A multistep damage recognition mechanism for global genomic nucleotide excision repair (original) (raw)
Abstract
A mammalian nucleotide excision repair (NER) factor, the XPC–HR23B complex, can specifically bind to certain DNA lesions and initiate the cell-free repair reaction. Here we describe a detailed analysis of its binding specificity using various DNA substrates, each containing a single defined lesion. A highly sensitive gel mobility shift assay revealed that XPC–HR23B specifically binds a small bubble structure with or without damaged bases, whereas dual incision takes place only when damage is present in the bubble. This is evidence that damage recognition for NER is accomplished through at least two steps; XPC–HR23B first binds to a site that has a DNA helix distortion, and then the presence of injured bases is verified prior to dual incision. Cyclobutane pyrimidine dimers (CPDs) were hardly recognized by XPC–HR23B, suggesting that additional factors may be required for CPD recognition. Although the presence of mismatched bases opposite a CPD potentiated XPC–HR23B binding, probably due to enhancement of the helix distortion, cell-free excision of such compound lesions was much more efficient than expected from the observed affinity for XPC–HR23B. This also suggests that additional factors and steps are required for the recognition of some types of lesions. A multistep mechanism of this sort may provide a molecular basis for ensuring the high level of damage discrimination that is required for global genomic NER.
Keywords: Nucleotide excision repair, damage recognition, xeroderma pigmentosum, XPC–HR23B complex
DNA acquires various structural modifications through its intrinsic instability, the action of endogenously produced agents, and environmental attacks involving radiation and chemicals. Such DNA damage can block DNA replication and transcription, which may lead to arrested cell growth and/or apoptosis. It can also induce mutations, the accumulation of which can cause cell malfunctions, carcinogenesis, and possibly aging. To circumvent these problems, a network of multiple DNA repair pathways has evolved. Nucleotide excision repair (NER) is a versatile repair pathway that can eliminate a wide variety of lesions, such as ultraviolet light (UV)-induced photolesions [including cyclobutane pyrimidine dimer (CPD) and (6-4) photoproduct (6-4PP)], intrastrand cross-links, and bulky adducts induced by various carcinogens (Friedberg et al. 1995). It has been shown that eukaryotic NER involves at least two distinct subpathways, global genomic repair (GGR) and transcription-coupled repair (TCR). The GGR subpathway can operate at any location in the genome, and its efficiency varies depending on the type of lesion; for instance, 6-4PPs are eliminated from the global genome much faster than are CPDs (Mitchell and Nairn 1989). In contrast, TCR specifically removes lesions that block RNA polymerases on the transcribed strands of active genes (Bohr et al. 1985; Mellon et al. 1987). Unlike GGR, this subpathway eliminates different lesions at similar rates (van Hoffen et al. 1995), and contributes to the rapid recovery of transcriptional activity after DNA has been damaged, ensuring the maintenance of normal cellular functions and survival (Hanawalt and Mellon 1993; Friedberg 1996).
The biological relevance of mammalian NER has been well documented, because several human genetic disorders, including xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD), are associated with impaired NER activity. So far, seven NER-deficient complementation groups have been identified for XP (XP-A to XP-G), two for CS (CS-A and CS-B), and one for TTD (TTD-A), and most of the responsible genes have been cloned (for review, see Bootsma et al. 1997). Among these complementation groups, patients belonging to XP group C are deficient only in GGR, whereas TCR still functions normally (Venema et al. 1990, 1991). The responsible gene product, the XPC protein, is tightly complexed with HR23B, one of the two mammalian homologs of the Saccharomyces cerevisiae RAD23 protein (Masutani et al. 1994; Shivji et al. 1994). This complex formation is important for the cell-free repair function of XPC (Sugasawa et al. 1996, 1997; Batty et al. 2000), at least partly because of the physical stabilization it imparts (M. Araki, C. Matsutani, M. Takemura, A. Uchida, K. Sugasawa, J. Kondoh, Y. Okuma, and F. Hanaoka, unpubl.). Although the XPC–HR23B complex exhibits strong binding affinity for single- and double-stranded DNA even without damage (Masutani et al. 1994; Shivji et al. 1994; Reardon et al. 1996; Wakasugi and Sancar 1998), it preferentially binds to various types of damaged DNA, where it initiates the cell-free NER reaction (Sugasawa et al. 1998; Batty et al. 2000). Furthermore, XPC–HR23B has specific binding affinity for certain defined lesions, including UV-induced 6-4PP and a cholesteryl moiety (Sugasawa et al. 1998; Kusumoto et al. 2001), indicating that XPC–HR23B is involved in the GGR-specific damage recognition machinery. In TCR, RNA polymerase II itself has been presumed to function as a damage detector when blocked by a lesion (Donahue et al. 1994; Tornaletti et al. 1999). Following the subpathway-specific damage recognition events, several common NER factors, including transcription factor IIH (TFIIH), XPG, XPA, and replication protein A (RPA), may be recruited to the lesion, leading to the local unwinding of DNA double helix (Evans et al. 1997a,b; Mu et al. 1997b; Wakasugi et al. 1997). The XPB and XPD helicases, both of which are essential components of TFIIH (Schaeffer et al. 1993, 1994), play a critical role in this open complex formation. Two structure-specific endonucleases, the XPF–ERCC1 complex and XPG, subsequently introduce single-strand breaks on the 5′ and 3′ sides of the lesion, respectively, resulting in the excision of the oligonucleotide containing the injured base(s) (O'Donovan et al. 1994; Matsunaga et al. 1995; Sijbers et al. 1996).
Although it has been well documented that XPC–HR23B plays a critical role in global genomic NER, several issues remain to be solved. First, we have shown recently that UV-induced CPDs are hardly recognized by XPC–HR23B (Kusumoto et al. 2001). Because CPDs are removed from the global genome in an XPC-dependent manner, this suggests that recognition of some types of lesions may require involvement of other factors in addition to XPC–HR23B. Second, dual incision by a defined NER system, reconstituted with all the necessary purified factors except for XPC–HR23B, can remove a lesion in a bubble-like structure (Mu and Sancar 1997) or an artificial cholesteryl moiety (Mu et al. 1996). Furthermore, although the XPC protein has been shown to interact with TFIIH and likely recruits it to a lesion (Drapkin et al. 1994; Li et al. 1998; Yokoi et al. 2000), the precise reaction mechanism following the initial damage recognition event remains to be established. To understand the functions of XPC–HR23B in more detail, we have undertaken an extended analysis of its binding specificity for DNA containing various lesions and structures. The results provide an important insight into the damage recognition strategy utilized by the eukaryotic NER machinery.
