Neurons Express Hemoglobin α- and β-Chains in Rat and Human Brains (original) (raw)
. Author manuscript; available in PMC: 2011 Jun 24.
Published in final edited form as: J Comp Neurol. 2009 Aug 10;515(5):538–547. doi: 10.1002/cne.22062
Abstract
Hemoglobin is the oxygen carrier in vertebrate blood erythrocytes. Here we report that hemoglobin chains are expressed in mammalian brain neurons and are regulated by a mitochondrial toxin. Transcriptome analyses of laser-capture microdissected nigral dopaminergic neurons in rats and striatal neurons in mice revealed the presence of hemoglobin α, adult chain 2 (Hba-a2) and hemoglobin β (Hbb) transcripts, whereas other erythroid markers were not detected. Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis confirmed the expression of Hba-a2 and Hbb in nigral dopaminergic neurons, striatal γ-aminobutyric acid (GABA)ergic neurons, and cortical pyramidal neurons in rats. Combined in situ hybridization histochemistry and immunohistochemistry with the neuronal marker neuronal nuclear antigen (NeuN) in rat brain further confirmed the presence of hemoglobin mRNAs in neurons. Immunohistochemistry identified hemoglobin α- and β-chains in both rat and human brains, and hemoglobin proteins were detected by Western blotting in whole rat brain tissue as well as in cultures of mesencephalic neurons, further excluding the possibility of blood contamination. Systemic administration of the mitochondrial inhibitor rotenone (2 mg/kg/d, 7d, s.c.) induced a marked decrease in Hba-a2 and Hbb but not neuroglobin or cytoglobin mRNA in transcriptome analyses of nigral dopaminergic neurons. Quantitative RT-PCR confirmed the transcriptional downregulation of Hba-a2 and Hbb in nigral, striatal, and cortical neurons. Thus, hemoglobin chains are expressed in neurons and are regulated by treatments that affect mitochondria, opening up the possibility that they may play a novel role in neuronal function and response to injury.
Keywords: hemoglobin, transcriptome analysis, neuronal expression, rotenone, in vivo, laser-capture microdissection
Vertebrate blood hemoglobin consists of a tetrameric structure including two α and two β hemoglobin chains (α2β2), each with its own heme moiety, which cooperate in binding and release of oxygen (Mills and Ackers, 1979). Monomeric globin chains containing a heme prosthetic group (Fe-protoporphyrin IX) can reversibly bind the gaseous ligands nitric oxide (NO), carbon monoxide (CO), and oxygen and are present in bacteria, fungi, plants, and invertebrate animals (Hardison, 1998). Recently, hemoglobin monomers have been found in vertebrate tissues other than blood, including macrophages (Liu et al., 1999), alveolar epithelial cells (Newton et al., 2006), the lens of the eye (Wride et al., 2003), and isolated myelin (Setton-Avruj et al., 2007), indicating that they are not restricted to the erythroid lineage, as previously assumed.
Neuronal tissue is highly metabolically active and generates high levels of nitrogen and oxygen-derived radicals (Lin and Beal, 2006). In view of their known role in blood, heme-containing globins might be involved in regulation of cellular respiration and oxidative stress in neurons. Neurons express important regulators of hemoglobin expression including the erythropoietin receptor, erythropoietin, and the hypoxiainducible factor HIF1α (Masuda et al., 1994; Digicaylioglu et al., 1995; Stroka et al., 2001). However, it is unknown whether they also express hemoglobin proteins. Here we demonstrate mRNA and protein expression of hemoglobin chains in neurons by conducting gene expression analyses of laser-captured neurons in rats and mice, combined with protein expression analyses in rat and human brain tissue. In addition, we show that hemoglobin gene expression is markedly decreased in neurons after in vivo treatment with low doses of the mitochondrial toxin rotenone. These findings reveal a previously unknown presence in neurons of a key blood component that has been shown to participate in maintaining the oxidant/antioxidant balance in cells (Cimen, 2008).
MATERIALS AND METHODS
Gene array analyses of neurons
Cell type-specific transcriptome analyses were conducted with laser-capture microdissected tyrosine hydroxylase (TH)-positive neurons of the substantia nigra pars compacta (SNC) in rats and TH-positive neurons of the SNC as well as parvalbumin (PV)-positive and -negative striatal neurons in mice. Adult male Lewis rats (Charles River Labs, Portage, MI) were used, and experiments were conducted in accordance with the United States Public Health Service Guide for the Care and Use of Laboratory Animals. Procedures were approved by the Institutional Animal Care and Use Committee at the US Army Medical Detachment Brooks City-Base, San Antonio, and University of California, Los Angeles. Animals received a continuous subcutaneous injection of either rotenone at 2 mg/kg/d or vehicle (dimethylsulfoxide and polyethyleneglycol) for 1 week via Alzet minipumps (Durect, Cupertino, CA). At the end of the treatment period animals were decapitated and the brain immediately frozen at −70°C. For laser-capture microdissection (LCM) of dopaminergic neurons, the midbrain area containing the substantia nigra was cut in 10-μm serial sections, quickly immunostained for TH, and counterstained with cresyl violet as described previously (Meurers et al., 2008).
In brief, following ethanol fixation for 1 minute, sections were incubated with a polyclonal primary and a biotinylated secondary antibody (Chemicon, Temecula, CA) for 5 minutes each. The Vectastain Elite ABC Kit and the DAB Substrate Kit (both from Vector, Burlingame, CA), which were applied to the sections for 3 minutes each, were used for chromogenic detection of antibody binding. Sections were rinsed in 1× phosphate-buffered saline (PBS) between each incubation step. For visualization of TH-negative neurons, sections were counterstained with cresyl violet for 1 minute. This step increases the selectivity of the LCM, as it decreases the probability of contaminating the TH-positive samples with TH-negative cells. The entire procedure was carried out under RNase-free conditions. Duplicate samples of 200 TH-positive neurons were collected from each animal. RNA extraction and the first round of T7-based mRNA amplification were performed as described previously (Kamme et al., 2004; Meurers et al., 2008) followed by a second round of amplification with the Affymetrix IVT labeling kit (Affymetrix, Santa Clara, CA) for Affymetrix oligonucleotide microarrays (whole genome array 230 or 430 2.0).
