GCView: the genomic context viewer for protein homology searches (original) (raw)
Abstract
Genomic neighborhood can provide important insights into evolution and function of a protein or gene. When looking at operons, changes in operon structure and composition can only be revealed by looking at the operon as a whole. To facilitate the analysis of the genomic context of a query in multiple organisms we have developed Genomic Context Viewer (GCView). GCView accepts results from one or multiple protein homology searches such as BLASTp as input. For each hit, the neighboring protein-coding genes are extracted, the regions of homology are labeled for each input and the results are presented as a clear, interactive graphical output. It is also possible to add more searches to iteratively refine the output. GCView groups outputs by the hits for different proteins. This allows for easy comparison of different operon compositions and structures. The tool is embedded in the framework of the Bioinformatics Toolkit of the Max-Planck Institute for Developmental Biology (MPI Toolkit). Job results from the homology search tools inside the MPI Toolkit can be forwarded to GCView and results can be subsequently analyzed by sequence analysis tools. Results are stored online, allowing for later reinspection. GCView is freely available at http://toolkit.tuebingen.mpg.de/gcview.
INTRODUCTION
In bacterial and archaeal genomes, about one half of all protein-coding genes are organized into operons. (1). But even for the other half, conservation of the genomic context i.e. the genes upstream and downstream on the chromosome, is observable between related species (2). The genomic context can provide important information about duplication, insertion, translocation or deletion events. While the past decades have equipped scientists with a broad range of excellent bioinformatics tools for analysis and comparison of single protein sequences, taking a step back and looking at the bigger genomic picture and comparing it between different organisms is still largely manual work. For many well annotated proteins and operons, databases like BioCyc (3), STRING (4), The SEED (5) or Ensembl Bacteria (6) can provide important information. However, looking beyond the content of those databases to extend the search into more genomes or investigating less well-characterized proteins can be challenging.
GCView, the Genomic Context Viewer for protein homology searches aims to ease and automate the manual process of extracting and comparing genomic regions of interest. It is integrated into the Bioinformatics Toolkit of the Max-Planck Institute for Developmental Biology (MPI Toolkit) (7) and can be accessed through a user-friendly web interface at http://toolkit.tuebingen.mpg.de/gcview. This website is free and open to all users and there is no login requirement.
GCView uses protein homology to assign corresponding genes. The underlying homology information is taken from standard protein homology search tools like BLASTp or PSI-BLAST (8). In contrast to the above mentioned databases such as STRING, the homology searches are not precomputed, giving the user full control over and insight into the processes leading to the final result.
GCView can integrate multiple searches (e.g. one for each component of an operon) and compile a comprehensive overview of the combinatorial variants found in different genomes. Genomes featuring the same number and order of genes of interest are grouped together.
The results can be mapped onto a taxonomy tree for a quick overview of the distribution of operon structures throughout all sequenced procaryotic organisms.
The output is a series of images showing the genomic regions that contain the genes of interest. Additionally, for each image a list of the encoded proteins is provided that contains additional information such as descriptions and database links. Hits from the underlying searches are colored in the output for easy identification.
The integration into the MPI Toolkit allows users to run homology search jobs independent of GCView, providing maximum control over the input parameters, and then to internally forward the results to GCView for integration. Consequently, the results from GCView can also be forwarded to other specialized tools for a more detailed analysis of subsets of proteins or genes. All results are stored on the server for 2 weeks and can be revisited and reviewed at a later time point. It is possible to create an account on the MPI Toolkit, which allows jobs to be bound to the account and saved for extended periods of time.
FUNCTIONALITY
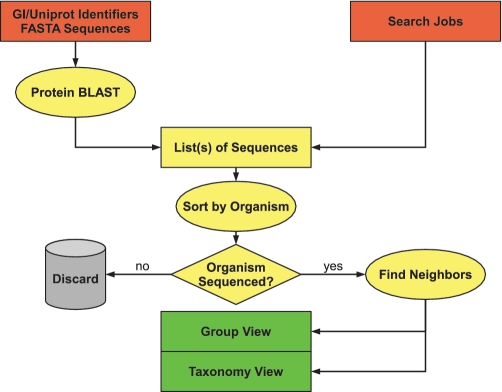
The design goal for GCView was to provide a quick and accurate overview of the combinatorial variants of operons in different genomes based on well established homology search methods accessible through a user-friendly straightforward web interface. The workflow of the tool is summarized in Figure 1.
Figure 1.