Results
Gel mobility shift assay for quantitative analysis of the binding affinity of XPC–HR23B
We have observed previously a specific damage-binding activity of XPC–HR23B by DNase I footprinting as well as by coimmunoprecipitation techniques (Sugasawa et al. 1998; Kusumoto et al. 2001). To examine the damage-binding specificity in more detail, a convenient, highly sensitive assay was developed by use of the gel mobility shift technique. Blunt-ended, radiolabeled DNA fragments (∼180–190 bp in length) containing a single lesion at a defined position were used for binding reactions with the purified recombinant human XPC–HR23B complex. After the binding reactions, the resulting protein–DNA complexes were fixed with glutaraldehyde before performing native polyacrylamide gel electrophoresis (PAGE). Because we noticed that complexes involving XPC–HR23B and monoadduct DNA are not completely stable during electrophoresis, the fixation step was applied to obtain more reproducible results. Although we have discovered recently that centrin 2 is an additional component of the XPC protein complex, its binding does not greatly affect either damage binding or the NER function of the complex, at least in the cell-free system (M. Araki, C. Matsutani, M. Takemura, A. Uchida, K. Sagasawa, J. Kondoh, Y. Okuma, and F. Hanaoka, unpubl.).
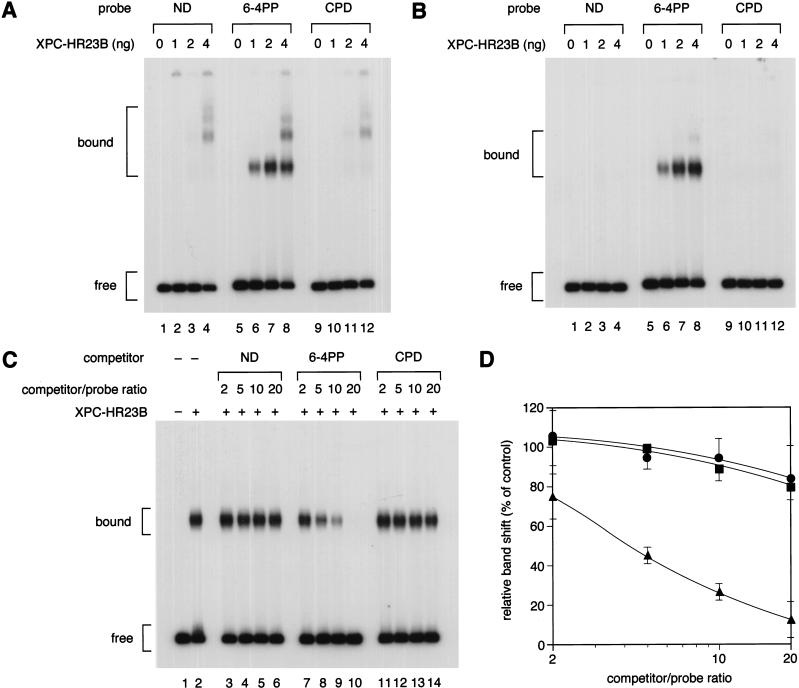
We first tested the binding of XPC–HR23B to two major UV-induced photolesions, 6-4PP and CPD (both involving TT dinucleotides; Fig. 1). When the 32P-labeled probes were incubated with XPC–HR23B in the absence of any other DNA, some shifted bands were observed even with the nondamaged DNA probe (Fig. 2A, lane 4). This nonspecific binding was abrogated by the addition of an appropriate amount of double-stranded plasmid DNA as a competitor (Fig. 2B, lanes 1–4). Under these conditions, the probe containing 6-4PP still gave rise to complexes containing bound XPC–HR23B (Fig. 2B, lanes 6–8), demonstrating that XPC–HR23B is capable of specifically binding 6-4PPs (note that the specific complexes are already visible without the competitor DNA; Fig. 2A, lanes 6–8). In marked contrast, little specific binding could be observed with the CPD probe (Fig. 2B, lanes 9–12), in agreement with our previous data from coimmunoprecipitation experiments (Kusumoto et al. 2001).
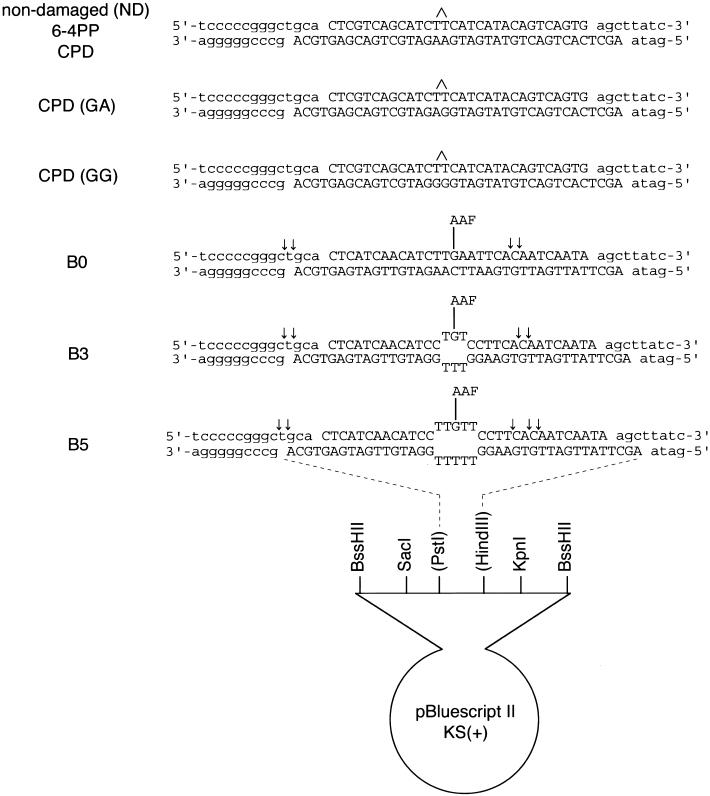
Figure 1.
Defined DNA substrates used in this study. Arrows indicate major incision sites in each AAF-damaged substrate, which were determined by the primer extension method.
Figure 2.
Gel mobility shift analysis of XPC–HR23B binding to a single UV photolesion. (A,B) Indicated amounts of purified XPC–HR23B were incubated with 3.5 fmole each of the 32P-labeled DNA fragment containing a 6-4PP or a CPD, or the cognate nondamaged (ND) control fragment. The binding reactions were performed in the absence (A) or presence (B) of a small amount (0.5 ng) of covalently closed circular plasmid DNA. The resulting DNA–protein complexes were fixed with glutaraldehyde and separated by PAGE. (C) A competition experiment using the labeled 6-4PP probe (3.5 fmole), which was incubated with (+) or without (−) 2 ng of XPC–HR23B and various amounts of cold competitor DNA fragments as indicated. (D) The amount of the labeled probe complexed with XPC–HR23B was calculated for each lane in C, and expressed as a percentage of the control (lane 2) without competitors. The mean values and standard errors were calculated from two or three independent experiments.(●) ND; (▴) 6-4PP; (▪) CPD.
