Par4 is a coactivator for a splice isoform–specific transcriptional activation domain in WT1 (original) (raw)
Abstract
The Wilms' tumor suppressor protein WT1 is a transcriptional regulator involved in differentiation and the regulation of cell growth. WT1 is subject to alternative splicing, one isoform including a 17–amino acid region that is specific to mammals. The function of this 17–amino acid insertion is not clear, however. Here, we describe a transcriptional activation domain in WT1 that is specific to the WT1 splice isoform that contains the 17–amino acid insertion. We show that the function of this domain in transcriptional activation is dependent on a specific interaction with the prostate apoptosis response factor par4. A mutation in WT1 found in Wilms' tumor disturbs the interaction with par4 and disrupts the function of the activation domain. Analysis of WT1 derivatives in cells treated to induce par4 expression showed a strong correlation between the transcription function of the WT1 17–amino acid insertion and the ability of WT1 to regulate cell survival and proliferation. Our results provide a molecular mechanism by which alternative splicing of WT1 can regulate cell growth in development and disease.
Keywords: WT1, par4, transcription, coactivator
Wilms' tumor is a pediatric malignancy of the kidneys that affects 1 in 10,000 children, making it the most common solid tumor in the young (for review, see Reddy and Licht 1996; Little et al. 1999). The Wilms' tumor suppressor protein, WT1, was cloned as a factor involved in the formation of Wilms' tumor, in which it is mutated in 10%–15% of cases. Wilms' tumor also is associated with a group of rare syndromes (Denys-Drash syndrome, Frasier syndrome, and WAGR syndrome), in which WT1 dysfunction appears to play a major role (for review, see Reddy and Licht 1996; Little et al. 1999).
WT1 initially was shown to be a transcription factor that is able to repress transcription of genes that are abnormally upregulated in Wilms' tumors, providing a molecular basis for the critical function of WT1 in this disease (Madden et al. 1991; Drummond et al. 1992; Gashler et al. 1992; Wang et al. 1992; Dey et al. 1994). Subsequent studies show that WT1 also can activate transcription in some instances (Reddy et al. 1993; Wang et al. 1993; Cook et al. 1996; Englert et al. 1997). Indeed, more recent studies indicate that activation of transcription by WT1 is likely to be the critical function of this tumor suppressor (English and Licht 1999). Only a few genes appear to be activated by WT1, suggesting a specific cellular context may be required to manifest its function as a transcriptional activator (Thate et al. 1998; Lee et al. 1999). Consistent with this, the WT1 transcriptional activation domain can be regulated by interactions with several other factors (Johnstone et al. 1996, 1998; Maheswaran et al. 1998a,b; McKay et al. 1999).
WT1 is subject to alternative splicing in two regions (for review, see Reddy and Licht 1996; Little et al. 1999). The best characterized alternative splice involves the insertion of the three amino acids, lysine-threonine-serine (KTS between the third and fourth zinc fingers of the DNA-binding domain at the C terminus of WT1. The inclusion of these amino acids drastically reduces the affinity of WT1 for a specific DNA sequence, and instead this isoform binds preferentially to RNA. Moreover, the +KTS, but not −KTS isoform of WT1, associates with the spliceosome and in immunofluorescence analysis shows a speckled pattern colocalizing with splicing factors (Larsson et al. 1995; Caricasole et al. 1996; Davies et al. 1998; Ladomery et al. 1999). Thus, it appears that WT1 may play a role in both transcription and RNA processing in a splice isoform–dependent manner (Englert 1998; Davies et al. 1999; Little et al. 1999).
The second alternative splice inserts 17 amino acids (17AAs) between the WT1 transcriptional activation domain and the zinc finger region. This splice variant, unlike the KTS insertion, is only found in mammalian WT1 (Kent et al. 1995). Studies of the effect of the +17AA form of WT1 have shown variable effects on the transcriptional function of WT1. In some cell types, it augments the transcriptional activation domain (Reddy et al. 1995; Moorwood et al. 1999), and in others it has been shown to constitute an independent transcriptional repression domain (Wang et al. 1995). The 17AA insertion of WT1 also has been linked to both the regulation of the cell cycle and apoptosis, although the molecular mechanisms that are involved are not clear (for review, see Reddy and Licht 1996).
Recent studies have shown that the relative level of the WT1 isoforms can vary during both development and in disease. For example, Frasier syndrome results from an imbalance of the +KTS/−KTS ratio (Barbaux et al. 1997). The relative level of the WT1 +17AA isoform also can be elevated in Wilms' tumors and leukemia (Pritchard-Jones and King-Underwood 1997; Renshaw et al. 1997; Iben and Royer-Pokora 1999). In addition, the +17AA variant is differentially expressed throughout development (Renshaw et al. 1997; Iben and Royer-Pokora 1999).
As mentioned above, the contribution of the 17AA alternative splice and surrounding region to the transcriptional function of WT1 is not clear. In this study, we set out to directly assess the effects of this region on transcription and how it is affected by the 17AA insertion. We show that this domain constitutes an independent transcriptional activation domain that requires the 17AA insert for maximal activity. The function of this activation domain was context specific in that it required the prostate apoptosis response protein 4 (par4) for its function. This was due to a direct interaction between this domain and par4. Underpinning the crucial function of this domain to normal WT1 function, we found that a mutation within the 17AA region isolated from a Wilms' tumor specimen failed to interact with par4 and consequently failed to activate transcription. Our results show that par4 can act as a coactivator for a splice isoform–specific transcriptional activation domain that plays a role in regulating cell growth.
Results
The 17AA alternative splice isoform of WT1 forms a transcriptional activation domain
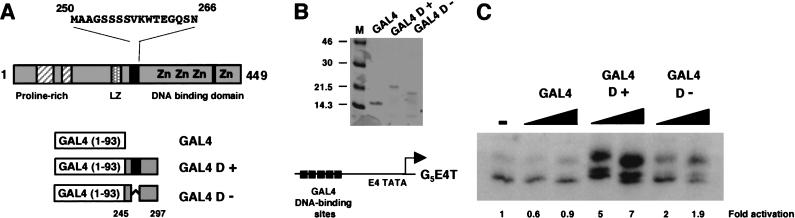
We sought to directly determine the transcriptional function of the region between the previously described transcriptional activation domain and the DNA-binding domain of WT1 (designated D domain, residues 245–297), which contains the 17AA alternative splice site (Fig. 1A). This region either lacking (D−) or containing (D+) the 17AA was fused to the GAL4 DNA-binding domain (residues 1–93), expressed in and purified from Escherichia coli (Fig. 1B). These proteins and a control GAL4 were used in an in vitro transcription assay with HeLa cell nuclear extract by using the adenovirus E4 (AdE4) promoter downstream from five GAL4 DNA-binding sites (G5E4T; Fig. 1C). The GAL4 DNA-binding domain alone had no effect on transcription. GAL4 D− activated transcription weakly, but with GAL4 D+ we observed a significantly greater level of transcriptional activation. Thus, WT1 contains an additional activation domain (D domain) that is dependent on the presence of the 17AA alternative splice for maximal activity.
Figure 1.
