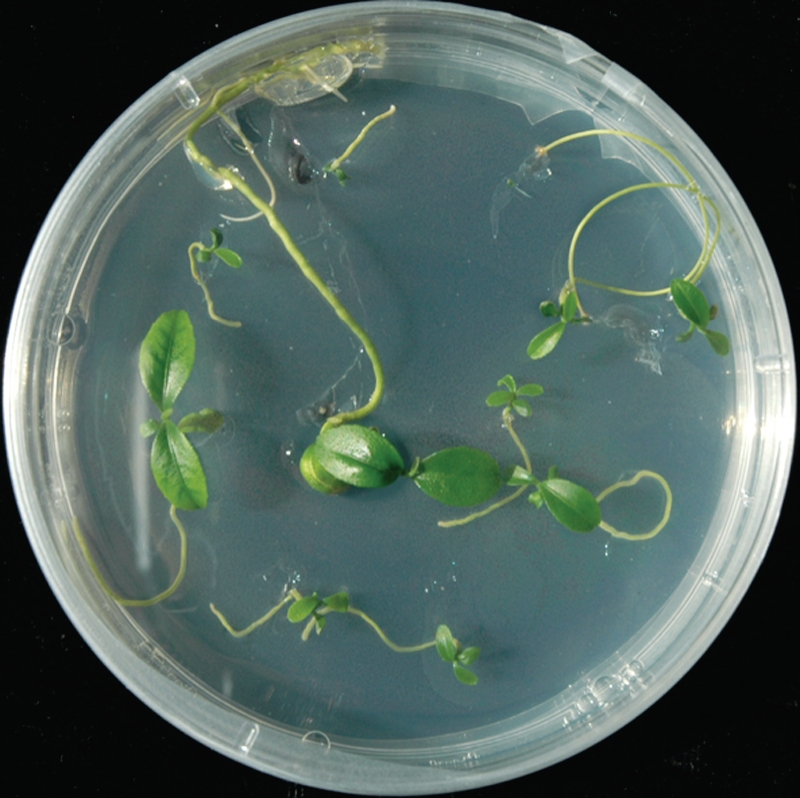
Plantlets obtained from embryos contained in one apomictic seed of ‘Tardivo di Ciaculli’ mandarin.
Fig. 5.
Distribution of the number of seeds according to the total number of recovered plants per seed after in vitro embryo rescue and the number of tetraploid plants (NT: 0, 1, 2 or 3) per seed in ‘Tardivo di Ciaculli’ mandarin.
Thirty-six diploid plants and ten tetraploid plants arising from eight seeds with different 2_x_/4_x_ ratio (4 : 1, 4 : 1, 4 : 1, 4 : 1, 4 : 1, 5 : 1, 3 : 2, 8 : 2) were analysed with seven SSR markers heterozygous for ‘Tardivo di Ciaculli’ mandarin (Ci02B07, mCrCIR03D12a, mCrCIR05A05, mest 15, mest 56, mest 123, mest 132). All the plants were identical to the parental diploid line, demonstrating they were all of nucellar origin. It is also clear that tetraploid plants were obtained from nucelli that predominantly comprised diploid cells.
Genotypic and inter-annual variation of the frequency of tetraploid seedlings
To analyse the importance of genetic and environmental factors, tetraploid seedling rates were evaluated with seeds harvested from the same trees of seven genotypes of the INRA/CIRAD Germplasm Bank over a period of three years (2002, 2003 and 2004). A total of 12 749 seedlings were analysed by flow cytometry (Table 2) and 580 tetraploid plantlets were identified. Systematic analysis of all tetraploid seedlings with five SSR markers (selected to be heterozygotic for each diploid parental genotype) confirmed their identity with their diploid parents (data not shown). Variability between genotypes for tetraploid production during the same year was highly significant. ‘Troyer’ and ‘Carrizo’ citranges produced the highest tetraploid rates, reaching 21 % for ‘Troyer’ in 2002. Significant variations were also observed between years for a single genotype, and consistent results were observed for all genotypes. Tetraploid rates were highest in 2002 and were lowest for all genotypes in 2004. Considering only the three varieties studied over the three years, average tetraploid rates were respectively 8·4, 3·9 and 1·4 % for 2002, 2003 and 2004. The high correlation coefficient (_R_2) between rates of tetraploid plant production by the different genotypes in successive years (0·893 and 0·871 for 2002–2003 and 2003–2004, respectively) suggests that environmental factors affect tetraploid plant production.
Variation in tetraploid frequencies in ‘Carrizo’ citrange seedlings from different geographical origins
Given the inter-annual variation of tetraploid rates and thus the impact of environmental conditions on tetraploidization events, we compared tetraploid rates between seed samples of a single genotype, harvested in different countries. ‘Carrizo’ citrange was chosen due to its high rate of tetraploid seedling production and the important variation observed between seed samples harvested in Corsica during three different years.
Significant differences were found for tetraploid production between the different geographical origins (Table 6). Highest rates of seeds producing tetraploid plants (19·9, 19·7 and 18·7 %) and tetraploid plants among all seedlings (19·2, 12·1 and 13·3 %) were found for those of Valencia, Corsica and Uruguay origins, respectively. Lower rates were found for Florida, Bahia and San Paulo, 4·2, 1·5 and 3·3 %, respectively, for seeds that produced 4_x_ plants and 2·5, 0·9 and 1·9 % for 4_x_ plants. Values for South Africa were intermediate, with 8·1 and 6·2 %, respectively, for seeds that produced 4_x_ seeds and 4_x_ plants. Rates for California were also intermediate but did not differ significantly from regions producing the highest rates (Valencia, Corsica and Uruguay) or from South Africa. Overall, it appears that tropical regions produced fewer tetraploid seedlings than the Mediterranean and sub-tropical regions. Moreover, tropical samples produced no more than one tetraploid plant per seed.