To obtain some quantitative information on the binding affinity, competition experiments were performed. Various amounts of nonradioactive DNA fragments with or without a single UV lesion were included in the XPC–HR23B binding reactions with a fixed amount of the 32P-labeled 6-4PP probe. As shown in Figure 2, C and D, the 6-4PP DNA fragment itself competed well with the 6-4PP probe for binding to XPC–HR23B; 50% inhibition of complex formation was achieved by adding a fourfold molar excess of the competitor. On the other hand, no significant difference was observed between the competition profiles of the nondamaged and CPD competitors, within the range of competitor concentrations used.
XPC–HR23B binds a small bubble structure regardless of the presence or absence of lesions
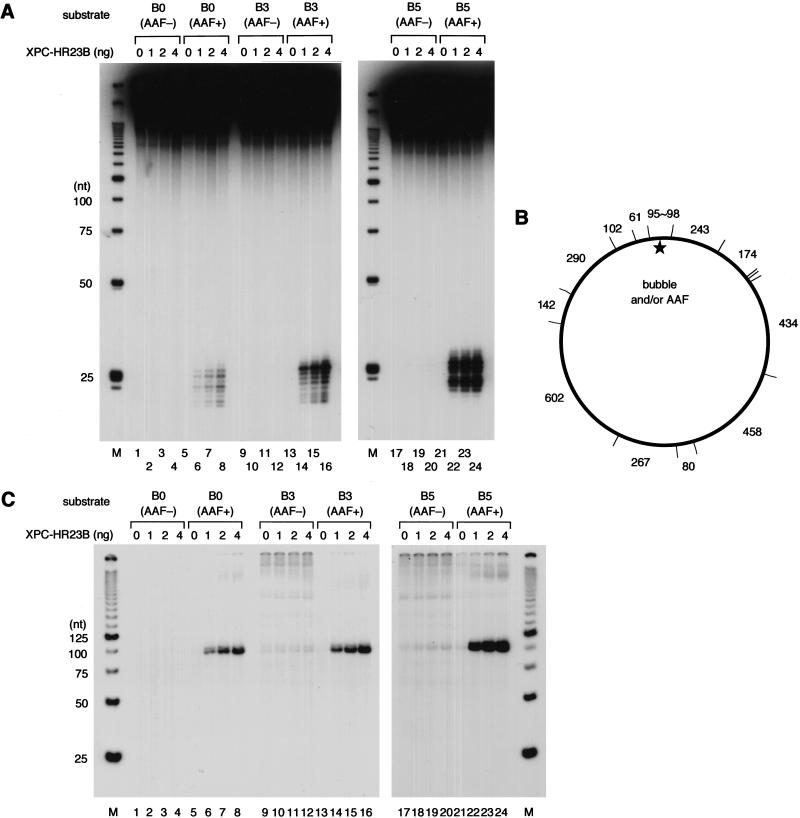
Structural analyses of DNA containing UV-induced photolesions have revealed that 6-4PP induces significant helix distortion, including disruption of base pairing, whereas the distortion induced by CPD is much less pronounced (Kim et al. 1995; McAteer et al. 1998). Furthermore, it has been reported recently that the mammalian NER machinery can be targeted in vitro to sites of defective Watson-Crick base pairing (Hess et al. 1998; Buschta-Hedayat et al. 1999). Taken together with our findings that CPD is not well recognized by XPC–HR23B, we have been interested in testing the binding of XPC–HR23B to double-stranded DNA fragments containing a small number of unpaired bases. For this purpose, several DNA substrates were prepared (B0, B3, and B5 in Fig. 1). A small bubble (3 or 5 bases) was created by inserting T residues in both strands as well as a G residue in the center of the bubble. This guanine residue allowed the site-specific modification of the DNA by _N_-acetoxy-2-acetylaminofluorene (AAF), resulting in an _N_-(guanin-8-yl)-AAF adduct, which is a known substrate for NER.
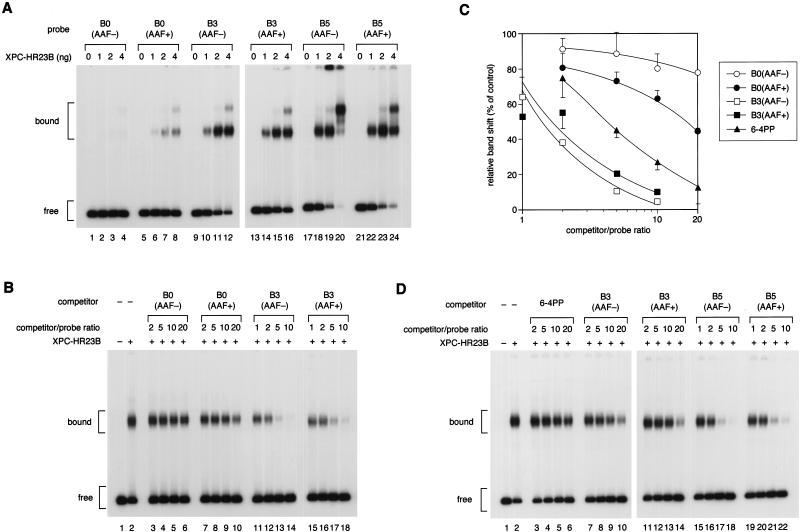
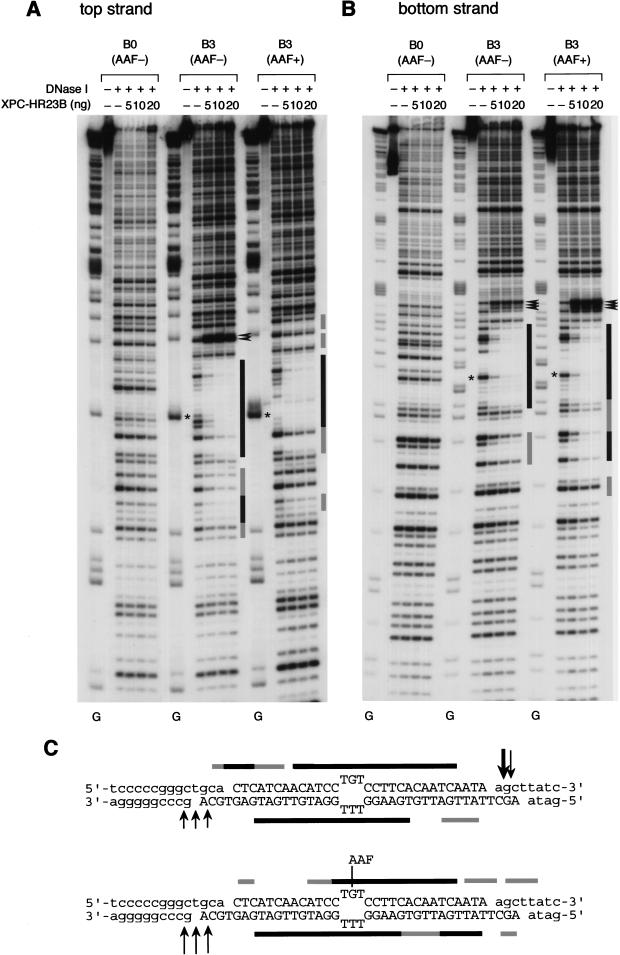
When non-bubbled (B0) control substrates were used for the gel mobility shift assay, specific complex formation could be observed in the presence of an AAF adduct, but not in its absence (Fig. 3A, lanes 1–8). Intriguingly, XPC–HR23B efficiently bound the substrates containing a 3-base (B3) or 5-base (B5) bubble, even in the absence of AAF damage (Fig. 3A, lanes 9–24). The binding properties were further characterized quantitatively by use of competition assays. The non-bubbled substrate significantly competed with the 6-4PP probe for binding XPC–HR23B in an AAF-lesion-dependent manner, although the observed affinity for the AAF adduct was lower than that for 6-4PP (Fig. 3B, lanes 3–10; see also C). Under the same conditions, the B3 substrates displayed a higher affinity for XPC–HR23B than 6-4PP, regardless of the presence or absence of the AAF lesion (Fig. 3C). Competition experiments using the AAF-damaged B5 probe revealed that XPC–HR23B binds the larger bubble with the higher affinity (Fig. 3D). The specificity of binding to the bubble structure was also examined by DNase I footprint analysis. As shown in Figure 4, A and B, the bubble site of the B3 substrate was well protected against DNase I attack by XPC–HR23B, either with or without the AAF adduct. However, a slight difference in the digestion pattern could be observed, as summarized in Figure 4C. For instance, a few phosphodiester bonds in the top strand on the 3′ side of the bubble became hypersensitive to DNase I upon XPC–HR23B binding to the nondamaged B3 substrate, whereas the 3′-hypersensitive sites were found mainly in the bottom strand when the B3 substrate contained the AAF adduct (the hypersensitive sites are indicated by arrowheads in Fig. 4, A and B, and by arrows in C). Thus, although the presence of the AAF adduct may somehow affect the precise positioning and/or binding mode, damage is not required for specific binding of XPC–HR23B when the DNA contains a small bubble. This indicates that the complex does not recognize the lesion itself, but rather the structural distortion of double-stranded DNA, as suggested previously (Batty et al. 2000).