Samples derived from mouse TH-positive neurons of the SNC were generated with a comparable protocol. PV-positive neurons and PV-negative medium-sized spiny neurons were captured from the striatum of mice expressing green fluorescent protein (GFP) under the control of the Gad1 (previously known as GAD67) promoter (Chattopadhyaya et al., 2004). As described in detail in Chattopadhyaya et al. (2004), _Gad1_-GFP mice were generated by using a cell type-specific promoter and bacterial artificial chromosome (BAC) engineering. A BAC clone containing the entire Gad1 gene and 60 kb of upstream and downstream regions was used for BAC modifications. GFP expression cassettes were inserted in the first coding exon at the translation initiation site of the gene of interest. From the resulting transgenic founders a line was chosen in which the GFP expression pattern was restricted to PV+ neurons in different brain regions including the striatum. Transcriptome analyses of GFP-positive neurons in these mice have confirmed the expression of the transgene selectively in neurons also expressing PV mRNA (Meurers and Chesselet, unpublished observations).
The scanned microarray image files were imported into the R statistical software package (The R Foundation for Statistical Computing, www.r-project.org). Within R (version 2.2.1) the Bioconductor package (version 1.6) was used to perform GCRMA background correction and quantile normalization of the data. Computation of the expression indices was based on the GCRMA method (Wu and Dewey, 2003). The gene expression values for the duplicate samples of each animal were averaged to one sample value. Differentially expressed genes were identified based on two criteria: a present-absent call ratio larger than 1 (>50% present), and a two-way analysis of variance (ANOVA) model with gene expression as the outcome and group, treatment, and group-by-treatment interaction as the terms of the model. Calculation of the false discovery rate (FDR) was based on the empirical Bayes methodology discussed in (Efron and Tibshirani, 2002). All statistical analyses were performed by using R.
Quantitative real-time PCR analyses of selected neuronal populations
Cell type-specific gene expression analyses were conducted with laser-capture microdissected neurons from different brain regions in rats. On average 150 cells were laser dissected for each gene and cell type, and nonamplified RNA samples were used for quantitative polymerase chain reaction (qPCR) experiments as described previously (Meurers et al., 2008). Samples contained TH-positive neurons from the SNC, cortical pyramidal neurons, or medium-sized spiny striatal γ-aminobutyric acid (GABA)ergic neurons from rats treated with rotenone or vehicle. qPCR experiments were carried out in an ABI Prism 7900HT sequence detection system (ABI, Foster City, CA) or a Roche LightCycler 480 by using the Roche LightCycler FastStart DNA MasterPLUS SYBR Green I mix (Roche Diagnostics, Mannheim, Germany). All samples including serial plasmid dilutions for generating standard curves were run in triplicate. Primers were added at a final concentration of 400 nM. Aldehyde reductase (Akr1a1) and cyclophilin A (Ppia) were used for normalization of RNA content. These genes were chosen because they are abundantly expressed and the results of the prior array analysis demonstrated a lack of treatment-dependent changes in expression levels.
For each gene standard curves were generated from 10-fold serial dilutions of plasmid clones. Average mRNA quantities of both genes were calculated for each sample and the combined values served as normalization factors. Calculation of absolute quantities was performed on the ABI Prism 7900HT or LightCycler 480 software. Statistical significance of group differences was determined by t-tests using P values < 0.05 as a cutoff. The specificity of the amplicons was determined by sequencing of the gel-purified qPCR products.
In situ hybridization histochemistry
Coronal sections (10 μm) were cut from frozen brains of untreated rats on a cryostat and mounted on charged glass slides. Experiments were performed on sections of the SNC (4.50–5.40 mm posterior to bregma) based on the atlas of Paxinos and Franklin (2001). Two non-overlapping hybridization probes coding for Hba-a2 and three different (two non-overlapping) hybridization probes coding for Hbb were generated from expressed sequence tag (EST) clones containing the 3′ ends of the respective genes or from PCR products that had been amplified with T3 and T7 sequences attached to the 5′ end of the gene-specific primers. Prior to synthesis of the RNA probes, all clone inserts and PCR products were sequence verified. The RNA probes were synthesized by appropriate transcription polymerases in the presence of a digoxigenin-substituted nucleotide (dig-UTP) and an inhibitor of RNAse according to standard protocols (incubation for 2 hours at 37°C). The labeled RNA was purified with the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer's protocol.
In situ hybridization histochemistry was performed as follows. Frozen sections were quickly dried on a metal surface at 37°C, fixed in 4% paraformaldehyde (PFA) for 20 minutes, equilibrated in 5× standard sodium citrate (SSC), and prehybridized in humid chambers for 2 hours at 58–62°C in hybridization buffer (50% formamide, 40 μg/ml salmon sperm DNA, 5× SSC) without riboprobes. Thereafter the sections were incubated with RNA probe diluted in hybridization buffer at a concentration of 1 ng/μl at 58 −70°C for 16 hours. Posthybridization treatments included two washes in 2× and 0.1× SSC at 65–70°C for 1 hour, incubation with anti-digoxigenin alkaline phosphatase (1:1,000) for 2 hours, and development with 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP) and nitro-blue tetrazolium chloride (NBT) at room temperature overnight. Finally the sections were rinsed in TE buffer (10 mM Tris-HCl and 1 mM EDTA), and the background was removed by incubation in 95% ethanol. After rehydration the sections were processed for a quick neutral red counterstaining and dehydrated in increasing concentrations of ethanol, cleared in xylene, and coverslipped with DPX (Sigma, St. Louis, MO).
For combined in situ hybridization histochemistry and neuronal nuclear antigen (NeuN) immunohistochemistry, sections were first processed for quick NeuN immunostaining (mouse anti-NeuN, 1:100, Millipore, Bedford, MA) as described above for the TH staining followed by in situ hybridization. The NeuN antibody from Millipore has been widely used and has been characterized as described previously (Mignon et al., 2005).
Immunohistochemistry
Untreated adult male Lewis rats were anesthetized with pentobarbital (100 mg/kg i.p.) and intracardially perfused with 0.1 M PBS followed by ice-cold 4% PFA. Brains were quickly removed, postfixed for 12 hours in 4% PFA, and rapidly frozen on dry ice. Forty-micrometer free-floating coronal sections were collected for immunostaining. In addition, paraffin-embedded human brain sections (3–4 μm; female control case, age 54 years) from selected brain regions were processed for immunohistochemistry. Sections were stained following standard protocols by using hemoglobin antiserum and antibodies against hemoglobin α-or β-chain (see Table 1 for the origins and concentrations of antibodies). Sections were washed in 0.1 M PBS (pH 7.6) and incubated in 2% hydrogen peroxide for 30 minutes to block endogenous peroxidases, rinsed, and then preincubated for 1 hour in a blocking solution (10 % normal serum, 0.3 % Triton X-100 in Tris-buffered saline [TBS; pH 7.6]). Thereafter, the sections were transferred into the primary antibody with 2% normal serum for 20 hours and after rinsing placed for 1 hour in conjugated secondary antibody (see Table 1). For biotin labeling the avidin-biotin complex method was used to detect the secondary antibody (ABC elite kit, Vector), and the reaction product was visualized with 3,3′-diaminobenzidine (DAB). The sections were mounted on glass slides, air-dried, dehydrated, and coverslipped with DPX. Human brain sections were counterstained with hematoxylin for nuclear staining.