A splice isoform–specific transcriptional activation domain in WT1. (A) Diagram of WT1 showing the DNA-binding domain, proline-rich region, and potential leucine zipper (LZ). The two alternate splice sites are shown in black: the 17AA alternative splice is shown with the amino acid sequence and is located between the putative leucine zipper and DNA-binding domain (designated D domain); the second alternative splice (amino acid sequence KTS) is located between zinc fingers three and four. The region of WT1 fused to GAL4 (1–93) are shown below (amino acids 245–297). These fusions included the +17AA form (GAL4 D+) and the –17AA form (GAL4 D−). (B) Analysis of purified GAL4 (1–93), GAL4 D+, and GAL4 D− by SDS-PAGE and Coomassie staining. Molecular weight markers (kD) are shown at left. (C) In vitro transcription assay using 200 and 400 ng of each GAL4 fusion protein with Hela cell nuclear extract and the G5E4T promoter (shown above). Transcripts were analyzed by primer extension and denaturing electrophoresis. (−)no addition of a GAL4 derivative. The signals were quantified by phosphorimager analysis and are presented as fold activation compared to the basal level of transcription (−).
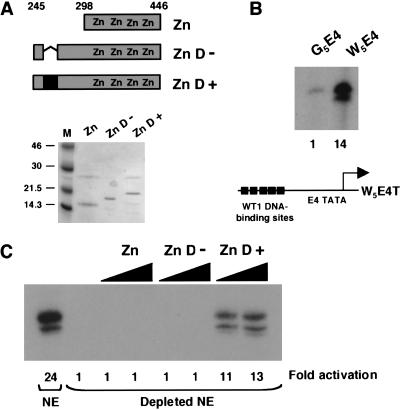
We next sought to obtain evidence that the 17AA region of WT1 can function as a transcriptional activation domain in the context of the natural WT1 DNA-binding domain. WT1 residues 245–446 either containing or lacking the 17AA alternative splice and the DNA-binding domain alone (residues 295–446) were expressed in and purified from E. coli (Fig. 2A). A transcription reporter DNA template was constructed containing five consensus WT1 DNA-binding sites upstream of the AdE4 promoter (W5E4T; Fig. 2B). Compared to G5E4T, W5E4T proved to be highly active in transcription assays with HeLa nuclear extract, indicating the presence of factors in the HeLa cell nuclear extract that bind to this site and activate transcription (Fig. 2B). We therefore fractionated HeLa nuclear extract over a column containing concatenated immobilized WT1 DNA-binding sites. This “depleted” HeLa nuclear extract showed a significantly reduced background transcription level (Fig. 2C). Addition of the recombinant WT1 derivatives to this extract with the W5E4T reporter shows clearly that the +17AA isoform of WT1 activated transcription, but the DNA-binding domain alone or the version lacking the 17AAs did not. Importantly, all of the WT1 derivatives interact with a WT1 DNA-binding site with equivalent affinity (data not shown). Thus, the +17AA insertion of WT1 bestows a transcriptional activation function both as a GAL4 fusion and in the context of the natural WT1 DNA-binding domain.
Figure 2.
The 17AA transcriptional activation region functions in the context of the natural WT1 DNA-binding domain. (A) Diagram indicating the regions of WT1 expressed as recombinant His-tagged proteins. The 17AA alternative splice is indicated in black fill. Recombinant proteins were analyzed by SDS-PAGE and Coomassie staining (M, molecular weight markers in kD, are shown at left). (B) In vitro transcription assay using the W5E4T (containing 5 × WT1 DNA-binding sites) and G5E4T promoters. Transcripts were detected by primer extension. Transcription levels (quantified by phosphorimager analysis) are presented relative to G5E4T. (C) In vitro transcription assays were performed and analyzed as for part B except that 200 and 400 ng of each recombinant His-tagged protein was added. A HeLa nuclear extract (NE) or HeLa nuclear extract that had been fractionated over a column containing WT1 DNA-binding sites (Depleted NE) was used as indicated. Transcript levels were quantified by phosphorimager analysis and are presented as fold activation relative to the transcriptional activity of the depleted extract in the absence of a WT1 derivative.
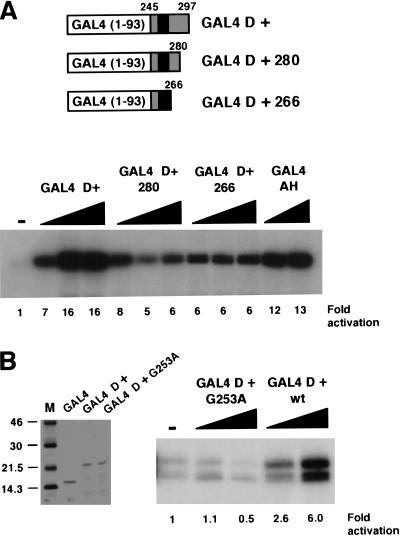
We next performed deletion mutagenesis to determine if the 17AAs alone were sufficient to activate transcription. Two C-terminal deletion mutants were constructed, one containing residues 245–280 and the other 245–266, and were expressed and purified as GAL4 fusion proteins (Fig. 3A). Analysis in transcription assays showed that GAL4 WT1 (245–266), which contains the 17AAs and five additional N-terminal residues, was sufficient for transcriptional activation (Fig. 3A). The entire D domain, however, was required for maximal transcriptional activation, indicating cooperation of the 17AAs with the remainder of the D domain. We therefore conclude that the WT1 17AA constitutes a splice isoform–specific transcriptional activation domain.
Figure 3.
The 17AA motif of WT1 is sufficient for transcriptional activation. (A) Deletion mutants of WT1 (245–297) were constructed as indicated. The purified proteins (200 and 400 ng) then were used in in vitro transcription assays with the G5E4T promoter and HeLa nuclear extract. (−) No recombinant protein was added, and fold activation is presented relative to this. (B) Recombinant His-tagged GAL4 fusions were purified and analyzed by SDS-PAGE and Coomassie staining. Molecular weight markers (kD) are shown at left. In vitro transcription assays were performed as for A.
A Wilms' tumor specimen has been reported that contains a mutation in WT1 (G253A), which is within the alternatively spliced 17AA (Schumacher et al. 1997). We produced this mutant as a GAL4 fusion protein and tested it in a transcription assay alongside GAL4 D+ (Fig. 3B). As before, GAL4 D+ elicited transcriptional activation. However, the GAL4 D+ derivative G253A failed to activate transcription. Thus, transcriptional activation by the 17AA alternative splice appears to be important for normal cellular function.
The 17AA WT1 activation domain is cell context specific
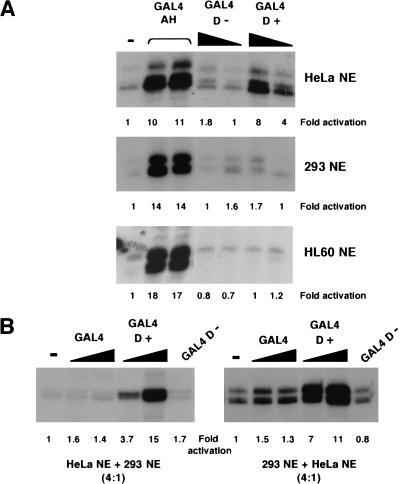
Our studies so far have shown that the 17AAs of WT1 form a transcriptional activation domain that functions both in the context of a GAL4 fusion, and importantly also in the context of the natural WT1 DNA-binding domain. We next determined if the 17AA activation domain of WT1 is cell type specific. We compared the transcriptional activity of GAL4 D+ and GAL4 D− in an embryonic kidney 293 and a HL60 nuclear extract alongside the HeLa nuclear extract from above (Fig. 4A). As before GAL4 D+, but not GAL4 D−, elicited strong transcriptional activation in the HeLa cell nuclear extract, as did the control synthetic activator GAL4– amphipathic helix (AH). However, in both the HL60 and 293 nuclear extracts, although GAL4 AH functioned similarly to that observed in HeLa nuclear extract, GAL4 D+ did not activate transcription. These results indicate a cell type–specific function of the WT1 17AA transcriptional activation domain that is due to either a positive-acting factor in the HeLa nuclear extract or a negative-acting factor in the 293 and HL60 nuclear extracts. To distinguish between these possibilities, we performed the following experiment; first, if the 293 nuclear extract contains a negative-acting factor, then the addition of 293 nuclear extract to a HeLa nuclear extract should reduce the level of transcriptional activation by GAL4 D+. However, this was not the case (Fig. 4B, left). Alternatively, if the HeLa nuclear extract contains a positive-acting factor, then the addition of HeLa nuclear extract to a 293 nuclear extract should result in the conditions required to support transcriptional activation by GAL4 D+. We found that this was indeed the case (Fig. 3B, right). Taken together, we conclude that a critical target of the 17AA motif is likely to be a factor that is not part of the general transcription machinery and is a coactivator with specific function.