Figure 3.
Damage-independent binding of XPC–HR23B to a small bubble. (A) Gel mobility shift assay using the 32P-labeled probe with or without the AAF adduct and a bubble. (B) A competition experiment involving the 32P-labeled 6-4PP probe (3.5 fmole) and 2 ng of XPC–HR23B. Various amounts of nonlabeled competitor DNA fragments were included in the binding reactions as indicated. (C) Quantitative representation of the competition profiles. Complex formation in each lane of B was quantified as in Fig. 2D. The mean values and standard errors were calculated from at least two independent experiments. The competition profile by 6-4PP obtained in Fig. 2D is superimposed. (○) B0(AAF−); (●) B0(AAF+); (□) B3(AFF−); (▪) B3(AFF+); (▴) 6-4PP. (D) A competition experiment using the 32P-labeled B5(AAF +) probe and 1 ng of XPC–HR23B.
Figure 4.
DNase I footprinting analysis of XPC–HR23B binding to a 3-base bubble. (A,B) The indicated substrates were 5′-end-labeled for top strands (A) or bottom strands (B), and subjected to the DNase I footprinting assay using various amounts of XPC–HR23B as indicated. The digested DNA samples were subjected to denaturing PAGE followed by autoradiography. The Maxam-Gilbert G ladder prepared from each probe was loaded in parallel, where indicated, below the gels. The position of the bubble in each substrate is shown by an asterisk. (C) A schematic representation of the protection patterns for the B3 substrates. In each panel, strongly and weakly protected regions are shown by solid and shaded bars, respectively. The sites that became hypersensitive to DNase I upon binding are indicated by arrowheads (A,B) or arrows (C). Size of the arrows in C corresponds to the observed degree of hypersensitivity.
Damage is required for the dual incision even after XPC–HR23B has bound to a bubble
To examine whether the binding of XPC–HR23B to the bubble leads to the following repair process, the substrates were tested for cell-free dual incision activity. Oligonucleotides with or without the AAF adduct (top strands shown by uppercase letters in Fig. 1) were 5′-end labeled with [γ-32P]ATP and used for the preparation of double-stranded closed circular DNAs. This allowed the generation of DNA substrates containing an internal 32P-label at nucleotide position 14 or 15 on the 5′ side of the AAF lesion. When the internally labeled B0 substrate containing the AAF adduct was incubated with the XP-C whole cell extract supplemented with purified XPC–HR23B, dual incision products could be detected by denaturing PAGE (Fig. 5A, lanes 6–8). These excised oligonucleotides were undetectable when XPC–HR23B was omitted from the reaction (lane 5), or when the nondamaged, control B0 substrate was used (lanes 1–4). XPC–HR23B-dependent dual incision was also detected with the AAF-damaged B3 and B5 substrates, which gave rise to increasing amounts of excised oligonucleotides, correlating with the observed affinity of XPC–HR23B for these substrates (Fig. 5, lanes 14–16 and 22–24). However, very few labeled oligonucleotides were released from the nondamaged B3 and B5 substrates (Fig. 5, lanes 9–12 and 17–20), although their affinity for XPC–HR23B was indistinguishable from that of their AAF-damaged counterparts.
Figure 5.
Binding of XPC–HR23B is not sufficient for the NER incision, but damage is also required. (A) The indicated substrates were internally labeled with 32P and used for the dual incision assay involving the XP-C whole cell extract and various amounts of XPC–HR23B. The DNA samples were subjected to denaturing PAGE followed by autoradiography. (M) 32P-labeled 25-bp ladder. (B) A map of _Hae_III-cutting sites in the DNA substrates, where the size of each fragment (in base pairs) is indicated. (C) The nonlabeled closed circular DNA substrates indicated were incubated in the cell-free NER reactions including the XP-C whole cell extract, various amounts of XPC–HR23B, and aphidicolin. The purified DNA samples were subjected to gap-filling DNA synthesis with T4 DNA polymerase and radiolabeled dNTPs, digestion with _Hae_III, and nondenaturing PAGE followed by autoradiography.