TABLE 1.
Primary and Secondary Antibodies Used
| Antigen | Immunogen | Manufacturer and source | Dilution |
|---|---|---|---|
| Hemoglobin α-chain (rat) | Synthetic peptide with sequence of aa 110–126 from mouse protein (P01942)1 | Santa Cruz Biotechnology (Santa Cruz, CA), goat IgG polyclonal, #31333 | 1:100–1:500 |
| Hemoglobin α-chain (human) | Synthetic peptide with sequence of aa 63–80 from human protein (P69905)1 | Santa Cruz Biotechnology, goat IgG polyclonal, #31110 | 1:100 |
| Hemoglobin β-chain (rat) | Synthetic peptide with sequence of aa 109–129 from mouse protein (P02089)1 | Santa Cruz Biotechnology, goat IgG polyclonal, #31116 | 1:100–1:500 |
| Hemoglobin β-chain (human) | Purified human hemoglobin β | Santa Cruz Biotechnology, mouse IgG1 monoclonal, #21757 | 1:100 |
| Hemoglobin protein | Purified human hemoglobin | Sigma-Aldrich (St. Louis, MO), rabbit whole antiserum, #H4890 | 1:500 |
| Neuronal nuclear antigen (NeuN) | Purified cell nuclei from mouse brain | Millipore (Billerica, MA), mouse monoclonal, #MAB377 | 1:100 |
| Tyrosine hydroxylase (TH) | Denatured tyrosine hydroxylase from rat pheochromocytoma | Millipore, rabbit polyclonal, #AB152 | 1:200 |
| Anti-goat IgG | Goat IgG | Millipore, donkey polyclonal, #AP180P | 1:600–1:10,000 |
| Anti-rabbit IgG | Rabbit IgG | Millipore, goat polyclonal, #AP132P | 1:20,000 |
Western blot
Whole rat brain tissue and postnatal midbrain primary cell cultures from rats were used for protein extraction. Brain tissue was taken from rats that were intracardially perfused with 0.1 M PBS to minimize blood pollution. Samples from a midbrain cell culture enriched for neurons were washed three times with 0.1 M phosphate buffer before harvesting. Red blood cells concentrated by centrifugation of a rat blood sample were used as positive controls. After protein extraction in warm extraction buffer (1% sodium dodecyl sulfate [SDS], protease inhibitor) the protein concentration was determined by the Bradford method (Bio-Rad, Hercules, CA) followed by denaturation in 1× Laemmli buffer at 100°C for 5 minutes. A 12% SDS-polyacrylamide electrophoresis gel was loaded with maximal 50 μg protein per lane and electrotransferred to PVDF membrane with a Semi-Dry Transfer Unit (Bio-Rad). The blot was incubated with a hemoglobin antiserum at 4°C overnight. Following repeated washes, the membrane was incubated with secondary, horseradish peroxidase-labeled antibody (Millipore, 1:20,000), washed, and developed with a chemiluminescent substrate (SuperSignal West Pico; Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Finally, an x-ray film was exposed to the lumines-cent membrane followed by development. Molecular masses were determined by using a parallel processed protein standard (Precision plus protein standards, Bio-Rad). Chromogenic images were acquired with a Zeiss Axioskop microscope (Zeiss, Gottingen, Germany) and a Spot digital camera (Diagnostic Instruments, Sterling Heights, MI) and adjusted for brightness and contrast by using Photoshop 6.0 (Adobe Systems, San Jose, CA) and CorelDRAW(R) Graphics Suite 12.0 (Corel, Mountain View, CA).
Antibody characterization
The antibody against rat hemoglobin α-chain (#31333, Santa Cruz Biotechnology, Santa Cruz, CA) used for immunohistochemical studies recognizes a single band of ~16 kDa, the MW of the monomeric hemoglobin α-chain, in Western blot analyses of mouse heart (manufacturer's datasheet). Specificity is further supported by a decrease in immunoblot signal detected with this antibody in mouse erythroleukemia (MEL) cells treated with an α-hemoglobin siRNA (Voon et al., 2008). In our study, a positive control for hemoglobin detection with this antibody was provided by staining of erythrocytes by immunohistochemistry in brain tissue sections, and specificity was confirmed by elimination of the staining by preincubation of the primary antibody with the corresponding blocking peptide (antigen from Santa Cruz Biotechnology) at fivefold excess (see Results). The labeling pattern was similar in cell bodies to that observed with two non-overlapping RNA probes complementary to rat hemoglobin α-chain (Hba-2a; see Results).
The antibody against rat hemoglobin β-chain (#31116, Santa Cruz Biotechnology) used for immunohistochemical studies recognizes a single band of ~16 kDa, the MW of the monomeric hemoglobin β-chain, in Western blot analyses of rat peripheral blood leukocyte (PBL) whole cell lysate (manufacturer's datasheet). Selectivity for the β-chain of hemoglobin is further supported by detection of an unchanged immunoblot signal in MEL cells treated with α-hemoglobin siRNA (Voon et al., 2008). In our study, a positive control for hemoglobin detection with this antibody was provided by staining of erythrocytes by immunohistochemistry in brain tissue sections, and specificity was confirmed by elimination of the staining by preincubation of the primary antibody with the corresponding blocking peptide (antigen from Santa Cruz) at fivefold excess (see Results). The labeling pattern was similar in cell bodies to that observed with two non-overlapping RNA probes complementary to rat hemoglobin β-chain (see Results).
The antibody against human hemoglobin α-chain (#31110, Santa Cruz Biotechnology) used for immunohistochemical studies recognizes a single band of ~16 kDa in Western blot analyses of TF-1, HEL 92.1.7, and K-562 whole cell lysates (manufacturer's datasheet) and specifically stained the typical ~16-kDa band of α-hemoglobin monomer in Western blots of total cellular extracts of cultured nucleated human erythro-blasts with high sensitivity (Suzuki et al., 2008). This antibody specifically stained hemoglobin proteins in Western blots of purified human hemoglobin (Newton et al., 2006). The presence of α-hemoglobin chains in the ~16-kDa gel band could be verified by sequencing using tandem mass spectrometry (Newton et al., 2006). In our study, a positive control for hemoglobin detection with this antibody was provided by staining of erythrocytes by immunohistochemistry in human brain tissue sections (see Results). As described in the Results section, this antibody showed a staining pattern in human brain tissue that was comparable to the specific staining we found with the α-hemoglobin antibody used in rat brain tissue.