Table 6.
Variation in tetraploid plant rates in seedlings of ‘Carrizo’ citrange from different geographical origin
| Location | No. of germinated seeds | No. of plants per seed | No. of plants analysed | No. of seeds producing 4_x_ plants | No. of 4_x_ plants | Percentage seeds producing 4_x_ plants | Percentage 4_x_ plants |
|---|---|---|---|---|---|---|---|
| Corsica | 234 | 1·80c | 421 | 46 | 51 | 19·7d | 12·1cd |
| California | 222 | 1·53b | 340 | 27 | 30 | 12·2cd | 8·8bc |
| Valencia | 191 | 1·31a | 229 | 38 | 44 | 19·9d | 19·2d |
| Uruguay | 219 | 1·77c | 383 | 41 | 51 | 18·7d | 13·3cd |
| South Africa | 295 | 1·69bc | 498 | 24 | 31 | 8·1bc | 6·2b |
| Florida | 236 | 1·73c | 408 | 10 | 10 | 4·2ab | 2·5a |
| San Paulo | 210 | 1·75c | 364 | 7 | 7 | 3·3a | 1·9a |
| Bahia | 203 | 1·58b | 321 | 3 | 3 | 1·5a | 0·9a |
Significant negative correlations (Pearson coefficient) were observed between the rates of seeds producing tetraploid plants and mean temperatures (R = –0·91), mean maximal temperatures (R = –0·94) and mean minimum temperatures (R = –0·84) of the site where the seed samples were harvested (the mean temperature covered the period of floral induction and blooming; see Materials and methods). Likewise, a significant negative correlation was found between the rate of tetraploid plants among all seedlings and the same temperature parameters (R = –0·86, –0·86 and –0·81, respectively).
DISCUSSION
Genetic structure and origin of citrus tetraploid seedlings
Tetraploid seedlings may result from chromosome doubling of somatic or zygotic tissue or from the fertilization of non-reduced ovules by diploid pollen. Seeds from open pollination were harvested in the Germplasm Banks of IVIA (Spain) and INRA/CIRAD (Corsica), where no tetraploid adult parents were present at this time. Non-reduced pollen seems to be extremely rare in citrus (Esen and Soost, 1977; Luro et al., 2004) so the probability of obtaining tetraploid zygotic embryos from two diploid parents can be considered as quasi null and has never been described in citrus. Therefore, the potential origin of the tetraploids obtained could be: (1) nucellar seedlings from fruits of tetraploid branches of the diploid mother tree produced by chromosome doubling in somatic tissues of the diploid parental tree, (2) nucellar seedlings from doubled primordium cells of nucellar tissue, (3) chromosome doubling during the nucellar embryo development, and (4) chromosome doubling of the zygotic embryo at early or later stages of development.
Flow cytometry analysis of numerous leaves and roots of tetraploid seedlings clearly demonstrated that these plants were not chimeric for ploidy. Thus the tetraploid plants did not originate by chromosome doubling at a later stage of embryo development.
Seventy-eight tetraploid plants arising from 22 diploid genotypes have been genotyped with 24 SSR markers and compared with the parental diploid genotype. Note that all sweet orange cultivars displayed identical profiles for all analysed markers, as well as the two grapefruits. These results are in agreement with previous molecular studies (Barkley et al., 2006; Luro et al., 2008), which have clearly shown that each of these apomictic taxa originated from a single hybrid and that within-taxa diversification was due to budsport mutation events without further sexual recombination. All the tetraploid plants displayed the same SSR profile as their diploid parents for all the heterozygous markers analysed. Such identity between tetraploid seedlings and diploid parents was also observed for the 580 tetraploid plants identified in the seedlings of seven citrus genotypes (including Poncirus and intergeneric hybrids) over three years. These results clearly demonstrate that potential origin by early endomitosis of zygotic embryos can be discarded. This identity of tetraploid seedlings with parental diploid genotypes is also consistent with the hypothesis that they do not arise from fertilization of a 2_n_ ovule by diploid pollen. From these results we conclude that tetraploid seedlings in apomictic genotypes arise from somatic chromosome doubling of maternal cells.
The fact that the tetraploid seedlings were obtained from seeds producing mostly diploid embryos is consistent with previous results (Cameron and Frost, 1968). It indicates that polyploid seeds do not arise from tetraploid sectors of the trees, or from fruits resulting from vegetative or floral bud solid tetraploidization. Indeed, in this case all the seeds from a single fruit would have the same ploidy level. Kobayashi et al. (1981) demonstrated that, in apomictic citrus genotypes, adventitious embryos originated from a single cell of the nucellus which they termed primordium cells. As observed by Hutchison and Barrett (1981), we have shown that diploid and tetraploid seedlings can arise from the same apomictic seed and, moreover, we have demonstrated by SSR analysis that the associated diploid seedlings were also of nucellar origin in ‘Tardivo di Ciaculli’ mandarin seedlings. Consequently, tetraploidization events occur in nucellar tissues that are mostly diploid. Our results demonstrate that the formation of tetraploid embryos is the consequence of chromosome doubling of individual nucellar primordium cells of the nucellus, each subsequently producing one solid tetraploid embryo.
Furthermore, in other experiments performed by our group, seeds of different tetraploid apomictic citrus genotypes always produced tetraploid seedlings, associated with octoploid plantlets in some cases (Schwarz, 2001). This is in agreement with the results obtained here.