For each AAF-damaged substrate, we determined the major incision sites using the primer extension methods described by others (Moggs et al. 1996), and confirmed that the 5′-incision took place mainly 5′ to the 32P-labeled site (indicated by arrows in Fig. 1). Although dual incision was not detected with the nondamaged bubble substrates, the possibility could not be excluded that the 5′-incision sites may have shifted to the 3′-side of the labeled site, or that the incision may have occurred on the other, unlabeled, strand. To test these possibilities, we utilized another type of assay. Cell-free NER reactions were carried out with nonlabeled, double-stranded circular DNA substrates in the presence of aphidicolin, which inhibits gap-filling DNA repair synthesis (Coverley et al. 1992). The purified DNA was then treated with T4 DNA polymerase and radioactive dNTPs to fill the gaps left behind and further digested with _Hae_III into 14 pieces (Fig. 5B). Upon this digestion, a fragment of 95–98 bp (depending on the bubble size) should contain the site of the AAF adduct and/or a bubble, and thus would be specifically labeled if dual incision had occurred around the site. As shown in Figure 5C, specific incorporation of radioactivity, which depended on the presence of XPC–HR23B, was observed in a fragment of the expected size with the AAF-damaged substrates (Fig. 5, lanes 5–8, 13–16, and 21–24). In contrast, the bubble substrates without the lesion failed to show specific incorporation into the corresponding fragment, although rather high background DNA synthesis may have occurred (Fig. 5, lanes 9–12 and 17–20). From these findings, we conclude that the binding of XPC–HR23B to a bubble lacking lesions does not lead to dual incision in the cell-free NER system.
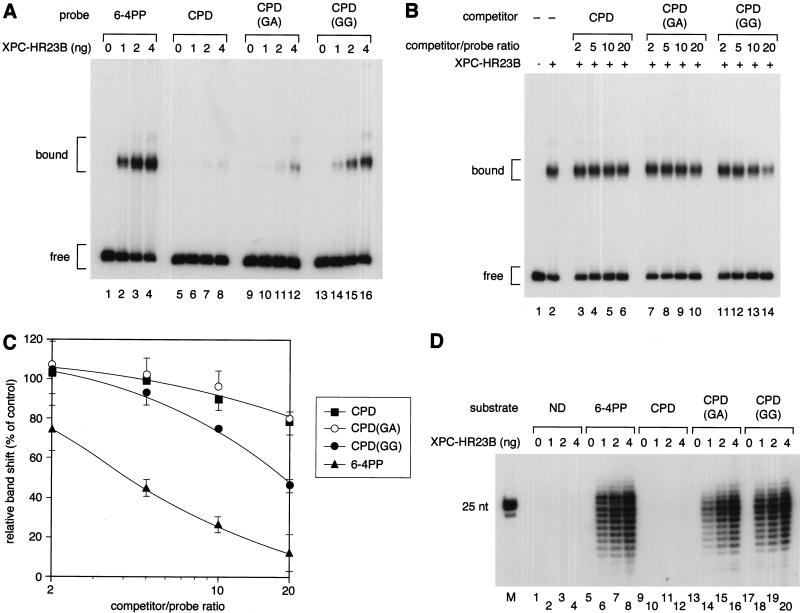
Mismatched bases opposite CPD potentiate the binding of XPC–HR23B and the in vitro repair efficiency
It has been documented that some compound lesions, such as CPDs opposite one or more mismatched bases, are more efficiently repaired in vitro than are lesions paired with correct bases (Moggs et al. 1997; Mu et al. 1997a). On the basis of our findings that XPC–HR23B recognizes a distorted helix, we postulated that efficient repair of such compound lesions could be explained by the increased affinity of XPC–HR23B, because one would expect that the presence of mismatched bases may enhance the level of helix distortion associated with the lesions. To test this possibility, a set of substrates was prepared that contained a site-specific CPD opposite GA or GG dinucleotides (Fig. 1). Gel mobility shift assays using these substrates revealed that the presence of the mismatched bases increased the specific affinity of XPC–HR23B for the CPD. Under conditions in which XPC–HR23B hardly bound to the correctly paired CPD, CPD opposite one mismatched G showed weak but nonetheless detectable binding activity, which was further increased by the presence of an additional mismatched G (Fig. 6A). Competition experiments indicate that the affinity of XPC–HR23B for CPD opposite GG, although higher than without the mismatches, is still similar to its affinity for the non-bubbled AAF lesion, and significantly lower than that for 6-4PP (Fig. 6B,C). Competition by CPD opposite GA was indistinguishable from CPD normally paired with AA, at least in the range of competitor concentrations used.
Figure 6.
Presence of mismatched bases opposite CPD enhances the binding affinity for XPC–HR23B as well as the damage excision efficiency. (A) Gel mobility shift assay using the indicated probes and various amounts of XPC–HR23B. (B) A competition experiment involving the 32P-labeled 6-4PP probe (3.5 fmole) and 2 ng of XPC–HR23B. Various amounts of nonlabeled competitor DNA fragments were included in the binding reactions as indicated. (C) Quantitative representation of the competition profiles. The mean values and standard errors were calculated from at least two independent experiments. The competition profile of 6-4PP is superimposed. (▪) CPD; (○) CPD(GA); (●) CPD(GG); (▴) 6-4PP. (D) The indicated, internally labeled substrates were assayed for the NER incision in the XP-C whole cell extract supplemented with various amounts of XPC–HR23B. A part of the autoradiograph showing the dual incision products is presented. (M) 32P-labeled 25-bp ladder.
The same set of substrates was subjected to the dual incision assay in the cell-free system (Fig. 6D). In agreement with our previous findings (Kusumoto et al. 2001), excision of CPDs was much less efficient compared with that of 6-4PPs (Fig. 6, lanes 5–12). Longer exposure of the gel shown in Figure 6D revealed a trace amount of excised CPD, which was at least 50-fold less than the amount of excised 6-4PP. Interestingly, CPDs opposite GA were excised quite efficiently (20- to 30-fold more than normally paired CPD; Fig. 6, lanes 13–16), although their affinity for XPC–HR23B was found to be only marginal. Furthermore, excision of CPDs opposite two mismatched Gs was at a level similar to that of 6-4PPs (Fig. 6, lanes 17–20), which was much more efficient than expected from the observed affinity for XPC–HR23B. In both cases, XPC–HR23B was absolutely necessary for the excision of CPDs. Thus, although the increased affinity for XPC–HR23B may contribute to efficient excision to some extent, other factors present in the extracts must be involved in the initial detection of the compound lesions, which may then somehow target XPC–HR23B.
Discussion
Importance of competitive reaction conditions for observing the damage specificity and the NER initiator function of XPC–HR23B
We have shown previously that the XPC–HR23B complex is capable of specific binding to some defined lesions using DNase I footprinting as well as coimmunoprecipitation techniques (Sugasawa et al. 1998; Kusumoto et al. 2001). Here we describe a more convenient gel mobility shift assay, which allowed us to detect the specific binding of XPC–HR23B, not only to the AAF adduct, but also to a bubble structure and to compound lesions involving CPD opposite mismatched base(s). As found in our previous studies (Sugasawa et al. 1998; Kusumoto et al. 2001), and as also discussed by Batty et al. (2000), inclusion of the appropriate competitor DNA in the binding reactions was necessary to absorb the nonspecific DNA binding of XPC–HR23B. Furthermore, special care was taken in the present study in the preparation of DNA fragments used for the gel mobility shift assay as well as the DNase I footprinting experiments, in that all of the probes and the competitors were treated with T4 DNA polymerase to make blunt-ended fragments. From our results showing that XPC–HR23B recognizes and binds to a small number of unpaired bases within a double-stranded region, one might anticipate that protruding termini generated by restriction enzyme digestion could affect the binding of XPC–HR23B. We have actually noticed that DNA fragments with 5′-protruding termini (with 4-base overhangs) competed more effectively for XPC–HR23B binding than the corresponding, blunt-ended fragments (data not shown). Therefore, the use of fragments with terminal overhangs may lead to the formation of undesirable complexes in which XPC–HR23B binds to the end of the DNA, and thus the apparent binding specificity for lesions could be significantly underestimated.