Figure 4.
The function of the 17AA activation domain is cell type specific. (A) Transcription assays were performed using 200 and 400 ng of recombinant His-tagged GAL4 fusions with the G5E4T promoter. Four hundred nanograms of the synthetic activator GAL4 AH was added where indicated. Assays contained either HeLa, embryonic kidney 293, or HL60 nuclear extract. (−) No recombinant protein was added to the assay. Transcripts were detected as previously, quantified, and presented as fold activation relative to the basal level (−) for each nuclear extract. (B) Transcription assays were performed as in A with HeLa:293 nuclear extract (ratio of 4:1) at left and 293:HeLa nuclear extract (ratio of 4:1) at right. Fold activation was determined as in A. (NE) Nuclear extract.
The 17AA activation domain interacts with par4
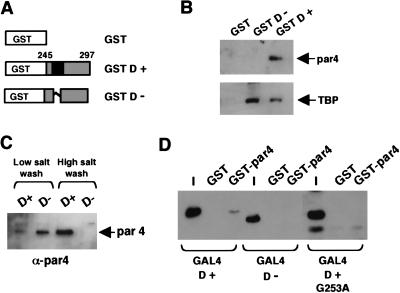
In a search for candidate factors that could perform this specific coactivator function, we first considered proteins that previously have been shown to associate with WT1 (for review, see Little et al. 1999). Protein affinity columns containing either GST, GST D+, or GST D− (Fig. 5A) were used to fractionate a HeLa nuclear extract; the columns were washed and the bound proteins were eluted. The eluates were resolved by SDS-PAGE and immunoblotted with antibodies against known WT1-interacting proteins. Neither E1b (which is present in 293 cells, but not HeLa cells), p53, nor Hsp70 were able to interact with the 17AA motif (data not shown). However, par4 (prostate apoptosis response factor 4), previously isolated in a yeast two hybrid screen by using WT1 as the bate (Johnstone et al. 1996), was able to associate with the 17AA motif of WT1 (Fig. 5B). Consecutive low- and high-salt washes showed that par4 interacts with the 17AA containing WT1 derivative with considerably greater affinity than the form lacking the 17AA (Fig. 5C). We also tested the ability of the GST D+ and GST D− columns to bind TFIID, a component of the general transcriptional machinery that has been proposed as a target of transcriptional activators (for review, see Roberts 2000). Unlike par4, TFIID from the HeLa nuclear extract was retained on both the GST D+ and GST D− columns (Fig. 5B). Thus, the D domain of WT1 can interact with both the general transcription factor TFIID and the prostate apotosis response factor par4. However, stable interaction with par4 is dependent on the 17AA insertion.
Figure 5.
Interaction partners for the WT1 17AA region. (A) Diagram of GST-WT1 fusion proteins (residues 245–297) either lacking (GST D−) or containing (GST D+) the 17AA motif. (B) Protein affinity chromatography of HeLa nuclear extract fractionated over a GST, GST D−, or GST D+ column. After extensive washing, the bound fraction was eluted and analyzed by immunoblottting with either anti-par4 (top) or anti-TBP (bottom) antibodies. (C) HeLa nuclear extract was fractionated over either a GST D− or a GSD D+ column, and after a brief low-salt wash (250 mM KCl) and a subsequent wash (1 M KCl), the presence of par4 was assessed in each fraction by immunoblotting with anti-par4 antibodies. (D) A GST pull-down assay was performed with immobilized GST-par4 and a bacterial lysate containing the recombinant GAL4 derivatives indicated. The bound fraction was analyzed by immunoblotting with anti-GAL4 antibodies. I is 10% of the input into each pull-down assay.
We next determined if the 17AA region of WT1 interacts directly with par4. A GST fusion of full-length par4 was purified from bacteria and immobilized on glutathione agarose beads. This then was incubated with bacterial lysate containing either GAL4 D+ or GAL4 D−. Stably bound proteins were eluted, subjected to SDS-PAGE, and immunoblotted with anti-GAL4 antisera (Fig. 5D). Consistent with the affinity chromatography data using HeLa nuclear extract, GAL4 D+ but not GAL4 D− was able to interact directly with GST-par4.
Our transcription experiments showed that the mutation G253A disrupted the function of the WT1 17AA transcriptional activation domain. We therefore tested the ability of GAL4 D+ (G253A) to interact with GST-par4 (Fig. 5D). The G253A mutant failed to interact with par4, providing strong evidence that the interaction between the 17AA activation domain of WT1 and par4 is required for transcriptional activation.
Par4 is a coactivator for the WT1 17AA activation domain
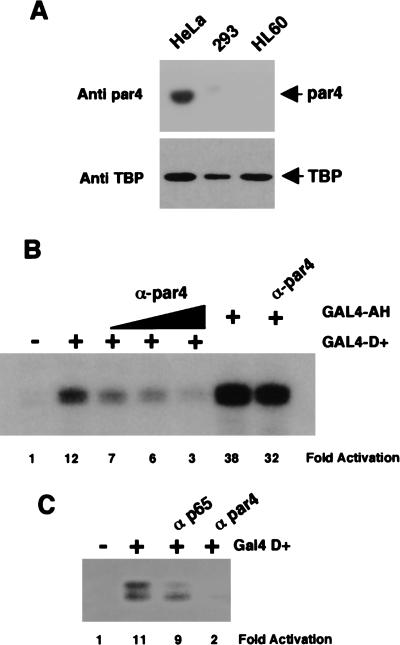
The transcription data of Figure 4 showed that the 17AA activation domain was functional in HeLa nuclear extracts, but not 293 or HL60 nuclear extracts. We therefore immunoblotted the HeLa, 293, and HL60 nuclear extracts with par4 antibodies to determine if the abundance of par4 in the nuclear extracts could explain this difference. As a control, we also immunoblotted the three extracts with antibodies against TATA-binding protein (TBP). Figure 6A shows that, although par4 was detected in the HeLa nuclear extract, the 293 and HL60 extracts contain negligible levels. In contrast, TBP was detected in all extracts.
Figure 6.