Frequency of tetraploid plants in seedlings of diploid apomictic citrus is dependent on genotype and environment
The global frequency of tetraploid seedlings among 30 analysed genotypes of Citrus cultivated in Valencia and Corsica was 1·8 % (varying from 0 to 9·7 % with cultivar and environmental conditions). This value is higher than that recorded by Barrett and Hutchison (1978) from seeds harvested in Florida (0·75 %) and California (0·9 %). In our analysis of 30 citrus genotypes we observed some variation between species as well as within species. However, the relatively low number of plants analysed for some genotypes, and the different geographical and annual origin of seeds does not allow us to draw any general conclusions on genotypic effect. This aspect was analysed further by study of tetraploid rates of seven genotypes from Corsica over three years. We found very high rates of tetraploid seedlings for ‘Troyer’ and ‘Carrizo’ citranges, reaching 20 % for some seed samples. As for Citrus species, these values are higher than those observed by Hutchison and Barrett (1981) in Florida (between 1 and 3 %). Such a discrepancy may be due to differences among the genotypes analysed and the geographical origin of the seeds harvested; however, they could also be due to differences in the tetraploid identification method, which in previous studies was based on visual observation of morphological traits. From a systematic search of autotetraploids in a wide range of apomictic genotypes, Barrett and Hutchison (1978) postulated that the ability to produce such tetraploid seedlings is a variable genetic trait present in apomictic citrus and related species. Moreover, Hutchison and Barrett (1981) concluded from a 3-year analysis of ‘Troyer’ and ‘Carrizo’ citranges from Florida that tetraploid seedling frequency is affected by environmental conditions. Analysis of seedlings of seven genotypes of Citrus, Poncirus and intergeneric hybrids harvested during three consecutive years in Corsica confirmed these assumptions. Indeed, both genotypic and inter-annual effects were significant. A higher incidence of tetraploid plants was found for two citranges, with rates varying between 3·5 and 21 % depending on the year, while the lowest rates were found for ‘Cleopatra’ mandarin, varying from 0·2 to 4 %. We found that the inter-annual effect influenced the different genotypes in the same way, as reported by Hutchison and Barrett (1981) but with a stronger impact. This could be related to the greater inter-annual climatic variability in the Mediterranean area than in Florida during floral induction and the blooming period.
We confirmed the importance of environmental conditions on tetraploidy rates by studying seeds of the same genotype (‘Carrizo’ citrange) harvested in different countries. Higher rates were observed for the Mediterranean than for tropical areas, and significant negative correlations were observed between tetraploid rates and mean temperatures during floral induction and the blooming period. These results are in agreement with the higher tetraploid rates found in our experiments compared with data for Florida (Barrett and Hutchison, 1978; Hutchison and Barrett, 1981) and they suggest that colder conditions favour tetraploidization events in citrus nucellar cells. In mango (Mangifera indica L.), another apomictic species, the influence of environmental conditions on spontaneous tetraploidy follows the same pattern as in citrus. In a small-scale study in the Canary Islands, spontaneous tetraploid apomictic seedlings were found on the relatively cold island of Tenerife, where temperatures are not suitable for commercial mango production, but they were not found in the same genotypes on the island of La Gomera, which has a tropical climate that supports commercial mango production (Galán-Sauco et al., 2001). Tetraploid seedlings of mango have not been reported from any of the tropical areas where the crop is produced.
The effect of low temperatures on polyploidization events appears to be a general rule, both in plants and in animals (Ramsey and Schemske, 1998; Otto and Whitton, 2000). Moreover, it has long been known that the frequency of polyploid taxa increases with latitude in the Northern Hemisphere. The possible reasons for this trend have been debated for many decades. Hagerup (1931) suggested that polyploids are better adapted than diploids to extreme climates. Manton (1950), Löve and Löve (1957), and Johnson et al. (1965) considered that polyploids have selective advantages where climatic fluctuations have been frequent and catastrophic, because they have great genetic variability resulting from recent hybridization. Stebbins (1950) argued that polyploids seem better suited than diploids to colonize newly deglaciated areas because of greater ecological adaptability. Recent studies have shown that important modifications in gene expression occur immediately after allopolyploidization events (Comai et al., 2000; Adams et al., 2004; Wang et al., 2004, 2006; Flagel and Wendel, 2010). Neoregulation of parental genome expression in allopolyploid plants would partially explain why they often give rise to new phenotypes, exceeding the variability range of the diploid gene pool and their higher adaptability (Osborn et al., 2003). Besides the adaptive advantage of polyploids, the higher rate of polyploidization events in a colder environment could be one of the factors underlying the increased frequency of polyploid species with latitude in the Northern Hemisphere. It could contribute to the overall increase in gametophytic apomictic taxa in northern regions (Asker and Jerling, 1992). Gametophytic apomixis is intimately related to polyploidy and it was proposed that apomixis was due to epigenetic variation associated with polyploidy or interspecificity (Koltunow and Grossniklaus, 2003). In angiosperms, apomixis is most likely to arise in areas and time periods influenced by glaciations of the Pleistocene, where range fluctuations of plants because of climatic oscillations offered the most frequent opportunities for interspecific hybridization and polyploidization events (Hörandl, 2006). In these areas with potentially higher rates of tetraploidization events, the establishment and persistence of these tetraploid cytotype was favoured by apomixis. Indeed, taxa with gametophytic apomixis have advantages in higher latitudes/altitudes over their sexual relatives due to (1) a shortening of the reproductive pathway and faster seed development in shorter vegetation periods, (2) reproductive assurance if pollinators and mating partners are rare and (3) potentially less pressure from predators because of reduced biotic interactions from colder climates (Hörandl, 2006).