The competitive conditions are also important for observing the initiator function of XPC–HR23B in the cell-free NER reaction. We have shown previously that preincubation of damaged DNA with XPC–HR23B is sufficient to target the whole NER machinery, which consequently induces preferential repair of the bound DNA over other damaged DNA that is not bound to XPC–HR23B (Sugasawa et al. 1998). In that study, the repair kinetics of two damaged DNAs could be compared feasibly, because both DNAs were eventually incubated in the same reaction. On the other hand, it has been reported recently that preincubation with XPC–HR23B inhibited damage excision, as compared with preincubation with XPA and RPA, or when all of the NER factors were added at the same time (Wakasugi and Sancar 1999). In the latter study, repair rates were compared among separately incubated reactions; under such conditions it cannot be excluded that the apparent inhibition was due, for instance, to heat inactivation or aggregation of XPC–HR23B during the preincubation.
Requirement of XPC–HR23B for damage excision from bubbled substrates
It has been reported that, when a lesion such as CPD resides within a bubble structure, XPC–HR23B is not necessary for damage excision in the cell-free NER reaction reconstituted with purified factors (Mu and Sancar 1997). From these results, it has been supposed that one of the roles of XPC–HR23B may be to create a small opening in the DNA strands around the lesion, which would allow subsequent entry of other NER factors, such as TFIIH, leading to full opening. This idea provides a plausible explanation for the absence of a requirement for XPC–HR23B in TCR, because the damaged site is already opened when RNA polymerase II encounters the lesion. On the other hand, we show here that the AAF adduct in a small bubble is still excised in an XPC–HR23B-dependent manner. Although one may argue that our bubble size (3 or 5 bases) might be too small to bypass the requirement for XPC–HR23B, we obtained the same results with a larger, 9-base bubble (data not shown). It is still possible that a trace level of excision of the AAF adduct may occur with our bubbled substrates in the absence of XPC–HR23B. Even if this is the case, however, the observed stimulation of the dual incision by XPC–HR23B may be due to its efficient recruitment of TFIIH (Li et al. 1998; Yokoi et al. 2000). Because TFIIH itself does not bind DNA very strongly or with any specificity for damage (Nocentini et al. 1997; Wakasugi and Sancar 1998; Yokoi et al. 2000), it seems to need protein–protein interactions with other factors for efficient binding to target sites. Nevertheless, we do not rule out the alternative possibility that some factors present in the cell extract might somehow suppress the XPC–HR23B-independent repair of the bubbled AAF substrates. Reconstitution of the dual incision reaction with our substrates and purified NER factors is now in progress.
Multistep damage recognition in NER
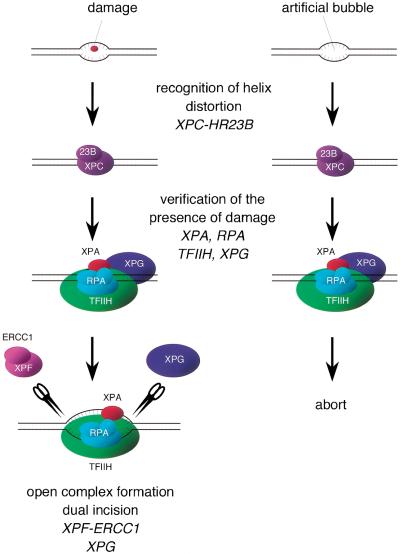
Because NER can handle a wide variety of structurally unrelated lesions, it has been supposed that the repair machinery does not recognize the lesion itself, but rather the distortion of the helical structure of the DNA induced by the presence of the lesion. Such helix distortion may include the disruption of base pairing as well as the bending of DNA strands. The importance of helix distortion in mammalian damage recognition has been shown experimentally recently by use of certain artificial adducts as well as cell-free NER reactions (Hess et al. 1998; Buschta-Hedayat et al. 1999). In those studies, it was shown that the NER system is efficiently recruited to sites in which Watson-Crick base pairing is weakened or disrupted. In accordance with this idea, XPC–HR23B is able to efficiently recognize and bind a small bubble involving unpaired bases, for which damage is not required at all. Although the specific binding of XPC–HR23B would normally initiate the NER reaction leading to excision of the lesion, dual incision hardly takes place in such bubbled substrates in the absence of damage. These findings indicate that, after XPC–HR23B binding, the repair reaction does not proceed to dual incision in a straightforward manner. Instead, the damage recognition is accomplished through at least two steps; XPC–HR23B first binds to a suspected damage site that has helix distortion, and then the presence of a lesion that is suitable for processing by the NER machinery is somehow verified (Fig. 7). One possibility is that XPC–HR23B itself can discriminate whether injured bases are present or absent; our footprinting analysis revealed that the existence of the AAF adduct in a bubble slightly alters the pattern of protection by XPC–HR23B, although the overall binding affinity is barely affected. Alternatively, and more likely, other NER factors, which are recruited after the binding of XPC–HR23B, may be responsible for damage verification. Because XPA and RPA have been shown to have some specific binding affinity for damaged DNA (Clungston et al. 1992; Jones and Wood 1993; Asahina et al. 1994; He et al. 1995; Burns et al. 1996), these factors may be among the candidates. Moreover, recent knowledge concerning the mechanism of TCR may provide an additional clue to understanding this key step. Not only are lesions normally processed by NER, but major oxidative damage, such as thymine glycol and 8-oxoguanine, has also been shown to be repaired in a transcription-dependent manner (Cooper et al. 1997; Le Page et al. 2000). This transcription-coupled base excision repair (BER) requires the functions of TFIIH and XPG, which are distinct from those required for NER, in addition to CSB (and probably also CSA), which is well known to be specifically involved in transcription-coupled NER, but not in global genomic NER. A model has been proposed in which RNA polymerase II stalling at the injured site may assemble a TCR complex involving at least TFIIH, XPG, and CSB, which may subsequently recruit an appropriate DNA glycosylase to initiate BER (Le Page et al. 2000). Because the TCR complex seems to be commonly utilized in transcription-coupled NER, it is possible that the complex may have the ability to somehow discriminate whether the lesion should be processed by NER or by BER. We reported recently that the XPC protein interacts with TFIIH and targets it for damaged DNA in vitro (Yokoi et al. 2000). It has been proposed by others (Naegeli 1999; Wood 1999) that a lesion may be sensed by the blockage of translocation by TFIIH helicases, although it has not yet been demonstrated experimentally. In analogy to the case of TCR, TFIIH, which is recruited by XPC–HR23B bound to the suspected site, may be involved in the verification of the presence of damage, likely together with other factors such as XPG.