Par4 is required for transcriptional activation by WT1 17AA. (A) Anti-par4 and anti-TBP immunoblots of SDS-PAGE resolved HeLa, 293, and HL60 nuclear extracts (10 μg each). (B) In vitro transcription assay showing the effects of increasing amounts of anti-par4 antibodies (100, 200, and 400 ng) on transcriptional activation by GAL4 D+ (200 ng). (−) No recombinant protein was added to assay. GAL4 AH was included as a control, with 400 ng of anti-par4 antibody added where indicated. Assays were performed using HeLa nuclear extract and transcripts analyzed as before. (C) In vitro transcription assay showing that anti-par4 antibodies but not a nonrelevant rabbit antibody (anti-p65 subunit of NF-κB) inhibits transcriptional activation by GAL4 D+.
To determine if the presence of par4 in the HeLa nuclear extract was critical for the function of the 17AA activation domain, we tested the effect of anti-par4 antibodies on transcriptional activation by GAL4 D+ in a HeLa nuclear extract (Fig. 6, B and C). As before, GAL4 D+ activated transcription in the HeLa nuclear extract. The addition of anti-par4 antibodies to the HeLa nuclear extract resulted in the inhibition of transcriptional activation by GAL4 D+ in a concentration-dependent manner. Transcriptional activation by GAL4 D+ was not significantly affected when a control rabbit antibody was added to the HeLa nuclear extract (Fig. 6C). Significantly, we observed only a small reduction in the level of transcriptional activation by GAL4 AH when the highest concentration of anti-par4 antibody was present (Fig. 6B). Thus, in agreement with the ability of the WT1 17AA to interact with par4, par4 is specifically required for the function of the WT1 17AA transcriptional activation domain.
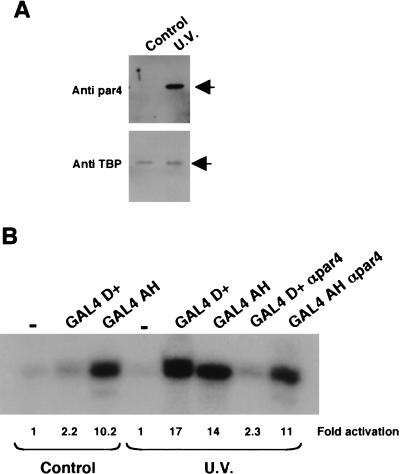
The HeLa nuclear extracts used in the studies above were purchased from a commercial source, whereas the 293 and HL60 nuclear extracts were prepared from cells grown in our laboratory. HeLa cells cultured in our own laboratory, under identical conditions to those used for the 293 cells, contained significantly lower levels of par4 (Fig. 7A). However, as expected, treatment of the HeLa cells with UV light induced par4, but the levels of TBP in the nuclear extracts remained constant. We therefore compared the ability of GAL4 D+ to elicit transcriptional activation in nuclear extracts derived from UV light–treated HeLa cells with untreated control HeLa cells (Fig. 7B). In the nuclear extracts derived from untreated HeLa cells, we observed a low level of transcriptional activation by GAL4 D+ but robust activation by GAL4 AH. In contrast, in nuclear extract made from UV light–treated HeLa cells, we observed a dramatic increase in the ability of GAL4 D+ to activate transcription, but GAL4 AH showed only a marginal increase in transcription potency. As we observed before, anti-par4 antibodies were able to inhibit the transcriptional activation elicited by GAL4 D+, but not by GAL4 AH. Thus, the 17AA activation domain of WT1 is only functional in nuclear extracts derived from cells that have been treated with proapoptotic stimuli to elevate nuclear par4 levels.
Figure 7.
Par4 expression is induced by UV irradiation and is directly linked to transcriptional activation by WT1 17AA. (A) Control and UV light–treated HeLa cells were used to prepare nuclear extract that was subsequently immunoblotted (10 μg per lane) with either anti-par4 (top) or anti-TBP (bottom) antibodies. (B) In vitro transcription assay showing the effect of UV light on the transcriptional activity of the GAL4 D+ fusion (200 ng) or GAL4 AH (200 ng) in assays using the G5E4T promoter. Anti-par4 antibodies (400 ng) were added as indicated. Transcripts were detected as before, quantified, and expressed relative to the basal level (−) in each extract.
Par4 mediates transcriptional activation by the WT1 17AA in vivo
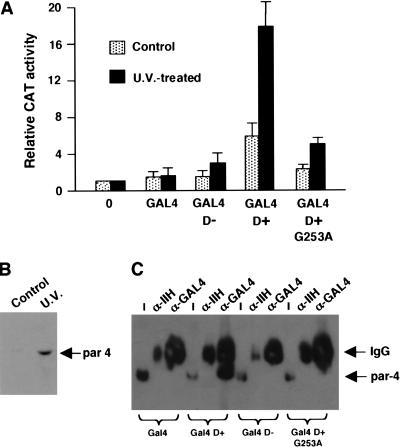
Our data so far indicate that the 17AA activation domain of WT1 can only function in cells that have been exposed to proapoptotic stimuli. We therefore sought to confirm our observations by transfection of plasmids expressing the GAL4 derivatives along with the G5E4CAT reporter into embryonic kidney 293 cells. Forty hours after transfection, the cells were treated with a lethal dose of UV radiation, and 6 h later, before the cells had lost adherence, CAT activity was measured. This activity was compared with that observed in control cells that were not treated with UV light and the results are presented graphically in Figure 8A. GAL4 D+ elicited only weak transcriptional activation in control cells. However, in the cells treated with UV light there was a significant increase in transcriptional activation mediated by GAL4 D+. GAL4 D− and the GAL4 D+ derivative G253A, however, showed a reduced ability to activate transcription in response to UV light treatment of the cells. As we had observed with HeLa cells, UV light treatment of 293 cells resulted in a significant increase in the level of par4 (Fig. 8B). These data agree well with our in vitro observations and confirm that the 17AA insertion of WT1 is a regulated transcriptional activation domain that requires par4 for its activity.
Figure 8.
WT1 17AA is a UV light responsive transcriptional activation domain in vivo. (A) Plasmids expressing GAL4, GAL4 D−, GAL4 D+, and GAL4 D+ G253A (4 μg each) were transfected into embryonic kidney 293 cells along with the reporter G5E4CAT (1 μg). Six hours after UV light treatment of the cells, CAT activity was measured and is presented relative to the level of CAT activity with empty pCDNA3 vector alone (0 lane). The results are the mean average of four independent experiments. (B) Immunoblot with anti-par4 antibodies showing that the level of par4 is elevated in 293 cells that have been treated with UV light. (C) Coimmunoprecipitation of par4 with the WT1 17AA motif. 293 cells were transfected with 4μg of each GAL4 derivative and after UV light treatment nuclear extracts were prepared. Immunoprecipitation was performed with either anti-GAL4 or anti-TFIIH antibodies and complexes harvested with protein G Sepharose. Par4 content was assessed by immunoblotting. Input (I) is 1% of the amount of nuclear extract used in each immunoprecipitation.
We next sought to determine if, after the induction of par4 in 293 cells by UV light there was a resulting association with the WT1 17AA motif. Cells were transfected with the GAL4 WT1 derivatives as in Figure 8A, and after UV light treatment of the cells, nuclear extract was prepared. The GAL4 WT1 derivatives were immunoprecipitated with anti-GAL4 antibodies, or control anti-TFIIH antibodies, and the association of par4 assessed by immunoblotting with anti-par4 antibodies (Fig. 8C). Par4 immunoprecipitated with GAL4 D+, but not with GAL4, GAL4 D−, or the point mutant GAL4 D+ G253A. Thus, the 17AA motif of WT1 complexes with par4 in UV light–treated 293 cells, consistent with the transcription effects that we observed.