In these areas with potentially higher rates of tetraploidization events and the insignificant role of polyploidy in citrus evolution
It should be noted that the rate of tetraploid plants found among greenhouse seedlings is an underestimation of the real rate of tetraploidization events per seed. Indeed, by in vitro rescue of all the embryos contained in seeds of ‘Tardivo de Ciaculli’ mandarin, we found that 42 % of the seeds contained at least one tetraploid embryo, while in the greenhouse seedlings, only 8 % of seeds of the same sample produced tetraploid plantlets. This is a consequence of the small size of many embryos that do not germinate under greenhouse conditions. Overall, even considering only tetraploid seedling rates in the greenhouse, successful polyploidization frequencies observed for citrus are very high compared with the mean value of 10−5 tetraploids per individual and generation estimated in plants by Ramsey and Schemske (1998). It is highly probable that adventitious embryony is favourable to polyploidization. Indeed, the same phenomenon has been observed in apomictic mango (Galán-Sauco et al., 2001). It is interesting to note that species having a sporophytic form of apomixis (adventitious embryony), such as citrus or mango, do not present the general link found between ploidy and apomixis (Asker and Jerling, 1992; Richards, 1996; Roche et al., 2001), but display repetitive tetraploidization events in primordium embryogenic cells, producing relatively high levels of tetraploid seedlings.
As with most angiosperms it is very probable that the citrus genome has undergone one or more very ancient whole genome duplication events (palaeopolyploidy; Adams and Wendel, 2005; Cui et al., 2006). However, despite frequent natural polyploidization events and the apomictic nature of most of the nucellar doubled-diploids, it appears that polyploids have played a minor role in more recent diversification of the Aurantioideae subfamily and evolution of the Citrus genus, as demonstrated by their scarcity in natural germplasm. One possible explanation could be that citrus originated and were domesticated in tropical areas of South-East Asia, and our study shows that the frequency of tetraploidization events is much lower in tropical regions than in cooler areas, such as the Mediterranean Basin. From an evolutionary perspective, the occurrence of tetraploidization events in the tropical areas where citrus originated was rarer than the rate we estimated from the germplasm banks in Spain and Corsica. Even the data from Florida germplasm genotypes growing in a much warmer environment (Barrett and Hutchison, 1978) give tetraploid seedling rates of between 0·2 and 3·3 %, with a 0·75 % average from the analysis of 32 citrus genotypes. These values are high compared with the tetraploidization rates of other plant species (Ramsey and Schemske, 1998).
Polyploidy has played an important role in the creation of plant diversity (Stebbins, 1971; Masterson, 1994) but several factors influence the successful establishment of a tetraploid compartment from rare tetraploid plants. For species with sexual mating, self-pollination and all modifications that favour the transfer of tetraploid plant pollen to other tetraploid plants (such as a change in the flowering time, in pollinator preferences, or limited pollen and seed dispersion) are favourable for tetraploid compartment establishment (Baack, 2005). Pollen competition in favour of 2_x_ pollen compared with 1_x_ pollen is also a major factor allowing minority tetraploid cytotypes to establish a tetraploid compartment in sympatric situations, as demonstrated in Chamerion angustifolium (Husband et al., 2002). Apomixis of the tetraploid cytotypes is generally considered as a favourable element for successful establishment of tetraploids (Schranz et al., 2005; Hufft-Kao, 2008). Consendai and Hörandl (2010) demonstrated that tetraploid apomicts colonize previously devastated and also distant areas via long-distance dispersal and they have the advantage of uniparental reproduction. For the diploid sexual cytotype, self-sterility and pollinator dependence may strongly limit range expansions.
As already mentioned, the better adaptability of allopolyploids could result from new genome regulation. Genome neo-regulation has been shown to be much more closely related to interspecificity than polyploidization itself. Indeed, several studies, such as in Spartina (Salmon et al., 2005), Helianthus (Lai et al., 2006) and Arabidopsis (Wang et al., 2006), reveal that neo-regulation of genome expression occurs in interspecific crosses within the same ploidy level, while chromosome doubling or autopolyploidization affects the genome (Wang et al., 2006; Stupar et al., 2007) or proteome (Albertin et al., 2005) expression only slightly. In addition to epigenetic changes induced by a ‘genomic shock’ (McClintock, 1984), an advantage of allopolyploidy was proposed to result from the juxtaposition and interaction of divergent homeologous regulatory networks, leading to new cis- and trans-acting effects on gene expression (Chen, 2007; Landry et al., 2007). In the case of chromosome doubling of fertile intra- or inter-specific diploid genotypes, as occurs for apomictic citrus, the previous arguments for the advantage of tetraploid over diploid parents are not valid. Cameron and Frost (1968) and further studies have shown that most of the doubled diploid citrus lines are less vigorous than their parental diploid lines, probably limiting their competitiveness. In addition, in citrus, polyembryony of most natural doubled-diploids strongly limits the potential for evolution and fitness of the tetraploid gene pool. In seeds of apomictic citrus, competition between the zygotic and nucellar embryos generally results in failed development of the zygotic embryo (Frost and Soost, 1968; Koltunow, 1993), hampering the contribution to the next sexual generation of these genotypes as female parents. Thus, tetraploid plants should mainly contribute to the sexual next generation as male parents, pollinating non-apomictic diploids and generally producing abnormal triploid seeds that are highly unlikely to germinate under natural conditions. Moreover, it has been shown that the degeneration of pollen mother cells is more frequent in doubled-diploid citrus than in their diploid parental genotypes (Frost and Soost, 1968). These authors also observed high variability in chromosome conjugation (quadrivalent, trivalent, bivalent and univalent) during metaphase I. Consequently, most autotetraploids generally produce few pollen grains with a normal chromosome complement (Frost and Soost, 1968) and have lower pollen viability than diploid parental lines. This lower pollen fertility for nucellar tetraploid plants might also have limited their contribution to generation of new sexual polyploids. Furthermore, in the case of successful triploid offspring production, the triploid hybrids have very low fertility and are virtually seedless, which precludes their perpetuation by natural means. During the domestication process, tetraploids were not selected by farmers owing to their thick peel and rough pulp (Cameron and Frost, 1968).