Figure 7.
A model for two-step damage recognition in global genomic NER. XPC–HR23B first recognizes the site where helix distortion is induced. Other NER factors, involving TFIIH, XPA, XPG, and RPA, may be then recruited to the suspected site, where they somehow verify the presence of a lesion that is suitable to be handled by NER. If there is a lesion (left), the pre-incision complex containing fully opened DNA would be assembled, leading to dual incision by XPF–ERCC1 and XPG. If there is not a lesion (right), the process may be cancelled at a certain stage prior to the open complex assembly.
Using the coimmunoprecipitation technique, we have shown recently that XPC–HR23B fails to recognize UV-induced CPD (Kusumoto et al. 2001). It has been also described that treatment of UV-irradiated DNA with CPD photolyase does not affect specific binding to XPC–HR23B (Batty et al. 2000). The gel mobility shift assay described here provided us with a much more sensitive method, which further confirmed the lack of specific binding affinity for the CPD. As suggested from the solution structure determined by nuclear magnetic resonance (Kim et al. 1995; McAteer et al. 1998), the DNA helix distortion induced by CPD may be too subtle to be recognized by XPC–HR23B. In fact, the introduction of one or two mismatched base(s) opposite the CPD, which one would expect to potentiate the helix distortion, significantly increases the affinity for XPC–HR23B. Because XPC–HR23B is necessary anyway for the excision of CPDs from the global genome in vivo as well as in the cell-free system, it has been proposed that other factors, such as the damaged DNA binding factor (DDB), must be required for the primary recognition of CPDs, and that they somehow target XPC–HR23B to the lesion (Hwang et al. 1999; Tang et al. 2000; Kusumoto et al. 2001). Intriguingly, the compound lesions involving CPD are excised much more efficiently in the cell-free extract than would be expected from their observed affinity for XPC–HR23B, indicating the involvement of other factors in the recognition of such compound lesions. It is possible that the same putative factors that may be required for normal CPD recognition are also responsible for the efficient recognition of the compound lesions. In addition, some mismatch repair factors might be involved in the recognition and repair of the compound lesions, although this has not yet been demonstrated (Moggs et al. 1997; Mu et al. 1997a). Although the GGR mechanism involving CPDs thus remains to be elucidated, our results strongly indicate that, unlike the case of 6-4PPs, an additional step is required for the recognition of CPDs, regardless of whether the CPD is paired with correct or mismatched bases. Furthermore, chromatin remodeling may have to precede the damage binding by XPC–HR23B and/or other factors, raising the possibility of the presence of even earlier steps in vivo. Because it must be tremendous work for cells to search throughout the huge genome for a small number of lesions, the multistep mechanism described here would be a crucial contribution to achieving the very high level of damage discrimination required in eukaryotic cells.
Materials and methods
Cell culture and extract preparation
A lymphoblastoid cell line (GM2248B) from the XP3BE (group C) patient was cultured in suspension with Dulbecco's modified Eagle medium (Nissui) supplemented with 15% fetal bovine serum. The whole cell extract for cell-free NER reactions was prepared according to the described method (Wobbe et al. 1985) with minor modifications. Briefly, cells were harvested by low-speed centrifugation and washed twice with ice-cold phosphate-buffered saline. The cell pellet was suspended in five times the volume of hypotonic buffer (20 mM Hepes-NaOH at pH 7.5, 5 mM KCl, 1.5 mM MgCl2, and 1 mM dithiothreitol) containing a mixture of protease inhibitors (50 μM EGTA, 0.25 mM PMSF, 0.2 μg/mL aprotinin, 0.2 μg/mL leupeptin, 0.1 μg/mL antipain, and 0.1 μg/mL pepstatin A). The cell suspension was kept on ice for 5 min and centrifuged for 10 min at 200_g_. The swollen cell pellet was homogenized by 20 strokes of an all-glass Dounce homogenizer (Kontes, pestle B), and NaCl was added to a final concentration of 0.2 M with gentle stirring. After 30 min of stirring on ice, the lysate was clarified by centrifugation (50,000_g_ for 30 min), and dialyzed against buffer A (20 mM Hepes-NaOH at pH 7.0, 50 mM NaCl, 0.1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, and 0.25 mM PMSF). Insoluble materials were removed by centrifugation (50,000_g_ for 30 min), and the resulting extract was aliquoted, frozen in liquid nitrogen, and stored at −80°C. The XP3BE cell extract obtained with this modified procedure was found to be very active in dual incision when supplemented with XPC–HR23B.
Protein preparation
Human XPC protein was expressed in the insect cell line Sf9 by infection with a recombinant baculovirus vVL.XPC, and purified as described previously (Sugasawa et al. 1996). Recombinant human HR23B protein, which was fused to the carboxy-terminal hexa-histidine tag (HR23BHis), was overexpressed in the Escherichia coli strain BL21(DE3) and purified as described (Masutani et al. 1997). To reconstitute the complex, 250 μg each of the recombinant XPC and HR23BHis proteins were mixed in buffer B (25 mM Tris-HCl at pH 7.5, 1 mM EDTA, 10% glycerol, 0.01% Nonidet P-40, 1 mM dithiothreitol, and 0.25 mM PMSF) containing 0.3 M NaCl and incubated on ice for 2 h. To remove unbound HR23BHis, the mixture was loaded onto a HiTrap-heparin column (Amersham Pharmacia Biotech; 1 mL) equilibrated with buffer B containing 0.3 M NaCl. After washing with the same buffer, bound proteins were eluted with buffer B containing 1 M NaCl. The eluate was dialyzed against buffer C (20 mM sodium phosphate at pH 7.8, 0.3 M NaCl, 10% glycerol, 0.01% Nonidet P-40, and 0.25 mM PMSF) and further loaded onto a HiTrap-chelating column (Amersham Pharmacia Biotech; 1 mL) that had been bound to nickel ions and then equilibrated with buffer C. The column was washed with the same buffer and eluted successively with buffer C containing 20, 100, and 250 mM imidazole-HCl (pH 7.8). The XPC–HR23BHis complex was recovered in the 100 mM imidazole fractions. The peak fractions were pooled, dialyzed against buffer D (25 mM Hepes-KOH at pH 7.9, 1 mM EDTA, 0.3 M KCl, 50% glycerol, 1 mM dithiothreitol, and 0.25 mM PMSF) and stored at −80°C. Protein concentration was determined according to the Bradford method by use of reagents from Bio-Rad and bovine serum albumin as a standard.