GCView workflow. Input: red; processing: yellow and results: green.
Input
GCView accepts several different types of input: FASTA protein sequences, protein GI or UniProt identifiers and forwarded homology search jobs. Currently GCView is limited to protein homology searches or protein sequences as input, mostly due to the higher sensitivity of protein searches compared to DNA searches. The inclusion of DNA searches (BLASTn) is planned for a future version. It is possible to use not only full protein sequences, but also single domains as query for the search. Genes containing multiple domains will be labeled accordingly in the output.
Primarily, homology search jobs can be forwarded to GCView within the MPI Toolkit. If, alternatively, FASTA sequences or protein identifiers are provided, GCView internally executes a PSI-BLAST run for each sequence or identifier provided and analyzes the results. Additional input parameters are the size of the genomic region to be displayed and the _E_-value cutoffs for the results to be included in the output. The size of the genomic region is interpreted as the number of genes to be extracted before the first hit and after the last hit in any genome.
Note that the quality of the GCView results strongly depends on the underlying homology search being exhaustive, i.e. containing results at least up to the _E_-value cutoff specified for GCView. This is especially important in Group View: only exhaustive searches lead to a maximum of labeled operon components. Operons with unlabeled components lead to additional groups, which would not be observed after an exhaustive search. For the same reason, caution is advised when using BLAST databases prefiltered at a certain sequence similarity cutoff.
For technical reasons, it is only possible to use BLAST databases, which contain GI or UniProt identifiers. Using a database which does not provide appropriate identifiers in the output will not give any results in GCView.
Processing
From each input homology search, a list of protein GI numbers is extracted along with the exact region and degree of similarity. The lists are filtered for proteins with _E_-values below the threshold specified in the input and for proteins from organisms which have not been fully sequenced. The backend database of sequenced genome data is built from the genomes found in NCBI GenBank (9) (ftp://ftp.ncbi.nih.gov/genomes/Bacteria) and comprises fully sequenced bacterial and archaeal genomes.
For each hit the genes upstream and downstream of the hit are extracted from the database, resulting in one genome chunk for each hit. The number of genes extracted depends on the range set in the input parameters. Overlapping regions from the same genome are subsequently merged. This implies that an operon which has been duplicated in a genome can show up as one or two chunks, depending on the distance between the duplicates and the range settings. After merging, the resulting regions are grouped by the number and order of genes of interest.
OUTPUT
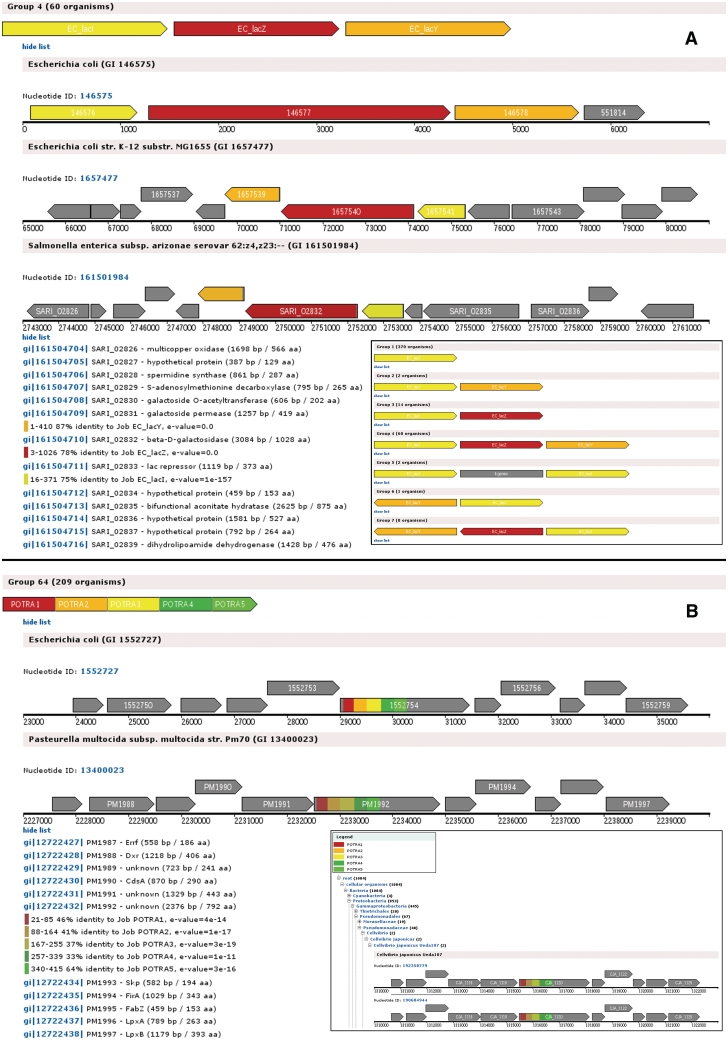
GCView generates two different views for the results: the Group View and the Taxonomy View. Both views contain the same information the difference is in the sorting. Figure 2 shows example outputs for both views for two different runs of GCView.