Tetraploids for citrus breeding
Scion breeding
We have discussed here the selection of tetraploid plants of sweet oranges (‘Sanguinelli’, ‘Pineapple’, ‘Boukhobza’, ‘Tarocco Rosso’, ‘Cara Cara’, ‘Moro’, ‘Shamouti’ and ‘Parson Brown’), mandarins (‘Anana’, ‘Kinnow’, ‘Kara’, ‘Tardivo di Ciaculli’ and ‘Willow leaf’), grapefruits (‘Duncan’ and ‘Star Ruby’), tangors (‘Murcott’, ‘Nadorcott’ and ‘Ortanique’) and tangelos (‘Mapo’, ‘Minneola’, ‘Page’, ‘Sunburst’ and ‘Faichild’). In addition, tetraploid lines of C. aurantifolia (IVIA-490), ‘Eureka Frost’ lemon (IVIA-495), ‘Orlando’ tangelo (IVIA-492) and ‘Dweet’ tangor (IVIA-496) have been selected by the IVIA (Spain). All these new tetraploid plants have been included in the IVIA and CIRAD/INRA Germplasm Banks for use as male parents in triploid breeding programmes. Several thousand triploid hybrids have already been produced at the IVIA by 2_x_ × 4_x_ crosses with these new tetraploid parents. With the demonstration that tetraploid seedlings from apomictic genotypes are doubled-diploid, between 66 % (under the tetrasomic random chromosome segregation model; Muller, 1914; Marsden et al., 1987) and 56 % (tetrasomic maximum equational segregation model; Mather, 1936) of the diploid parental heterozygosity is transmitted to the triploid progeny. In comparison with triploids obtained from 2_n_ gametes in 2_x_ × 2_x_ crosses and in agreement with the hypothesis that 2_n_ gametes in citrus arise from second division restitution (SDR) (Esen et al., 1979; Luro et al., 2000; Cuenca et al., 2011) it should be assumed that the phenotypic traits of the diploid parent from which the diploid gamete originated will be transmitted to the triploid progenies at higher frequencies in 2_x_ × 4_x_ crosses than in 2_x_ × 2_x_ crosses. Moreover, less polymorphism can be expected in triploid progenies arising from 2_x_ × 4_x_ crosses than from 2_n_ SDR gametes (Ollitrault et al., 2008).
Rootstock breeding
Recent work on citrus tetraploid rootstocks suggests that they could be more tolerant to salt and water stress than their parental diploids (Saleh et al., 2008), probably due to modified abscisic acid constitutive synthesis (Allario et al., 2009). Furthermore, tetraploid rootstocks generally reduce canopy size, which is a desirable trait in modern orchards (Barrett and Hutchison, 1978; Lee, 1988). The selection of doubled-diploid lines from seedlings of traditional rootstocks could be an interesting way to improve tolerance to abiotic stress without modifying allelic constitution, with high probabilities of transmitting traits related to disease resistance. Here we have described the selection of tetraploid lines of sour oranges, ‘Cleopatra’ mandarin, ‘Pomeroy’ Poncirus and citranges (‘Carrizo’, ‘Troyer’ and ‘C.35′). In addition to these genotypes, tetraploid nucellar plants were obtained for C. macrophylla Wester (IVIA-518), C. volkameriana Ten. & Pasq (SRA-729 and IVIA-500), C. limonia Osb. (SRA-777 and IVIA-516), C. jambhiri (IVIA-498), ‘Gou Tou’ and ‘Sevillano’ sour oranges (SRA-506 and IVIA-497), Poncirus (‘Rubidoux’ IVIA-504 and ‘Flying Dragon’ IVIA-499), C. reticulata × P. trifoliata (ICVN0110155) and C. paradisi × P. trifoliata (‘4475’ ICVN0110140 and ‘CP4475’ IVIA-501). All these tetraploid genotypes, with potential interest for use as rootstocks, have been included in the IVIA and INRA/CIRAD Germplasm Banks. Agronomical trials are currently underway to evaluate the agronomic performance of some of these tetraploid rootstocks under abiotic stress conditions.
Conclusions
Tetraploidization is a frequent event in seeds of diploid apomictic Citrus, Poncirus and their intergeneric hybrids. Genotypic and environmental factors affect the rate of tetraploidy. The higher frequencies of tetraploidy were observed for intergeneric hybrids of sweet orange and Poncirus (‘Carrizo’ and ‘Troyer’ citranges) cultivated in the Mediterranean area, where tetraploid seedling frequencies could exceed 20 %. The colder conditions of marginal production areas would seem to favour tetraploidization. Even in tropical areas, the rates of tetraploidy in seedlings of diploid apomictic genotypes are higher than the frequency of polyploidization events generally reported in the literature for other species. Therefore, the factors limiting the evolutionary role of polyploidization in citrus are probably related to the problems of establishment and persistence of new polyploids. Molecular marker analysis definitively demonstrates that the tetraploids found in seedlings of apomictic diploid citrus arise from chromosome doubling of nucellar cells. Given the high frequency of tetraploids in seedlings of apomictic diploid citrus, chromosome doubling cannot be considered a negligible mechanism of polyploidization in plants. However, it is probably less favourable for successful establishment and persistence of a new tetraploid compartment. It would be interesting to investigate its occurrence in other species with a similar reproductive system (apomixis by adventitious nucellar embryony). In the near future, ploidy manipulation associated with new breeding strategies aiming to develop triploid seedless easy-to-peel fruits and tetraploid rootstock should lead to a dramatic change in cultivated citrus, moving from the cultivation of diploid to polyploid plants. The tetraploid genotypes arising from chromosome doubling of nucellar cells of apomictic citrus could play a significant role in the evolution of cultivated forms, as parents in interploid crosses or directly as new rootstocks.