The antibody against human hemoglobin β-chain (#21757, Santa Cruz Biotechnology) used for immunohistochemical studies recognizes a single band of ~16 kDa in Western blot analyses of TF-1 whole cell lysate (manufacturer's datasheet) and specifically stained the typical ~16-kDa band of β-hemoglobin monomer in Western blots with total cellular extracts of cultured nucleated human erythroblasts with high sensitivity (Suzuki et al., 2008). This antibody was successfully used to confirm different expression levels of β-hemoglobin RNA in several cancer cell lines by immunohistochemical analyses (Onda et al., 2005). In addition, the hemoglobin β-chain antibody was shown to specifically identify erythrocytes in flow cytometry (manufacturer's datasheet). In our study, a positive control for hemoglobin detection with this antibody was provided by staining of erythrocytes by immunohistochemistry in human brain tissue sections (see Results). As described in the Results section, this antibody showed a staining pattern in human brain tissue that was comparable to the specific staining we found with the β-hemoglobin antibody used in rat brain tissue.
The anti-hemoglobin antibody used for immunoblotting (#H4890, Sigma) labeled a single band at ~16 kDa in immunoblots of crude human cerebellar proteins (Slemmon et al., 1994) and red blood cells (Dupuy and Engelman, 2008) and showed strong reactivity with human hemoglobin at 1 mg/ml (Sigma Product Information Sheet no. H4890). In our study, staining of the positive control (protein extracted from rat blood) resulted in strong and highly selective bands for the hemoglobin tetramer (~64 kDa), dimer (~32 kDa), and monomer (~16 kDa) (see Results, Fig. 3 discussion). Staining of rat brain sections with this antibody showed a staining pattern similar to the patterns achieved with the specific anti-rat α-and β-hemoglobin antibodies (data not shown).
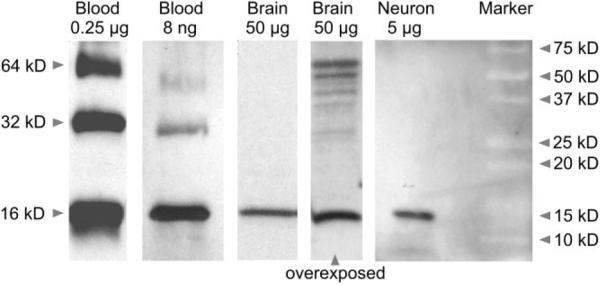
Figure 3.
Western blot with extracted protein from rat blood (0.25 μg and 8 ng), brain (50 μg), and primary midbrain cultures enriched for neurons (“Neuron” 5 μg) probed with whole hemoglobin antiserum. A strong band at 16 kDa is present in all lanes, which is indicative of the presence of the hemoglobin chains monomers. Rat blood shows additional bands at 32 kDa (dimer) and 64 kDa (tetramer) with higher protein concentrations. Comparable-sized bands also become visible in the brain sample after overexposure of the blot.
The NeuN and TH antibodies from Millipore have been widely used and characterized as described previously for NeuN in Mignon et al. (2005) and for TH in Meurers et al. (2008). The NeuN antibody MAB377 (Millipore) has been extensively characterized and recognized the 46- and 48-kDa isoforms of NeuN as shown by Western blot and electro-phoretic separation of crude nuclear fraction of adult mouse brain (Lind et al., 2005). To ensure the specificity of the anti-body against TH (#AB152, Millipore), we laser-capture micro-dissected neurons from the substantia nigra that have been labeled with this antibody and obtained a sample of neurons that express TH but not GAD1 as verified by PCR (Meurers et al., 2008). Staining for NeuN and TH in our study gave similar results to those documented in the literature (Zhu et al., 2004; Mignon et al., 2005; Meurers et al., 2008).
RESULTS
Hemoglobin α- and β-chain mRNAs are present in neurons
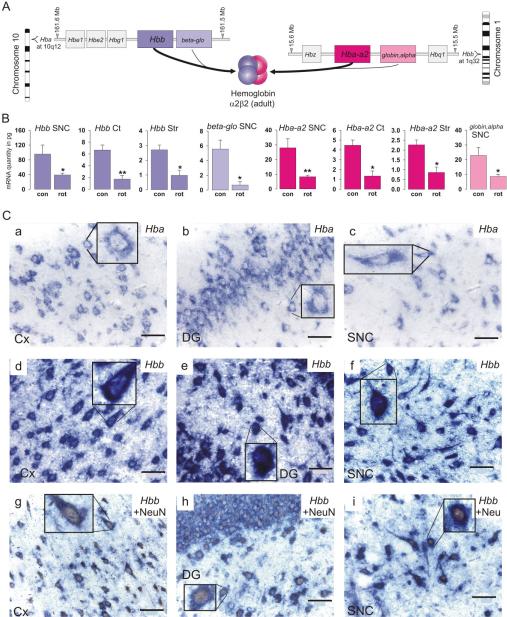
Evidence for hemoglobin gene expression was an unexpected discovery in cell type-specific transcriptome analyses conducted with laser-capture microdissected neurons from different brain regions in rats (microarray data published in supplemental information in Meurers et al., 2008) and mice. Gene expression profiles from TH-positive neurons of the SNC neurons of the rat revealed the presence of mRNA for hemoglobin alpha, adult chain 2 (Hba-a2) and globin, alpha, as well as hemoglobin, beta (Hbb) and beta-glo (Table 2). Rat Hba-a2 and globin, alpha have a nearly identical sequence and are located side by side on the α-globin gene cluster, showing that Hbb and beta-glo are highly homologous members of the β-globin gene cluster (Fig. 1A). Hemoglobin transcripts were also expressed by neurons from mouse SNC and striatum (data not shown). The absence of the erythroid marker GATA binding protein 1 (Gata1) and erythroid-associated factor (Eraf) confirms that hemoglobin gene expression in nigral neurons was most likely not the result of blood contamination. However, mRNAs for transcription factors related to hemoglobin expression such as Hif1 α, Gata2, and Gata3 were present in our array data (Table 2).
TABLE 2.