The 17AA containing isoform of WT1 regulates the response to proapoptotic signals
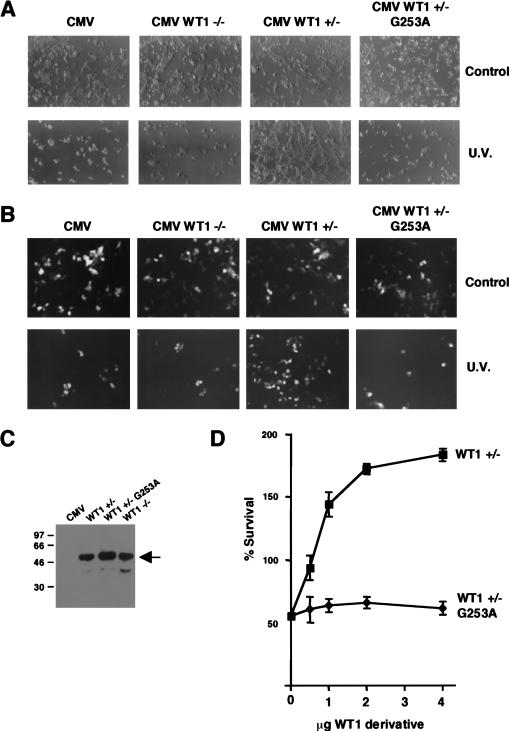
The in vitro and in vivo transcription studies have shown that the WT1 17AA activation domain is only functional under proapoptotic conditions. We therefore sought to determine the effect of the WT1 17AA on the cellular response to lethal UV light treatment. 293 cells, in which we routinely obtain a very high transfection efficiency, were transfected with full-length WT1 derivatives followed by lethal UV light treatment. Cells were transfected with either empty CMV expression vector or the same expression vector driving the expression of either WT1 −/− (lacking the 17AA insertion) or WT1 +/− (containing the 17AA insertion). In untreated transfected cells, we did not observe any phenotypic differences that resulted from the presence of either of the two WT1 isoforms (Fig. 9A). However, 16 h after UV light treatment, the cells transfected with the control CMV vector died, as did the cells transfected with CMV WT1 −/−. Strikingly, the cells transfected with CMV +/− not only remained viable, but reproducibly showed proliferation. Thus, the 17AA insertion confers a survival function to WT1 in UV light–treated 293 cells. Significantly, the WT1 +/− derivative G253A failed to rescue the UV light–treated 293 cells from death. Comparable results were obtained when we treated the 293 cells with the proapoptotic drug etoposide (data not shown). We next performed the same experiment as above, but cotransfected green fluorescent protein so that only the transfected cells would be observed under fluorescence (Fig. 9B). As before, WT1 +/− significantly increased the number of transfected cells that survived UV light treatment, but the WT1 −/− isoform or the WT1 +/− mutant G253A did not. Immunoblotting showed the equivalent expression of all the WT1 isoforms (Fig. 9C). To confirm and extend these data, we tested the dose dependence of the survival/proliferation effects with wild-type WT1 +/− and the mutant G253A. After UV light treatment, cells were counted and are plotted as a percentage of the cell number in a control experiment in which empty CMV expression vector was transfected. Increasing, levels of WT1 +/− elicited a dose-dependent cell survival effect after UV treatment of the 293 cells. In addition, transfection of WT1 +/− resulted in almost double the number of cells compared to control cells that had not undergone UV light treatment. In contrast, the WT1 +/− mutant G253A had no effect at any of the levels of expression plasmid. The WT1 −/− isoform also had no effect in a dose-dependence assay (data not shown). Taken together, these results show that WT1 +/− confers a cell survival/proliferation effect in cells that have been subject to lethal UV light treatment. Importantly, these data parallel both the transcription function of the WT1 derivatives and their ability to interact with par4. Thus, par4 acts as a coactivator for WT1 +17AA that is likely to lead to a transcription cascade to prevent apoptosis.
Figure 9.
The WT1 17AA motif confers a survival function in cells exposed to UV light. (A) Embryonic kidney 293 cells were transfected with either empty CMV expression vector or full-length WT1 (−/−), WT1 ( +/−), or WT1 (+/−) G253A under the control of a CMV promoter (4 μg in each case). Sixteen hours after UV light treatment (where indicated), the cells were photographed. (B) Cells were transfected as in A, but along with 1 μg of an expression vector containing green fluorescent protein. Cells were photographed under fluorescence to visualize only the transfected cells. (C) Immunoblot with anti-WT1 antibodies showing that the WT1 +/−, −/−, and +/− G253A derivatives are expressed at equivalent levels in 293 cells. (D) Survival curves to quantify the effect of WT1 +/− and the mutant G253 on cell number after UV light treatment. Each WT1 derivative (0.5, 1, 2, and 4 μg; balanced with empty CMV vector) was transfected into 293 cells, and 16 h after UV light treatment the cells were harvested from the plate and counted. Percentage survival is relative to a control plate of 293 cells transfected with CMV, but not subject to UV light treatment. Each point is the mean average of three independent transfection experiments with standard deviation.
Discussion
In this study, we have shown that the 17AA alternative splice of WT1 constitutes a transcriptional activation region both in the context of the natural WT1 DNA-binding domain and as a GAL4 fusion protein. The function of the WT1 17AA activation domain required a direct interaction with the prostate apoptosis response factor par4 and correlated with the rescue of cells from lethal UV light treatment. Our data shed light on the cell context–specific transcription function of WT1 and provide a transcription-mediated mechanism for the effects of WT1 on cell growth and survival.
The 17AA alternative splice isoform of WT1 previously has been shown to have effects on both cell proliferation and apoptosis (Englert et al. 1995; Johnstone et al. 1996; Menke et al. 1997; Murata et al. 1997; Mayo et al. 1999). However, the mechanisms underlying these effects have not been clear with conflicting results obtained in different cell lines and under different conditions. In addition, previous studies of the WT1 17AA have reported both transcriptional activation and repression function (Reddy et al. 1995; Wang et al. 1995; Moorwood et al. 1999). Our present data provide a framework to understand how different cellular contexts can determine the transcriptional function of the 17AA motif. Specifically, under resting conditions, WT1 +17AA is transcriptionally inactive or perhaps may act as a transcriptional repressor. However, under conditions in which nuclear par4 levels are elevated, WT1 +17AA converts to a transcriptional activator. Indeed, it is also possible that other aspects can affect this mechanism, such as cell type and promoter context.
A central function of WT1 in kidney development is in the metanephric mesenchyme (MM) to epithelium transition leading to the formation of the nephron (Reddy and Licht 1996). In the WT1 knockout mouse, the MM apoptoses, suggesting that WT1 has a survival role in these cells (Kreidberg et al. 1993). Interestingly, analysis of the relative level of WT1 splice variants in the human kidney has shown that the ratio of the +17AA form increases throughout fetal development (Renshaw et al. 1997; Iben and Royer-Pokora 1999). It therefore will be interesting to determine if the +17AA splice variant is responsible for the prevention of MM apoptosis. Alterations in the ratio of WT1 splice variants also have been observed in human tumors. Significantly, the +17AA isoform of WT1 is overrepresented in at least some Wilms' tumors and also in leukemia (Pritchard-Jones and King-Underwood 1997; Renshaw et al. 1997; Iben and Royer-Pokora 1999). The results we present here provide a direct role for the WT1 +17AA in circumventing proapoptotic signals, which is entirely consistent with tumorogenesis. Taken together, these observations add weight to an oncogenic role for WT1 in addition to its better defined function as a tumor suppressor. As only 10%–15% of Wilms' tumors contain mutant WT1, it is possible that aberrant levels of the +17AA isoform contribute to oncogenesis in at least some of the other cases.