SUPPLEMENTARY DATA
Supplementary Data
ACKNOWLEDGEMENTS
This work was jointly financed by the AGL2008-00596-MCI and Prometeo 2008/121 Generalidad Valenciana projects and supported by the European Commission, under the FP6-2003- INCO-DEV-2 project CIBEWU (no. 015453). We thank J. A. Pina for growing plants in the greenhouse and in the field.
LITERATURE CITED
- Adams KL, Wendel J. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Adams KL, Percifield R, Wendel JF. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–2226. doi: 10.1534/genetics.104.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allario T, Brumos J, Colmenero JM, et al. Autotetraploid Citrus limonia rootstocks are more tolerant to water deficit than parental diploids. International Conference on Polyploidy, Hybridization and Biodiversity. 2009:98. May 17–20, 2009, Saint-Malo, France, program and abstracts. Rennes: Université de Rennes 1. [Google Scholar]
- Albertin W, Brabant P, Catrice O, et al. Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics. 2005;5:2131–2139. doi: 10.1002/pmic.200401092. [DOI] [PubMed] [Google Scholar]
- Aleza P, Juárez J, Ollitrault P, Navarro L. Production of tetraploid plants of non apomictic citrus genotypes. Plant Cell Reports. 2009;28:1837–1846. doi: 10.1007/s00299-009-0783-2. [DOI] [PubMed] [Google Scholar]
- Aleza P, Juárez J, Cuenca J, Ollitrault P, Navarro L. Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2x × 2x sexual hybridisation and its application to extensive breeding programs. Plant Cell Reports. 2010a;29:1023–1034. doi: 10.1007/s00299-010-0888-7. [DOI] [PubMed] [Google Scholar]
- Aleza P, Juárez J, Ollitrault P, Navarro L. Polyembryony in non-apomictic citrus genotypes. Annals of Botany. 2010b;106:533–545. doi: 10.1093/aob/mcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker S, Jerling L. Apomixis in plants. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- Baack EJ. Ecological factors influencing tetraploid establishment in snow buttercups (Ranunculus adoneus, Ranunculaceae): minority cytotype exclusion and barriers to triploid formation. American Journal of Botany. 2005;92:1827–1835. doi: 10.3732/ajb.92.11.1827. [DOI] [PubMed] [Google Scholar]
- Bacchi O. Cytological observations in citrus III. Megaesporogenesis, fertilization and polyembryony. Botanical Gazzete. 1943;105:221–225. [Google Scholar]
- Barkley NA, Roose ML, Krueger RR, Federici CT. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs) Theoretical and Applied Genetics. 2006;112:1519–1531. doi: 10.1007/s00122-006-0255-9. [DOI] [PubMed] [Google Scholar]
- Barrett HC, Hutchison DJ. Spontaneous tetraploidy in apomictic seedlings of Citrus. Economic Botany. 1978;32:27–45. [Google Scholar]
- Benbouzas H, Jacquemin JM, Baudoin JP, Mergeai G. Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSr markers in polyacrylamide gels. Biotechnology, Agronomy, Society and Environment. 2006;10:77–81. [Google Scholar]
- Bretagnolle F, Thompson JD. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytologist. 1995;129:1–22. doi: 10.1111/j.1469-8137.1995.tb03005.x. [DOI] [PubMed] [Google Scholar]
- Cameron JW, Frost HB. Genetic, breeding and nucellar embryony. In: Reuther W, Batchelor LD, Webber HJ, editors. The citrus industry. Vol. 1. Riverside, CA: University of California; 1968. pp. 325–370. [Google Scholar]
- Cameron JW, Soost RK. Characters of new populations of Citrus polyploids, and the relation between tetraploidy in the pollen parent and hybrid tetraploid progeny. 1969:199–205. Chapman HD, ed. Proceedings of the International Citric Symposium, Vol. 1, University of California at Riverside. [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, et al. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–1567. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consendai A, Hörandl E. Cytotype stability, facultative apomixis and geographical parthenogenesis in Ranunculus kuepferi (Ranunculaceae) Annals of Botany. 2010;105:457–470. doi: 10.1093/aob/mcp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca J, Froelicher Y, Aleza P, Juárez J, Navarro L, Ollitrault P. Multilocus half tetrad analysis and centromere mapping in Citrus; evidence of SDR mechanism for 2n megagametophyte production and partial chromosome interference in mandarin cv Fortune. Heredity. 2011 doi: 10.1038/hdy.2011.33. (in press) doi:10.1038/hdy.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta J, Hicks JB. A plant DNA minipreparation: version II. Plant Molecular Biology Reports. 1983;1:19–21. [Google Scholar]
- Esen A, Soost RK. Unexpected triploids in Citrus: their origin, identification and possible use. Journal of Heredity. 1971;62:329–333. [Google Scholar]
- Esen A, Soost RK. Tetraploid progenies from 2x × 4x crosses of Citrus and their origin. Journal of the American Society for Horticultural Science. 1972;97:410–414. [Google Scholar]
- Esen A, Soost RK. Precocious development and germination of spontaneous triploid seeds in Citrus. Journal of Heredity. 1973;64:147–154. [Google Scholar]
- Esen A, Soost RK. Proceedings del Primer Congreso Mundial de Citricultura. Valencia: International Society of Citriculture; 1977. Relation of unexpected polyploids to diploid megagametophytes and embryo: endosperm ploidy ratio in Citrus; pp. 53–63. [Google Scholar]
- Esen A, Soost RK, Geraci G. Genetic evidence for the origin of diploid megagametophytes in Citrus. The Journal of Heredity. 1979;70:5–8. [Google Scholar]
- Flagel LE, Wendel JF. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytologist. 2010;186:184–193. doi: 10.1111/j.1469-8137.2009.03107.x. [DOI] [PubMed] [Google Scholar]