Hemoglobin-Related Genes (Microarray) in Rat Dopaminergic Neurons of the Substantia Nigra Pars Compacta (SNC)
| Gene title | Gene symbol | Gene ID | Presence in SNC neurons | Fold change after rotenone treatment |
|---|---|---|---|---|
| Globin family | ||||
| hemoglobin alpha, adult chain 2 | Hba-a2 | 25632 | Present | −17.66 (P = 0.0006) |
| globin, alpha | LOC287167 | 287167 | Present | |
| hemoglobin, beta | Hbb | 24440 | Present | −13.99 (P = 0.0008) |
| beta-glo | MGC72973 | 361619 | Present | |
| hemoglobin, alpha | Hbg1 | 94164 | ||
| hemoglobin, epsilon 1 | Hbe1 | 293267 | Absent | |
| hemoglobin, zeta | Hbz | 287168 | ||
| hemoglobin, theta 1 | Hbq1 | 303007 | ||
| neuroglobin | Ngb | 85382 | Present | 1.17 (P = 0.57) |
| cytoglobin | Cygb | 170520 | Present | 1.19 (P = 0.42) |
| myoglobin | Mb | 59108 | Absent | |
| Related transcription factors | ||||
| hypoxia-inducible factor 1, alpha | Hif1a | 29560 | Present | 1.12 (P = 0.30) |
| GATA binding protein 2 | Gata2 | 25159 | Present | 1.44 (P = 0.68) |
| GATA binding protein 3 | Gata3 | 85471 | Present | 1.13 (P = 0.43) |
| heme oxygenase | Hmox1 | 362454 | Present | 1.06 (P = 0.28) |
| Erythroid cell marker genes | ||||
| erythroid-associated factor | Eraf | 293522 | Absent | |
| GATA binding protein 1 | Gata1 | 25172 | Absent |
Figure 1.
Hemoglobin α- and β-chain mRNA expression. A: Arrangements of genes in the rat α- and β-globin gene cluster (not to scale) with the respective ideogram. Adult α- and β-globin-like gene products assemble to α2β2 hemoglobin in erythroid cells. Positions were taken from the MapViewer (RGSC v3.4) at The National Center for Biotechnology Information (NCBI) website. B: mRNA levels (qPCR) of α- and β-globin gene expression in dopaminergic neurons of the SNC, cortical pyramidal neurons, and striatal projection neurons. Con and rot indicate samples from control (n = 6) and rotenone-treated (n = 6) animals, respectively. Data are means ± SEM (*, P < 0.01; **, P < 0.001). C: In situ hybridization immunohistochemistry with DIG-labeled RNA probes shows intense blue staining for Hba-a2 (a–c) and Hbb (d–f) in the cortex (Cx; a,d), dentate gyrus of the hippocampus (DG; b,e), and substantia nigra pars compacta (SNC; c,f). Cells containing hemoglobin β-chain mRNA (blue) and the neuronal marker NeuN (brown nuclei) in the (g) cortex, (h) dentate gyrus, and (i) SNC. Scale bar = 50 μm in a–i (insets are 2.5-fold magnifications).
Real time qPCR analysis of nonamplified mRNA samples from recaptured nigral dopaminergic neurons from rats, cortical pyramidal neurons, and striatal GABAergic projection neurons confirmed the expression of hemoglobin mRNAs in neurons (Fig. 1B). The specific primers (Supplementary Table S1) distinguished between the homologous members of the α-globin or β-globin gene cluster as ensured by sequencing of the qPCR product. Table 3 summarizes the specificity of the different techniques used in this study for homologous genes and their products.
TABLE 3.
Specificity of Techniques Used in This Study for Different Globin mRNAs and Proteins1
| mRNA/proteins | ||||
|---|---|---|---|---|
| Technique | Hbb | beta-glo | Hba-a2 | globin-alpha |
| Microarray | + | + | ||
| Polymerase chain reaction (PCR) | + | + | + | + |
| In situ hybridization | + | + | ||
| Immunohistochemistry | + | + | ||
| Western blot | + |
In situ hybridization histochemistry confirmed the presence of hemoglobin mRNAs in neurons of all layers of the rat cerebral cortex and hippocampus, and in the SNC (Fig. 1C, a–f). Combined in situ hybridization histochemistry and immunohistochemistry for the neuronal marker NeuN showed cells clearly double labeled for NeuN and hemoglobin chain mRNAs (Fig. 1C, g–i). The negative controls with sense RNA probes were devoid of specific hybridization signal, as shown under identical experimental conditions for the hippocampus and the SNC (Supplementary Fig. S1). To further control the specificity of the in situ hybridization signal for hemoglobin chain mRNAs, we created a second RNA probe for Hba-a2 and two further RNA probes for non-overlapping regions of Hbb. A positive signal was observed with these probes, as shown in the hippocampus and SNC for Hbb, with no signal detected with the corresponding sense probes (Supplementary Fig. S2).
Hemoglobin chains are present in rat and human neurons
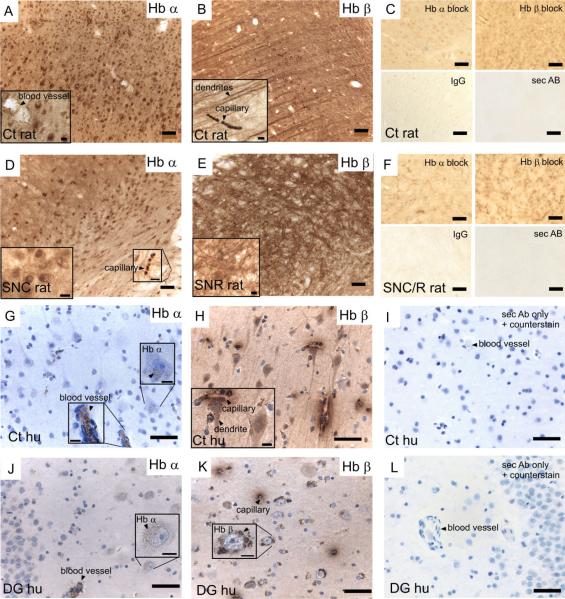
Immunohistochemistry with polyclonal antibodies that are specific for rat hemoglobin α-chain or rat hemoglobin β-chain revealed strong labeling for both hemoglobin chains in rat neurons (Fig. 2A–F). Hemoglobin chains were also expressed in human neurons, as shown by immunohistochemistry using polyclonal primary antibodies specifically raised against human α-hemoglobin chains or human β-hemoglobin chains (Fig. 2G–L). Erythrocytes that remained after perfusion in brain tissue provided a positive control for hemoglobin proteins. Although hemoglobin α-chains seemed to be mostly present in the cell body and nucleus (Fig. 2A,D,G,J), hemoglobin β-chains were also present in cellular processes (Fig. 2B,E,H,K). In human brain sections the staining was not uniformly distributed in the cytoplasm but formed a granular pattern. As a result, staining was inconclusive in the SNC, as hemoglobin chain staining could not be distinguished from the granular neuromelanin present in the dopaminergic neurons of the SNC (data not shown). Further analyses of the signal distribution revealed strong positive staining for hemoglobin α-chains in the cortex, basal ganglia structures, hippocampus, and hypothalamus and for hemoglobin β-chains in cortical and thalamic dendrites, hippocampal cells and processes, and substantia nigra pars reticulata of rat and human brains. Control sections with omitted primary antibody, normal serum IgG, or primary antibody blocked with the specific blocking peptide did not show a comparable staining pattern (Fig. 2C,F,I,L).