Our studies showed that the G253A mutation isolated from a Wilms' tumor specimen disrupted the ability of the 17AA insertion to rescue cell death. This correlated with a defect in the ability of the mutant to interact with par4 and to activate transcription. The antisurvival effect of the mutant seems somewhat paradoxical and may reflect either cell type–specific effects or a more complex pathway to the formation of Wilms' tumor. The G253A mutation was in the germ line of the patient, but the wild-type allele was still observed in the DNA isolated from a nonmicrodissected tumor sample. If the wild-type allele is indeed present in the tumor and wild-type mRNA is produced, this would suggest that the WT1 17AA motif is dose dependent or that the G253A mutant acts in a dominant fashion. Importantly, this mutation has not been observed in any other Wilms' tumor patient (Schumacher et al. 1997; Diller et al. 1998; Jeanpierre et al. 1998). Furthermore, this mutation was not seen in 50 non-Wilms' tumor controls (data not shown) nor in any of the non-Wilms' tumor patients studied for WT1 alterations by several groups (Algar et al. 1996; King-Underwood and Pritchard-Jones 1998). Taken together, these results indicate that G253A is not a polymorphism. However, the role of this mutation in Wilms' tumorigenesis remains to be elucidated.
Par4 originally was cloned in a study to find genes that are specifically upregulated by proapoptotic signals in prostate cells (for review, see Rangnekar 1998). Par4 is ubiquitously expressed as RNA, but when cells are subjected to stimuli that activate the cell death pathways, par4 RNA is rapidly translated. Par4 subsequently is found in both the nucleus and cytoplasm and “primes” cells for apoptosis, making them more sensitive to further stimuli. At least part of this effect has been proposed to result from interactions with Bcl-2, caspases, and protein kinase C ζ. The original identification of par4 as a WT1-interacting protein proposed that par4 enhances transcriptional repression by WT1 (Johnstone et al. 1996). In addition, this previous study showed that the zinc fingers of WT1 mediated the interaction with par4. This contrast with our findings can be explained if there are two independent binding sites within WT1 for par4, the 17AA motif present in only specific splice isoforms of WT1 and the zinc finger motif. Perhaps par4 can mediate both positive and negative interactions with WT1 via these domains that lead to either transcriptional activation or repression. Indeed, other cofactors such as MDM2 have been shown to act as both a coactivator and corepressor (Martin et al. 1995). It also will be interesting to determine if par4 can act as a coactivator for other transcriptional regulatory proteins that exert a specific function under proapoptotic conditions. The existence of two distinct domains within a transcription factor capable of interacting with the same cofactor also is not without precedence. NF-κB has two binding surfaces for the coactivator CBP, the availability of which is regulated by NF-κB phosphorylation (Zhong et al. 1998). Also, thyroid hormone receptor β (THRβ) can interact with two independent domains within the general transcription factor TFIIB (Baniahmad et al. 1993). Interestingly, this ligand-dependent switch in interaction surface converts THRβ from a transcriptional repressor into an activator.
Our interaction data showed that both the + and −17AA isoforms of WT1 can interact with the general transcription factor TFIID. Thus, unlike the interaction with par4, the interaction with TFIID is not mediated by the +17AA motif. Consistent with this, although the 17AA motif elicited transcriptional activation in isolation, the entire D domain was required for full activity. How par4 functions as a coactivator remains to be determined. It is possible that par4 provides a bridging function for the 17AA with the RNA polymerase II holoenzyme. It is also possible that par4 mediates contact with other coactivator components of the transcription machinery. In this regard, it is noteworthy that the +17AA, but not the −17AA form of WT1 elutes as part of a 700-kD complex in gel filtration chromatography (Iben and Royer-Pokora 1999). It will be interesting to determine the other components of this complex and if indeed par4 is contained within the complex.
Recent DNA micoarray analysis to identify WT1 target factors failed to find genes that can be repressed by WT1 (Lee et al. 1999). However, a few genes were found to be activated. Such studies indicate that the WT1 transcriptional activation function is highly cell context specific. Our present work is certainly in agreement with this notion. Future experiments to analyze the transcriptional effects of the 17AA domain under proapoptotic conditions should yield valuable information about WT1 target genes and how this leads to WT1 +17AA-mediated rescue from cell death.
Materials and methods
Plasmids and DNA analysis
The G5E4T and G5E4CAT transcription reporter templates have been described previously (Hawkes and Roberts 1999). W5E4T was constructed by the insertion of five copies of a double stranded oligonucleotide (5′-AGCTCGGGTTGCGG GGGCGGGCCGG GGGAAGCTTGGT-3′) upstream of the adenovirus E4 core promoter in pGEM3 (Promega). The region of WT1 encoding residues 245–297 was amplified by PCR from plasmids containing full-length WT1 that either lacks, CMV WT1 (−/−), or contains, CMV WT1 ( +/−), the WT1 17AA alternative splice. The PCR products were cloned as _Bam_HI/_Eco_RI fragments into pRSETA (Invitrogen) containing DNA sequence encoding GAL4 (residues 1–93). Fragments then were cloned into pCDNA3 (Invitrogen) containing GAL4 (residues 1–93). Deletion mutants of the WT1 D domain were constructed by PCR amplification of the appropriate regions to produce _Bam_HI/_Eco_RI fragments. To produce the WT1 DNA-binding domain derivatives, we used PCR amplification, and the DNA fragments were cloned into pRSET A. The WT1 G253A mutation was constructed using the Quickchange site directed mutagenesis kit (Invitrogen). SSCP analysis of WT1 exon 5 was performed as described previously (Schumacher et al. 1997).
Cell culture and transfection
Human embryonic kidney 293 cells and HeLa cells were cultured as monolayers in Dulbecco's modified eagle medium (DMEM) containing 10% fetal calf serum, 5 mM L-glutamine, 100 mg/mL streptomycin, and 100 U/mL penicillin. HL60 cells were grown in flasks in RPMI medium containing 10% fetal calf serum, 5 mM glutamine, 100 mg/mL streptomycin, and 100 U/mL penicillin. 293 cells were transfected in 90-mm dishes at 50% confluency by using calcium phosphate as described previously (Hawkes and Roberts 1999). Forty hours after transfection, media was removed from plates, and the cells were exposed to 13 mJ of UV radiation. Fresh media then was applied to the plates, and the cells were left for another 6 h before harvesting. CAT assays were performed and quantified by phosphorimager analysis. HeLa cells were exposed to UV light as described for 293 cells, and nuclear extracts were prepared as described below. Cells were counted by hemocytometry. For Western blot, either whole cell or nuclear extract (where indicated in the figure legends) was subjected to SDS-PAGE. After electrophoresis, proteins were transferred to Immobilon P membrane (Millipore). Immunoblotting was performed with anti-par4 (R-334) and anti-WT1 (C-19) from Santa Cruz Biotechnology Inc., anti-GAL4 (Scottish Antibody Production Unit) or anti-TBP (Lin et al. 1991). Detection was via chemiluminescence (ECL, Amersham-Pharmacia). Santa Cruz Biotechnology Inc. anti-par4 (R-334), anti-GAL4 (sc-577), anti-TFIIH (sc-292), and anti-NF-κB p65 (sc-372) were used for immunoprecipiation.