- Froelicher Y, Dambier D, Bassene JB, et al. Characterization of microsatellite markers in mandarin orange (Citrus reticulata Blanco) Molecular Ecology Resources. 2008;8:119–122. doi: 10.1111/j.1471-8286.2007.01893.x. [DOI] [PubMed] [Google Scholar]
- Frost HB. Polyembryony, heterozygosis and chimeras in Citrus. Hilgardia. 1926;1:365–402. [Google Scholar]
- Frost HB, Soost RK. Seed reproduction, development of gametes and embryos. In: Reuther W, Batchelor LD, Webber HB, editors. The citrus industry. Vol. 2. Berkeley: University of California; 1968. pp. 290–324. [Google Scholar]
- Galán-Sauco V, Grajal Martín MJ, Fernández Galván D, Coello Torres A, Juárez J, Navarro L. Occurrence of spontaneous tetraploid nucellar mango plants. HortScience. 2001;36:755–757. [Google Scholar]
- Geraci G, Esen A, Soost RK. Triploid progenies from 2x – 2x crosses of Citrus cultivars. Journal of Heredity. 1975;66:177–178. [Google Scholar]
- Grant V. Plant speciation. New York: Columbia University Press; 1981. [Google Scholar]
- Grosser JW, Ollitrault P, Olivares O. Somatic hybridization in Citrus: an effective tool to facilitate variety improvement. In vitro Cellular and Development Biology – Plant. 2000;36:434–449. [Google Scholar]
- Grosser WJ, Hyum JA, Calovic M, et al. Production of new allotetraploid and autotetraploid Citrus breeding parents: focus on zipperskin mandarins. HortScience. 2010;45:1160–1163. [Google Scholar]
- Hagerup O. Über Polyploidie in Beziehung zu Klima, Ökologie und Phylogenie. Hereditas. 1931;16:19–40. [Google Scholar]
- Harlan JR, De Wet JM. On Ö. Winge and a prayer: the origins of polyploidy. The Botanical Review. 1975;41:361–390. [Google Scholar]
- Hörandl E. The complex causality of geographical parthenogenesis. New Phytologist. 2006;171:525–538. doi: 10.1111/j.1469-8137.2006.01769.x. [DOI] [PubMed] [Google Scholar]
- Hufft-Kao R. Origins and widespread distribution of co-existing polyploids in Arnica cordifolia (Asteraceae) Annals of Botany. 2008;101:145–152. doi: 10.1093/aob/mcm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC, Schemske DW, Goodwillie C, Burton TL. Pollen competition as a unilateral mechanism of reproductive isolation between diploid and tetraploid Chamerion angustifolium. Proceedings of the Royal Society, London. 2002;269:2565–2571. doi: 10.1098/rspb.2002.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison DJ, Barrett HC. Proceedings of 4th International Citrus Congress. Tokyo: International Society of Citriculture; 1981. Tetraploid frequency in nucellar seedlings from single trees of Carrizo and Troyer Citrus hybrids; pp. 27–29. [Google Scholar]
- Iwamasa M, Nito N, Ling JT. Proceedings of 6th International Citrus Congress. Balaban, PA: International Society of Citriculture; 1988. Intra and intergeneric hybridization in the orange subfamily, Auranthioideae; pp. 123–130. [Google Scholar]
- Johnson AW, Packer JG, Reese G. Polyploidy, distribution, and environment. In: Wright HE Jr, Frey DG, editors. The Quaternary of the United States. Princeton, NJ: Princeton University Press; 1965. pp. 497–507. [Google Scholar]
- Kamiri M, Stift M, Srairi I, et al. Evidence for non-disomic inheritance in a Citrus interspecific tetraploid somatic between C. reticulata and C. lemon hybrid using SSR markers and cytogenetic analysis. Plant Cell Reports. 2011 doi: 10.1007/s00299-011-1050-x. doi:10.1007/s00299-011-1050-x. [DOI] [PubMed] [Google Scholar]
- Kijas JMH, Thomas MR, Fowler JCS, Roose ML. Integration of trinucleotide microsatellites into a linkage map of Citrus. Theoretical and Applied Genetics. 1997;94:701–706. [Google Scholar]
- Kobayashi S, Ieda I, Nakatani M. Proceedings of 4th International Citrus Congress. 1981. Role of the primordium cell in nucellar embryogenesis in citrus. Tokyo: International Society of Citriculture; pp. 44–48. [Google Scholar]
- Koltunow AM. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell. 1993;5:1425–1437. doi: 10.1105/tpc.5.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U. Apomixis, a developmental perspective. Annual Review of Plant Biology. 2003;54:547–574. doi: 10.1146/annurev.arplant.54.110901.160842. [DOI] [PubMed] [Google Scholar]
- Krug CA. Chromosome numbers in the subfamily Aurantioideae with special reference to the genus Citrus. Botanical Gazette. 1943;48:602–611. [Google Scholar]
- Lai Z, Gross BL, Zou Y, Andrews J, Rieseberg LH. Microarray analysis reveals differential gene expression in hybrid sunflower species. Molecular Ecology. 2006;15:1213–1227. doi: 10.1111/j.1365-294X.2006.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Hartl DL, Ranz JM. Genome clashes in hybrids: insights from gene expression. Heredity. 2007;99:483–493. doi: 10.1038/sj.hdy.6801045. [DOI] [PubMed] [Google Scholar]
- Lapin WK. Investigations on polyploidy in Citrus. USSR all-Union Science Research Institute Humid Subtropics Works. 1937;1:1–68. [Google Scholar]
- Lee LS. Citrus polyploidy. Origins and potential for cultivar improvement. Australian Journal of Agricultural Research. 1988;39:735–747. [Google Scholar]
- Longley AE. Polycary, polyspory and polyploidy in Citrus and Citrus relatives. Journal of the Washington Academy of Science. 1925;15:347–351. [Google Scholar]
- Löve Á, Löve D. Arctic polyploidy. Proceedings of the Genetics Society of Canada. 1957;2:23–27. [Google Scholar]