Figure 2.
Hemoglobin α-or β-chains on 40-μm rat brain sections (A–F) and 4-μm human brain sections (G–L). Hemoglobin α-chains (Hb α; A,D,G,J) and hemoglobin β-chains (Hb β; B,E,H,K) are present in neurons, as shown in the rat and human (hu) cortex (Cx), the rat substantia nigra pars compacta/substantia nigra pars reticulata (SNC/R), and the human dentate gyrus of the hippocampus (DG), visible as brown staining. C, F, I, and L show negative controls with blocked primary antibodies, IgG control, or secondary antibody only. Scale bar = 50 μm in A–L; 10 μmin insets to A,B,D,E,G,H,J,K.
Western blots from whole rat brain tissue and postnatal midbrain cell cultures enriched in neurons confirmed the presence of a hemoglobin protein with the correct molecular weight in neurons. The use of cultured neurons excluded the possibility that the hemoglobin protein detected could be derived from blood contamination. The blot was probed with whole hemoglobin antiserum. For positive control, protein extracted from rat blood was loaded in different concentrations, resulting in strong bands for the hemoglobin tetramer, dimer, and monomer, as expected from the high concentration of hemoglobin in blood (Fig. 3). Figure 3 shows the typical 16-kDa band of the monomeric hemoglobin α-or β-chains in protein extracts from whole brain tissue and in cultured neuronal cells. In blots from whole brain tissue that were overex-posed, but not in blots from cultured neuronal cells, weak bands of higher molecular mass were also visible and might account for dimeric (~32 kDa) or tetrameric (~64 kDa) hemoglobin proteins.
Hemoglobin chain mRNA expression is downregulated in response to mitochondrial inhibition
Rats were treated systemically with low doses (2 mg/kg/d) of rotenone, an inhibitor of the complex I of the mitochondrial respiratory chain (Zhu et al., 2004; Meurers et al., 2008). Global gene expression analyses in nigral dopaminergic neurons revealed that Hba-a2/globin, alpha and Hbb/beta-glo but not neuroglobin or cytoglobin were markedly decreased after low-dose rotenone treatment compared with vehicle controls (Table 2). In fact, hemoglobin genes showed the highest fold change in response to the toxin treatment in our microarray data (for regulation of other genes, see Meurers et al., 2008). Subsequent qPCR experiments confirmed a downregulation of these genes in dopaminergic neurons of the SNC as well as in cortical pyramidal neurons and in GABAergic projection neurons of the striatum in response to rotenone treatment (Fig. 1B).
DISCUSSION
Our results demonstrate mRNA and protein expression of α- and β-globin chains in neurons. We show that both hemoglobin chains are expressed in several brain regions and, furthermore, that their levels of expression are regulated in neurons in vivo in response to treatment with a mitochondrial toxin.
Hemoglobin chains are expressed in neurons
Gene expression profiling of laser-captured neurons of nigral, cortical, and striatal neurons from rat brains clearly demonstrated the expression of the adult hemoglobin α- and β-chain mRNAs. Product sequencing and specifically designed primers even enabled us to distinguish between highly homologous genes present in the α-globin or β-globin gene cluster. In erythroid cells, Hba-a2 and Hbb have a higher expression level than globin, alpha and beta-glo (Paunesku et al., 1990; Satoh et al., 1999). Our data support the expression of all four genes in neurons but do not determine whether their expression levels differ in a manner comparable to that of erythroid cells. Contamination of laser-capture microdissected samples with reticulocytes, the only RNA-containing red blood cells (Riley et al., 2001), is unlikely but cannot be totally excluded. However, the absence of the two erythroid markers Gata1 and Eraf (Kihm et al., 2002; Ohneda and Yamamoto, 2002) in our microarray data and a clearly intra-cellular signal in our in situ hybridization histochemistry data eliminate this possibility.
Immunohistochemistry of rat and human brain sections confirmed the presence of hemoglobin chains in brain neurons, with both chains present in the cell soma, and β-chains in dendrites. Co-localization in the cell soma might support an assembly of hemoglobin α- and β-chains into a hemoglobin tetramer (2α2β) as in blood cells. On Western blots, the strongest signal was obtained for the typical ~16-kDa monomer, but bands with a low signal in the molecular weight of dimers (~32 kDa) and tetramers (~64 kDa) were also present with longer exposure, supporting the possibility of assembly of hemoglobin chains in neurons. The presence of the monomeric hemoglobin chains in blood-free midbrain cell cultures, which were enriched for neurons, further confirmed that the detected hemoglobin protein was not derived from contamination with blood. We did not detect higher molecular bands in this preparation. An assembly, however, is not necessary for a function of hemoglobin monomers in neurons. Indeed, the affinity for oxygen of the α-or β-hemoglobin monomers is much higher compared with that of the α2β2 tetramer (P50 monomer, ~1 torr/P50 tetramer, ~27 torr; Mills and Ackers, 1979; Wittenberg and Wittenberg, 1990), and hemoglobin monomers are capable of binding CO and NO with very high affinity (Bellelli et al., 2006).
These binding affinities and related enzymatic activities account for the functions of monomeric hemoglobin in plants, bacteria, and invertebrates and might be important in neurons as well. Furthermore, isolated hemoglobin β-chains form stable tetramers (β4) with very high oxygen binding affinity, and hemoglobin α-chains form toxic precipitates if present in excess (Valdes and Ackers, 1978; Kihm et al., 2002). It is conceivable that β-chains bind to free α-chains in the cell soma and that the excess forms hemoglobin β-tetramers that are also present in cell processes, a strategy that would avoid toxic α-chain precipitates.
Regulation of hemoglobin monomers in the brain
NO is the strongest known ligand of the ferrous heme iron of hemoglobin, with a 500,000 times higher affinity than oxygen (Bellelli et al., 2006). All heme-containing globin monomers discovered also function in binding products of NO metabolism and might therefore protect cells against oxidative and nitrosative stress (Wilson and Reeder, 2008) or, like myoglobin, might reduce nitrite to NO under hypoxia, for which high NO levels are beneficial (Hendgen-Cotta et al., 2008). A possible role of hemoglobin chains in mitochondrial respiration and regulation of the redox system is supported by our finding that hemoglobin gene expression is strongly downregulated in neurons of rats treated with low doses of rotenone. Treatment with rotenone has been described to result in inhibition of the mitochondrial complex I and elevation of oxidative stress (Sherer et al., 2003).