Figure 2.
Example output. (A) Using GCView to look at different operon components. The lac Operon (Demo Data) is shown in Group View with one group expanded. Insert: Group View Overview for the same run. (B) Using GCView to look at single domains in different contexts. POTRA domains from Omp85 and related proteins (10) in different organisms shown in Group View. Insert: Taxonomy View for the same run.
The Group View presents an overview of the results. A group comprises all organisms which contain a specific number and order of the genes of interest.
A schematic image of each group summarizes which of the genes of interest can be found in the group and in which order they appear in the genome. Each query gene is represented by a colored arrow. The colors are explained in the legend, which is displayed on the top of the page. Additionally, the identifier of the input query is indicated on each arrow. The arrows in the Group View are not to scale and the colors do not indicate the degree of identity between query and hit sequences. Fused arrows indicate that multiple query sequences were mapped onto one gene.
Gray boxes represent one or multiple genes that are not homologous to any of the query sequences but located between genes of interest. A number indicates how many genes are represented by the corresponding box. The groups can be expanded to view the detailed genomic context for each organism in the respective group.
The Taxonomy View maps all results onto a taxonomy tree. The numbers next to the organism names represent the number of hits in this taxon and its sub-groups. Branches of the tree can be collapsed or expanded as required. The detailed information for each hit can be viewed at the leaves of the tree.
The detail representation of every genomic region is identical in both views. Each representation contains a genome ID, indicating the nucleotide GI number of the genome from which the corresponding region was extracted. In the case that genes of interest are located in several non-overlapping regions of the same genome (e.g. due to operon duplication), multiple representations with the same nucleotide ID are shown, one for each region.
A schematic image of the region shows the genetic neighborhood of the genes of interest. Protein-coding genes are shown as arrows. Regions of homology to the genes of interest are highlighted in the corresponding colors, which are indicated in the legend. In contrast to the Group View, the intensity of the color corresponds to the identity score of the hit and the arrow length correlates with the length of the gene. Please note that the scale may differ between different images. The ruler at the bottom of each image shows the position in the genome. Each section of the ruler corresponds to 1000 bp. Various details for each gene (description, precise location, length, distance to neighboring genes) can be viewed by hovering the mouse over the arrows.
Clicking on an arrow expands a detailed list of the genes in the image and the search hits therein. The selected gene is highlighted in the list. A clipboard widget located in the right corner of the screen can be used to pick genes from the output. These genes can be forwarded to sequence retrieval tools for further analysis or used in another GCView run for an iterative expansion of the set of analyzed genes.
CONCLUSIONS
We present GCView, an interactive web tool for automated retrieval and comparison of the genomic context of protein-coding genes. The underlying homology searches use protein sequences instead of DNA for higher sensitivity. Compared to classical databases like The SEED or BioCyc, the advantages of GCView are: (i) a greater focus on the query, as only the homologs of the input proteins are highlighted, and the degree of similarity is easily visible from the output; (ii) interactivity, as the query can iteratively be extended by more proteins of interest; (iii) transparency, as the user can have full control over the parameters of the underlying homology search; and (iv) flexibility, as e.g. single domains can be used as query, revealing different domain contexts. GCView is embedded into the MPI Toolkit, which allows users to save their GCView runs for later reinspection and directly analyze the genes found by GCView using a broad range of sequence and structure analysis tools.
FUNDING
Funding for the project as well as for open access charge: Departmental funding of the Max Planck Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank the people involved in the maintenance of the MPI Toolkit, especially Christina Wassermann, Vikram Alva, and Andé Noll, and furthermore Andrei Lupas for continuing support.
REFERENCES
- 1.Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLoS Genet. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nature Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 3.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold T, Zeth K, Linke D. Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition. J. Biol. Chem. 2010;285:18003–18015. doi: 10.1074/jbc.M110.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]