In vitro transcription assays
HeLa nuclear extracts were either purchased from Computer Cell Culture Centre or prepared as described previously from cells grown in monolayer (Lee et al. 1988). Nuclear extracts were prepared from human embryonic kidney 293 and HL60 cells in an identical fashion. In vitro transcription assays were performed as described previously (Lin and Green 1991). 6His GAL4 fusion proteins and 6his-tagged WT1 derivatives were prepared as described (Reece et al. 1993). Where indicated, antibodies were added to nuclear extract on ice for 30 min before the transcription reaction.
Binding assays
GST fusion proteins were prepared as described previously (Lin and Green 1991). HeLa nuclear extract was dialyzed into buffer D (20 mM HEPES at pH 8.0, 20% [v/v] glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT, 0.2 mM PMSF). Two milliliters of dialyzed HeLa nuclear extract (10 mg/mL) was fractionated over columns containing 0.25 mL of glutathione agarose on which 100 μg of GST fusion protein was immobilized. The columns then were washed with 10 column volumes of buffer D (0.1 M KCl), and then the bound fraction was eluted with buffer D (0.5 M KCl). For binding assays with GST-par4, 25 μL of glutathione agarose beads containing either 1 μg of GST or GST-par4 was incubated in 0.6 mL of binding buffer (40 mM HEPES at pH 8.0, 10% [v/v] glycerol, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.2 mM PMSF, 0.05% NP40) with 10 μL of bacterial lysate containing the GAL4 derivatives indicated. The samples were incubated at 4°C for 1 h with gentle rocking. After extensive washing, the bound fraction was eluted with SDS-PAGE loading dye, subjected to electrophoresis, and immunoblotted with anti-GAL4 antibody.
Immunoprecipitation assays were performed with nuclear extracts prepared from transfected cells in buffer D containing 150 mM KCl and 0.05% NP40. Protein G Sepharose was used to collect the immune complexes and washes performed in the same buffer. Bound proteins were eluted with SDS-PAGE loading dye, resolved by electrophoresis, and probed with anti-par4 antibodies.
Acknowledgments
We thank Neil Perkins and members of the Roberts lab for comments on the manuscript. We are also grateful to Lynn McKay for help during the initial stages of this work. Also, thanks to Rachel Davies, Nick Hastie, and Yang Shi for providing plasmids. This work was funded by the Cancer Research Campaign [CRC] (SP2410/0101). S.G.E.R. is a Wellcome Trust Senior Research Fellow (061207/Z/00/Z/CH/TG/dr).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL S.G.E.Roberts@dundee.ac.uk; FAX 01382-348072.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.185901.
References
- Algar E, Blackburn D, Kromykh T, Taylor G, Smith P. Mutation analysis of the WT1 gene in sporadic childhood leukaemia. Leukemia. 1996;11:110–113. doi: 10.1038/sj.leu.2400521. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai MJ, O'Malley BW. Interaction of human thyroid hormone receptor b with transcription factor TFIIB can mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaux S, Niaudet P, Gubler MC, Grunfeld JP, Jaubert F, Kuttenn F, Fekete CN, Souleyreau-Therville N, Thibaud E, Fellous M, et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Duarte A, Larsson SH, Hastie ND, Little M, Holmes G, Todorov I, Ward A. RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc Natl Acad Sci. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DM, Hinkes MT, Bernfield M, Rauscher FJ. Transcriptional activation of the syndecan-1 promoter by the Wilms' tumor protein WT1. Oncogene. 1996;13:1789–1799. [PubMed] [Google Scholar]
- Davies RC, Calvio C, Bratt E, Larsson SH, Lamond AI, Hastie ND. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes & Dev. 1998;12:3217–3225. doi: 10.1101/gad.12.20.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R, Moore A, Schedl A, Bratt E, Miyahawa K, Ladomery M, Miles C, Menke A, van Heyningen V, Hastie N. Multiple roles for the Wilms' tumor suppressor, WT1. Cancer Res. 1999;59:1747S–1750S. [PubMed] [Google Scholar]
- Dey BR, Sukhatme VP, Roberts AB, Sporn MB, Rauscher FJ, Kim SJ. Repression of the transforming growth-factor- β-1 gene by the Wilms-tumor suppressor WT1 gene-product. Mol Endocrinol. 1994;8:595–602. doi: 10.1210/mend.8.5.8058069. [DOI] [PubMed] [Google Scholar]
- Diller L, Ghahremani M, Morgan J, Grundy P, Reeves C, Breslow N, Green D, Neuberg D, Pelletier J, Li FP. Constitutional WT1 mutations in Wilms' tumour patients. J Clin Oncol. 1998;16:3634–3640. doi: 10.1200/JCO.1998.16.11.3634. [DOI] [PubMed] [Google Scholar]
- Drummond IA, Madden SL, Rohwernutter P, Bell GI, Sukhatme VP, Rauscher FJ. Repression of the insulin-like growth factor-II gene by the Wilms-tumor suppressor WT1. Science. 1992;257:674–678. doi: 10.1126/science.1323141. [DOI] [PubMed] [Google Scholar]
- Englert C. WT1—More than a transcription factor? Trends Biochem Sci. 1998;23:389–393. doi: 10.1016/s0968-0004(98)01277-8. [DOI] [PubMed] [Google Scholar]
- Englert C, Hou X, Maheswaran S, Bennett P, Ngwu C, Re GG, Garvin AJ, Rosner MR, Haber DA. WT1 suppresses synthesis of the epidermal growth-factor receptor and induces apoptosis. EMBO J. 1995;14:4662–4675. doi: 10.1002/j.1460-2075.1995.tb00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert C, Maheswaran S, Garvin AJ, Kreidberg J, Haber DA. Induction of p21 by the Wilms' tumor suppressor gene WT1. Cancer Res. 1997;57:1429–1434. [PubMed] [Google Scholar]
- English MA, Licht JD. Tumor-associated WT1 missense mutants indicate that transcriptional activation by WT1 is critical for growth control. J Biol Chem. 1999;274:13258–13263. doi: 10.1074/jbc.274.19.13258. [DOI] [PubMed] [Google Scholar]
- Gashler AL, Bonthron DT, Madden SL, Rauscher FJ, Collins T, Sukhatme VP. Human platelet-derived growth factor A chain is transcriptionally repressed by the Wilms tumor suppressor WT1. Proc Natl Acad Sci. 1992;89:10984–10988. doi: 10.1073/pnas.89.22.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes NA, Roberts SGE. The role of human TFIIB in transcription start site selection in vitro and in vivo. J Biol Chem. 1999;274:14337–14343. doi: 10.1074/jbc.274.20.14337. [DOI] [PubMed] [Google Scholar]
- Iben S, Royer-Pokora B. Analysis of native WT1 protein from frozen human kidney and Wilms' tumors. Oncogene. 1999;18:2533–2536. doi: 10.1038/sj.onc.1202618. [DOI] [PubMed] [Google Scholar]
- Jeanpierre C, Beroud C, Niaudet P, Junien C. Software and database for the analysis of mutations in the human WT1 gene. Nucleic Acids Res. 1998;26:271–274. doi: 10.1093/nar/26.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, See RH, Sells SF, Wang J, Muthukkumar S, Englert C, Haber DA, Licht JD, Sugrue SP, Roberts T, et al. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms' tumor suppressor WT1. Mol Cell Biol. 1996;16:6945–6956. doi: 10.1128/mcb.16.12.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Wang J, Tommerup N, Vissing H, Roberts T, Shi Y. Ciao 1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J Biol Chem. 1998;273:10880–10887. doi: 10.1074/jbc.273.18.10880. [DOI] [PubMed] [Google Scholar]