Hemoglobin gene expression in erythroid cells is dependent on the activity of the α-subunit of hypoxia-inducible factor 1 (HIF1α) (Wang and Semenza, 1993). Importantly, neurons express and regulate HIF1α, erythropoietin, and the erythropoietin receptor (Digicaylioglu et al., 1995; Stroka et al., 2001). Although HIF1α mRNA levels were unchanged in our experimental conditions, inhibition of mitochondrial complex I under physiological levels of NO may result in inactivation of HIF1α, which could then lead to the pronounced downregulation of hemoglobin mRNA caused by rotenone in this study (Agani et al., 2000; Mateo et al., 2003). Another possible mechanism of hemoglobin regulation might result from reduction of free heme, which is a strong activator of globin-chain transcription (Tsiftsoglou et al., 2006). Heme synthesis takes place in the mitochondria and is facilitated by ferrochelatase, which inserts the ferrous iron molecule into a tetrapyrrole (Ajioka et al., 2006). Inhibition of mitochondrial function might therefore lead to decreased heme synthesis.
Considering recent evidence that cells derived from the brain and from bone marrow share common features and regulating factors (Brazelton et al., 2000; Goolsby et al., 2003), and that the brain is the organ most sensitive to oxygen deprivation (LaManna, 2007), the expression of hemoglobin chains in neurons, although novel, is not totally unexpected. Indeed, hemoglobin-derived peptides have been described in the brain and there is ongoing debate as to how hemoglobin or these metabolites can pass the blood-brain barrier. These so-called hemorphines are ligands for opioid and AT4 receptors in the brain, with functional implications that include analgesia but also memory retention and retrieval (Nyberg et al., 1997). Our data support the hypothesis that neuronal hemoglobin is the precursor for hemorphins. In summary, hemoglobin chains may have an unsuspected role in neuronal survival and function and our findings should lead to further investigation of their implications in a wide range of experimental situations.
Supplementary Material
Sup Fig 1
Sup Fig 2
Sup material
ACKNOWLEDGMENTS
We thank Drs. Harry Vinters and Peter Shintaku, UCLA, for supply and assistance with immunostaining of human brain sections, Dr. Nigel Maidment, UCLA, for mesencephalic cell cultures, and Dr. Z.J. Huang at Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, for the gift of GAD1-GFP mice. The initial transcriptome data (published in Meurers et al., 2008) were obtained in a collaborative study with Drs. Cheryl DiCarlo (Southwest National Primate Research Center, Southwest Foundation for Biomedical Research, San Antonio, TX), Ronald Seaman (McKesson BioServices at US Army Medical Research Detachment, Brooks City-Base, TX), and Myung Oh and David Elashoff (UCLA, Los Angeles, CA).
Grant sponsor: the Alexander von Humboldt Foundation; Grant number: U54 ES012078 and a Fellowship to F.R.; Grant sponsor: US Army MRMC (for original transcriptome data): Grant number: DAMD17-94-C-4069; Grant sponsor: Public Health Service (for original transciptome data); Grant numbers: U54 ES12078 and P50 NS38367.
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Agani FH, Pichiule P, Chavez JC, LaManna JC. The role of mitochondria in the regulation of hypoxia-inducible factor 1 expression during hypoxia. J Biol Chem. 2000;275:35863–35867. doi: 10.1074/jbc.M005643200. [DOI] [PubMed] [Google Scholar]
- Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bellelli A, Brunori M, Miele AE, Panetta G, Vallone B. The allosteric properties of hemoglobin: insights from natural and site directed mutants. Curr Protein Pept Sci. 2006;7:17–45. doi: 10.2174/138920306775474121. [DOI] [PubMed] [Google Scholar]
- Brazelton TR, Rossi FM, Keshet GI, Blau HM. From marrow to brain: expression of neuronal phenotypes in adult mice. Science. 2000;290:1775–1779. doi: 10.1126/science.290.5497.1775. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, Huang ZJ. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimen MY. Free radical metabolism in human erythrocytes. Clin Chim Acta. 2008;390:1–11. doi: 10.1016/j.cca.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Digicaylioglu M, Bichet S, Marti HH, Wenger RH, Rivas LA, Bauer C, Gassmann M. Localization of specific erythropoietin binding sites in defined areas of the mouse brain. Proc Natl Acad Sci U S A. 1995;92:3717–3720. doi: 10.1073/pnas.92.9.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AD, Engelman DM. Protein area occupancy at the center of the red blood cell membrane. Proc Natl Acad Sci U S A. 2008;105:2848–2852. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Empirical Bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- Goolsby J, Marty MC, Heletz D, Chiappelli J, Tashko G, Yarnell D, Fishman PS, Dhib-Jalbut S, Bever CT, Jr, Pessac B, Trisler D. Hematopoietic progenitors express neural genes. Proc Natl Acad Sci U S A. 2003;100:14926–14931. doi: 10.1073/pnas.2434383100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- Hendgen-Cotta UB, Merx MW, Shiva S, Schmitz J, Becher S, Klare JP, Steinhoff HJ, Goedecke A, Schrader J, Gladwin MT, Kelm M, Rassaf T. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2008;105:10256–10261. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamme F, Zhu J, Luo L, Yu J, Tran DT, Meurers B, Bittner A, Westlund K, Carlton S, Wan J. Single-cell laser-capture microdissection and RNA amplification. Methods Mol Med. 2004;99:215–223. doi: 10.1385/1-59259-770-X:215. [DOI] [PubMed] [Google Scholar]
- Kihm AJ, Kong Y, Hong W, Russell JE, Rouda S, Adachi K, Simon MC, Blobel GA, Weiss MJ. An abundant erythroid protein that stabilizes free alpha-haemoglobin. Nature. 2002;417:758–763. doi: 10.1038/nature00803. [DOI] [PubMed] [Google Scholar]
- LaManna JC. Hypoxia in the central nervous system. Essays Biochem. 2007;43:139–151. doi: 10.1042/BSE0430139. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lind D, Franken S, Kappler J, Jankowski J, Schilling K. Characterization of the neuronal marker NeuN as a multiply phosphorylated antigen with discrete subcellular localization. J Neurosci Res. 2005;79:295–302. doi: 10.1002/jnr.20354. [DOI] [PubMed] [Google Scholar]