- Luro F, Maddy F, Ollitrault P, Rist D. Identification of 2n gamete parental origin and mode of nuclear restitution of spontaneous triploid Citrus hybrids. Proceedings of 9th International Citrus Congress. 2000:168–169. December 3–7, Orlando, Florida: International Society of Citriculture. [Google Scholar]
- Luro F, Maddy F, Jacquemond C, et al. Identification and evaluation of diplogyny in clementine (Citrus clementina) for use in breeding. In: XI Eucarpia Symposium on Fruit Breeding and Genetics. Acta Horticulturae. 2004;663:841–847. [Google Scholar]
- Luro F, Costantino G, Terol JF, et al. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics. 2008;9:287. doi: 10.1186/1471-2164-9-287. doi:10.1186/1471-2164-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton I. Problems of cytology and evolution in the Pteridophyta. Cambridge: Cambridge University Press; 1950. [Google Scholar]
- Marsden JE, Schwager SJ, May B. Single-locus inheritance in the tetraploid treefrog Hyla versicolor with an analysis of expected progeny ratios in tetraploid organisms. Genetics. 1987;116:299–311. doi: 10.1093/genetics/116.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- Mather K. Segregation and linkage in autotetraploids. Journal of Genetics. 1936;32:287–314. [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Muller HJ. A new mode of segregation in Gregory's tetraploid primulas. American Naturalist. 1914;48:508–512. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–479. [Google Scholar]
- Ollitrault P, Dambier D, Luro F, Froelicher Y. Ploidy manipulation for breeding seedless triploid citrus. Plant Breeding Reviews. 2008;20:323–354. [Google Scholar]
- Ollitrault P, Terol J, Chen C, et al. A Reference linkage map of C. clementina based on SNPs, SSRs and indels. Plant & Animal Genomes XIX Conference. 2011 January 15–19, 2011, San Diego, California. http://www.intl-pag.org/19/abstracts/P05h\_PAGXIX\_477.html. [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, et al. Understanding mechanisms of novel gene expression in polyploids. Trends in Genetics. 2003;19:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP. DARwin software. 2006 Available at http://darwin.cirad.fr/darwin . [Google Scholar]
- Perrier X, Flori A, Bonnot F. Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC, editors. Genetic diversity of cultivated tropical plants. Montpellier: Enfield, Science Publishers; 2003. pp. 43–76. [Google Scholar]
- Raghuvanshi SS. Cytologenetical studies in genus Citrus IV. Evolution in genus Citrus. Cytologia. 1962;27:172–188. [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plant. Annual Review of Ecology and Systematics. 2002;33:589–639. [Google Scholar]
- Richards AJ. Why is gametophytic apomixis almost restricted to polyploids? The gametophyte- expressed lethal model. Apomixis Newsletter. 1996;9:1–3. [Google Scholar]
- Roche D, Hanna WW, Ozias-Akins P. Is supernumerary chromatin involved in gametophytic apomixis of polyploid plants? Sexual Plant Reproduction. 2001;13:343–349. [Google Scholar]
- Saleh B, Allario T, Dambier D, Ollitrault P, Morillon R. Tetraploid citrus rootstocks are more tolerant to salt stress than diploid. Comptes rendus biolgies. 2008;331:703–710. doi: 10.1016/j.crvi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Salmon A, Ainouche ML, Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Molecular Ecology. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Dobes C, Koch MA, Mitchell-Olds TM. Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae) American Journal of Botany. 2005;92:1797–1810. doi: 10.3732/ajb.92.11.1797. [DOI] [PubMed] [Google Scholar]
- Schwarz SF. Autotetraploides espontáneos en patrones de cítricos: incidencia, características y comportamiento en vivero y campo. 2001 PhD thesis, Universidad Politécnica de Valencia, Spain. [Google Scholar]
- Soltis DE, Soltis PS. The dynamic nature of polyploid genomes. Proceedings of the National Academy of Sciences of the USA. 1993;92:8089–8091. doi: 10.1073/pnas.92.18.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soost RK. Breeding citrus-genetics and nucellar embryony. In: Abbott AJ, Atkin RK, editors. Improving vegetatively propagated crops. London: Academic Press; 1987. pp. 83–110. [Google Scholar]
- Stebbins GL. Variation and evolution in plants. New York: Columbia University Press; 1950. [Google Scholar]
- Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold Ltd; 1971. [Google Scholar]
- Stupar RM, Bhaskar PB, Yandell BS, et al. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics. 2007;176:2055–2067. doi: 10.1534/genetics.107.074286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle WT, Reece PC. The botany of Citrus and its wild relatives. In: Reuther W, Batchelor LD, Webber HJ, editors. The citrus industry. Vol. 1. Riverside, CA: University of California; 1967. pp. 190–430. [Google Scholar]
- Tachikawa T, Tanaka Y, Hara S. Investigation on the breeding of Citrus trees. Study on the breeding of triploid Citrus varieties. Bulletin of the Shizuoka Prefectural Citrus Experiment Station. 1961;4:33–44. [Google Scholar]
- Thompson JD, Lumaret R. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in Ecology & Evolution. 1992;7:302–307. doi: 10.1016/0169-5347(92)90228-4. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Tian L, Lee HS, et al. Genome wide non additive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J, Doyle J. Polyploidy and evolution in plants. In: Henry RJ, editor. Plant diversity and evolution. Genotypic and phenotypic variation in higher plants. Wallingford, UK: CAB International; 2005. pp. 97–117. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data