- Kent J, Coriat AM, Sharpe PT, Hastie ND, Vanheyningen V. The evolution of WT1 sequence and expression pattern in the vertebrates. Oncogene. 1995;11:1781–1792. [PubMed] [Google Scholar]
- King-Underwood L, Pritchard-Jones K. Wilms' tumour (WT1) gene mutations occur mainly in acute myeloid leukemia and may confer drug resistance. Blood. 1998;91:2961–2968. [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Ladomery MR, Slight J, McGhee S, Hastie ND. Presence of WT1, the Wilms' tumor suppressor gene product, in nuclear poly(A)(+) ribonucleoprotein. J Biol Chem. 1999;274:36520–36526. doi: 10.1074/jbc.274.51.36520. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, Vanheyningen V, Hastie ND. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- Lee KAW, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-messenger RNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- Lee SB, Huang K, Palmer R, Truong VB, Herzlinger D, Kolquist KA, Wong J, Paulding C, Yoon SK, Gerald W, et al. The Wilms' tumor suppressor WT1 encodes a transcriptional activator of amphiregulin. Cell. 1999;98:663–673. doi: 10.1016/s0092-8674(00)80053-7. [DOI] [PubMed] [Google Scholar]
- Lin YS, Green MR. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Lin YS, Ha I, Maldonado E, Reinberg D, Green MR. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Little M, Holmes G, Walsh P. WT1: What has the last decade told us? Bioessays. 1999;21:191–202. doi: 10.1002/(SICI)1521-1878(199903)21:3<191::AID-BIES3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Madden SL, Cook DM, Morris JF, Gashler A, Sukhatme VP, Rauscher FJ. Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991;253:1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Englert C, Lee SB, Ezzel RM, Settleman J, Haber DA. E1B 55K sequesters WT1 along with p53 within a cytoplasmic body in adenovirus-transformed kidney cells. Oncogene. 1998a;16:2041–2050. doi: 10.1038/sj.onc.1201741. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Englert C, Zheng G, Lee SB, Wong J, Harkin DP, Bean J, Ezzell R, Garvin AJ, McCluskey RT, et al. Inhibition of cellular proliferation by the Wilms tumor suppressor WT1 requires association with the inducible chaperone Hsp70. Genes & Dev. 1998b;12:1108–1120. doi: 10.1101/gad.12.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Trouche D, Hagemeier C, Sorensen TS, Lathangue NB, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by Mdm2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- Mayo MW, Wang CY, Drouin SS, Madrid LV, Marshall AF, Reed JC, Weissman BE, Baldwin AS. WT1 modulates apoptosis by transcriptionally upregulating the bcl-2 proto-oncogene. EMBO J. 1999;18:3990–4003. doi: 10.1093/emboj/18.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LM, Carpenter B, Roberts SGE. Regulation of the Wilms' tumour suppressor protein transcriptional activation domain. Oncogene. 1999;18:6546–6554. doi: 10.1038/sj.onc.1203046. [DOI] [PubMed] [Google Scholar]
- Menke AL, Shvarts A, Riteco N, van Ham RCA, van der Eb AJ, Jochemsen AG. Wilms' tumor 1-KTS isoforms induce p53-independent apoptosis that can be partially rescued by expression of the epidermal growth factor receptor or the insulin receptor. Cancer Res. 1997;57:1353–1363. [PubMed] [Google Scholar]
- Moorwood K, Salpekar A, Ivins SM, Hall J, Powlesland RM, Brown KW, Malik K. Transactivation of the WT1 antisense promoter is unique to the WT1[+/−] isoform. FEBS Lett. 1999;456:131–136. doi: 10.1016/s0014-5793(99)00944-8. [DOI] [PubMed] [Google Scholar]
- Murata Y, Kudoh T, Sugiyama H, Toyoshima K, Akiyama T. The Wilms tumor suppressor gene WT1 induces G1 arrest and apoptosis in myeloblastic leukemia M1 cells. FEBS Lett. 1997;409:41–45. doi: 10.1016/s0014-5793(97)00477-8. [DOI] [PubMed] [Google Scholar]
- Pritchard-Jones K, King-Underwood L. The Wilms tumour gene WT1 in leukaemia. Leuk Lymphoma. 1997;27:207–220. doi: 10.3109/10428199709059677. [DOI] [PubMed] [Google Scholar]
- Rangnekar VM. Apoptosis mediated by a novel leucine zipper protein Par-4. Apoptosis. 1998;3:61–66. doi: 10.1023/a:1009666705875. [DOI] [PubMed] [Google Scholar]
- Reddy JC, Licht JD. The WT1 Wilms' tumor suppressor gene: How much do we really know? Biochim Biophys Acta. 1996;1287:1–28. doi: 10.1016/0304-419x(95)00014-7. [DOI] [PubMed] [Google Scholar]
- Reddy JC, Morris JC, English MA, Haber DA, Luo XN, Atweh G, Licht JD. The Wilms-tumor (WT1) gene-product both activates and represses transcription. Blood. 1993;82:A324. [Google Scholar]
- Reddy JC, Morris JC, Wang J, English MA, Haber DA, Shi Y, Licht JD. WT1-mediated transcriptional activation is inhibited by dominant-negative mutant proteins. J Biol Chem. 1995;270:10878–10884. doi: 10.1074/jbc.270.18.10878. [DOI] [PubMed] [Google Scholar]
- Reece RJ, Rickles RJ, Ptashne M. Overproduction and single-step purification of Gal4 fusion proteins from Escherichia coli. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- Renshaw J, King-Underwood L, Pritchard-Jones K. Differential splicing of exon 5 of the Wilms tumour (WTI) gene. Genes Chromosomes Cancer. 1997;19:256–266. doi: 10.1002/(sici)1098-2264(199708)19:4<256::aid-gcc8>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Roberts SGE. Mechanisms of transcriptional activation and repression. Cell Mol Life Sci. 2000;57:1149–1160. doi: 10.1007/PL00000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher V, Schneider S, Figge A, Wildhardt G, Harms D, Schmidt D, Weirich A, Ludwig R, Royer-Pokora B. Correlation of germ-line mutations and two-hit inactivation of the WT1 gene with Wilms tumors of stromal-predominant histology. Proc Natl Acad Sci USA. 1997;94:3972–3977. doi: 10.1073/pnas.94.8.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thate C, Englert C, Gessler M. Analysis of WT1 target gene expression in stably transfected cell lines. Oncogene. 1998;17:1287–1294. doi: 10.1038/sj.onc.1202055. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Madden SL, Deuel TF, Rauscher FJ. The Wilms tumor gene product, WT1, represses transcription of the platelet-derived growth-factor A-chain gene. J Biol Chem. 1992;267:21999–22002. [PubMed] [Google Scholar]
- Wang ZY, Qiu QQ, Deuel TF. The Wilms tumor gene product WT1 activates or suppresses transcription through separate functional domains. J Biol Chem. 1993;268:9172–9175. [PubMed] [Google Scholar]
- Wang ZY, Qiu QQ, Huang J, Gurrieri M, Deuel TF. Products of alternatively spliced transcripts of the Wilms tumor suppressor gene, WT1, have altered DNA-binding specificity and regulate transcription in different ways. Oncogene. 1995;10:415–422. [PubMed] [Google Scholar]
- Zhong HH, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]