- Liu L, Zeng M, Stamler JS. Hemoglobin induction in mouse macrophages. Proc Natl Acad Sci U S A. 1999;96:6643–6647. doi: 10.1073/pnas.96.12.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Okano M, Yamagishi K, Nagao M, Ueda M, Sasaki R. A novel site of erythropoietin production. Oxygen-dependent production in cultured rat astrocytes. J Biol Chem. 1994;269:19488–19493. [PubMed] [Google Scholar]
- Mateo J, Garcia-Lecea M, Cadenas S, Hernandez C, Moncada S. Regulation of hypoxia-inducible factor-1alpha by nitric oxide through mitochondria-dependent and -independent pathways. Biochem J. 2003;376:537–544. doi: 10.1042/BJ20031155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurers BH, Zhu C, Fernagut PO, Richter F, Hsia YC, Fleming SM, Oh M, Elashoff D, Dicarlo CD, Seaman RL, Chesselet MF. Low dose rotenone treatment causes selective transcriptional activation of cell death related pathways in dopaminergic neurons in vivo. Neurobiol Dis. 2008;33:182–192. doi: 10.1016/j.nbd.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignon L, Vourc'h P, Romero-Ramos M, Osztermann P, Young HE, Lucas PA, Chesselet MF. Transplantation of multipotent cells extracted from adult skeletal muscles into the subventricular zone of adult rats. J Comp Neurol. 2005;491:96–108. doi: 10.1002/cne.20685. [DOI] [PubMed] [Google Scholar]
- Mills FC, Ackers GK. Quaternary enhancement in binding of oxygen by human hemoglobin. Proc Natl Acad Sci U S A. 1979;76:273–277. doi: 10.1073/pnas.76.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton DA, Rao KM, Dluhy RA, Baatz JE. Hemoglobin is expressed by alveolar epithelial cells. J Biol Chem. 2006;281:5668–5676. doi: 10.1074/jbc.M509314200. [DOI] [PubMed] [Google Scholar]
- Nyberg F, Sanderson K, Glamsta EL. The hemorphins: a new class of opioid peptides derived from the blood protein hemoglobin. Biopolymers. 1997;43:147–156. doi: 10.1002/(SICI)1097-0282(1997)43:2<147::AID-BIP8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Ohneda K, Yamamoto M. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 2002;108:237–245. doi: 10.1159/000065660. [DOI] [PubMed] [Google Scholar]
- Onda M, Akaishi J, Asaka S, Okamoto J, Miyamoto S, Mizutani K, Yoshida A, Ito K, Emi M. Decreased expression of haemoglobin beta (HBB) gene in anaplastic thyroid cancer and recovery of its expression inhibits cell growth. Br J Cancer. 2005;92:2216–2224. doi: 10.1038/sj.bjc.6602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunesku T, Stevanovic M, Radosavljevic D, Drmanac R, Crkvenjakov R. Origin of rat beta-globin haplotypes containing three and five genes. Mol Biol Evol. 1990;7:407–422. doi: 10.1093/oxfordjournals.molbev.a040616. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2nd edition Academic Press; San Diego: 2001. [Google Scholar]
- Riley RS, Ben-Ezra JM, Goel R, Tidwell A. Reticulocytes and reticulocyte enumeration. J Clin Lab Anal. 2001;15:267–294. doi: 10.1002/jcla.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Inokuchi N, Nagae Y, Okazaki T. Molecular cloning and characterization of two sets of alpha-theta genes in the rat alpha-like globin gene cluster. Gene. 1999;230:91–99. doi: 10.1016/s0378-1119(99)00055-4. [DOI] [PubMed] [Google Scholar]
- Setton-Avruj CP, Musolino PL, Salis C, Allo M, Bizzozero O, Villar MJ, Soto EF, Pasquini JM. Presence of alpha-globin mRNA and migration of bone marrow cells after sciatic nerve injury suggests their participation in the degeneration/regeneration process. Exp Neurol. 2007;203:568–578. doi: 10.1016/j.expneurol.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Testa CM, Seo BB, Richardson JR, Kim JH, Miller GW, Yagi T, Matsuno-Yagi A, Greenamyre JT. Mechanism of toxicity in rotenone models of Parkinson's disease. J Neurosci. 2003;23:10756–10764. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemmon JR, Hughes CM, Campbell GA, Flood DG. Increased levels of hemoglobin-derived and other peptides in Alzheimer's disease cerebellum. J Neurosci. 1994;14:2225–2235. doi: 10.1523/JNEUROSCI.14-04-02225.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroka DM, Burkhardt T, Desbaillets I, Wenger RH, Neil DA, Bauer C, Gassmann M, Candinas D. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 2001;15:2445–2453. doi: 10.1096/fj.01-0125com. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Takeda Y, Ikuta T. Immunoblotting conditions for human hemoglobin chains. Anal Biochem. 2008;378:218–220. doi: 10.1016/j.ab.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiftsoglou AS, Tsamadou AI, Papadopoulou LC. Heme as key regulator of major mammalian cellular functions: molecular, cellular, and pharmacological aspects. Pharmacol Ther. 2006;111:327–345. doi: 10.1016/j.pharmthera.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Valdes R, Jr, Ackers GK. Self-association of hemoglobin betaSH chains is linked to oxygenation. Proc Natl Acad Sci USA. 1978;75:311–314. doi: 10.1073/pnas.75.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon HP, Wardan H, Vadolas J. siRNA-mediated reduction of alpha-globin results in phenotypic improvements in beta-thalassemic cells. Haematologica. 2008;93:1238–1242. doi: 10.3324/haematol.12555. [DOI] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MT, Reeder BJ. Oxygen-binding haem proteins. Exp Physiol. 2008;93:128–132. doi: 10.1113/expphysiol.2007.039735. [DOI] [PubMed] [Google Scholar]
- Wittenberg JB, Wittenberg BA. Mechanisms of cytoplasmic hemoglobin and myoglobin function. Annu Rev Biophys Biophys Chem. 1990;19:217–241. doi: 10.1146/annurev.bb.19.060190.001245. [DOI] [PubMed] [Google Scholar]
- Wride MA, Mansergh FC, Adams S, Everitt R, Minnema SE, Rancourt DE, Evans MJ. Expression profiling and gene discovery in the mouse lens. Mol Vis. 2003;9:360–396. [PubMed] [Google Scholar]
- Wu X, Dewey TG. Cluster analysis of dynamic parameters of gene expression. J Bioinform Comput Biol. 2003;1:447–458. doi: 10.1142/s0219720003000307. [DOI] [PubMed] [Google Scholar]
- Zhu C, Vourc'h P, Fernagut PO, Fleming SM, Lacan S, Dicarlo CD, Seaman RL, Chesselet MF. Variable effects of chronic subcutaneous administration of rotenone on striatal histology. J Comp Neurol. 2004;478:418–426. doi: 10.1002/cne.20305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup Fig 1
Sup Fig 2
Sup material