Preparation of defined DNA substrates containing a single lesion and/or a bubble
Phagemid DNAs harboring the bottom strand sequences shown in Figure 1 were generated by ligating the corresponding complementary synthetic oligonucleotides to pBluescript II KS(+) (Stratagene) digested by _Pst_I and _Hin_dIII (restriction enzymes used in this study were purchased from Takara Shuzo, unless indicated). An E. coli strain XL-1 Blue was transformed with the resulting phagemids, and single-stranded circular DNAs (corresponding to the bottom strands) were recovered by infecting the bacterial culture with VCSM13 helper phage (Stratagene).
The oligonucleotides containing a single, UV-induced 6-4PP or CPD were prepared as described previously (Murata et al. 1990; Iwai et al. 1996). To introduce an AAF adduct, 1.25 nmole of each oligonucleotide containing a single G residue (the top strands shown by uppercase letters in Fig. 1) was incubated at 37°C for 3 h in a 50-μL reaction including 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.1 mM _N_-acetoxy-2-acetylaminofluorene (Chemsyn). The AAF-modified oligonucleotides were separated from unmodified ones by a high-performance liquid chromatography system (Beckman, System Gold) by use of a C18-reversed phase column (Beckman Ultrasphere 5 μm; φ4.6 × 250 mm). The column was pre-equilibrated with a mixture of 0.1 M triethylamine acetate (pH 7.0) and acetonitrile (95:5), and the bound oligonucleotides were eluted by linearly increasing the acetonitrile concentration up to 20%. The top-strand oligonucleotides (60 pmole) with or without damage were phosphorylated with T4 polynucleotide kinase (Takara Shuzo) and annealed to 30 μg (∼30 pmole) of the corresponding single-stranded circular DNA. The partially double-stranded DNA was incubated at 25°C overnight in a 200-μL reaction containing 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 1 mM ATP, and 600 μM each of dNTPs, bovine serum albumin (20 μg), T4 DNA polymerase (40 units; New England Biolab) and T4 DNA ligase (36 Weiss units; New England Biolab). Covalently closed circular DNA was purified by CsCl-ethidium bromide density gradient centrifugation. Internally 32P-labeled DNA substrates were prepared in essentially the same manner, except that the phosphorylation of the oligonucleotides was carried out using [γ-32P]ATP, the annealing and DNA synthesis reactions were scaled down, and covalently closed circular DNA was purified by agarose gel electrophoresis in the presence of ethidium bromide.
Gel retardation assay
For the preparation of radioactive probes, the covalently closed circular DNA containing a single lesion and/or a bubble (2 μg) was digested with _Bss_HII. The digested DNA was then incubated at 20°C for 15 min in a 50-μL reaction containing 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, and 200 μM each of dATP, dGTP, and dTTP, 1.85 MBq of [α-32P]dCTP (110 TBq/mmole; Amersham Pharmacia Biotech), and T4 DNA polymerase (6 units). After the first incubation, cold dCTP was added to a final concentration of 200 μM and further incubated at 20°C for 15 min to make blunt-ended DNA fragments. The smaller DNA fragments (∼180 bp) were purified by nondenaturing PAGE. Non-radioactive competitor DNAs were prepared in basically the same manner, except that 200 μM cold dCTP was substituted for [α-32P]dCTP in the 3′-end-filling reactions.
Binding reactions (10 μL) were carried out at 30°C for 30 min in mixtures including 20 mM sodium phosphate (pH 7.4), 5 mM MgCl2, 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol, 5% glycerol, 0.01% Triton X-100, bovine serum albumin (1 μg), 32P-labeled probe DNA (3.5 fmole), covalently closed circular plasmid DNA (0.5 ng), and the indicated amount of the recombinant XPC–HR23BHis complex. Appropriate competitor DNAs were also included where indicated. The reactions were then chilled on ice and the resulting DNA–protein complexes were fixed by addition of 1 μL of 2.5% glutaraldehyde. The mixtures were directly loaded on 4% nondenaturing polyacrylamide gels (acrylamide:bis-acrylamide, 37.5:1) containing 2.5% glycerol and TGE buffer (25 mM Tris, 192 mM glycine, and 1 mM EDTA) and electrophoresed at 7 V/cm in a cold room. The gels were dried and exposed to X-ray film (Fujifilm, New RX) at −80°C with intensifying screens. The percentage of the probe bound to XPC–HR23BHis was quantified using the BAS2500 bioimaging analyzer (Fujifilm).
DNase I footprinting assay
For labeling the damaged strands (top strands shown in Fig. 1), the closed circular substrates were digested by _Not_I, dephosphorylated by bacterial alkaline phosphatase (Takara Shuzo), and incubated with T4 polynucleotide kinase and [γ-32P]ATP. The labeled DNAs were further digested by _Vsp_I and subsequently treated with T4 DNA polymerase in the presence of cold dNTPs to make blunt-ended fragments. The bottom strands were labeled in essentially the same manner, except that _Acc_65I (New England Biolab) and _Pvu_I were used for the first and second digestions, respectively. The 5′-end labeled, ∼270-bp DNA fragments containing the damaged site were purified by nondenaturing PAGE.
Binding reactions were carried out under the same conditions as used for the gel mobility shift assay, except that the reactions were scaled up to 25 μL in which 10 fmole of the end-labeled DNA fragments were included. After incubation at 30°C for 30 min, the reactions were diluted with 25 μL of a 5 mM CaCl2 solution and then digested at 30°C for 2 min with DNase I (0.004 unit; Takara Shuzo). The purified DNA samples were subjected to 10% denaturing PAGE followed by autoradiography.
Nucleotide excision repair assays
For detection of the dual incision products, the internally labeled, double-stranded circular DNA substrates (2 × 105 cpm, ∼25 ng) were incubated at 30°C for 1 h in 25-μL reactions containing 40 mM Hepes-KOH (pH 7.8), 5 mM MgCl2, 0.5 mM dithiothreitol, 70 mM NaCl, 6.6% glycerol, 0.5 mM EDTA, 2 mM ATP, and 20 μM each of dATP, dGTP, and TTP, 8 μM dCTP, 22.5 mM creatine phosphate (di-Tris), phosphocreatine kinase (1.25 μg), bovine serum albumin (9 μg), the XP3BE whole cell extract (100 μg protein), and the indicated amount of the XPC–HR23BHis complex. The reactions were stopped by addition of EDTA to a final concentration of 10 mM, and DNA was purified and subjected to 10% denaturing PAGE.
To quantify the repair gaps created in the template DNAs, 100 ng of the cold double-stranded circular DNA substrate was incubated under the same condition, except that 20 μg/mL aphidicolin was included to inhibit the gap-filling DNA repair synthesis. The purified DNA was treated with T4 DNA polymerase in the presence of [α-32P]dCTP as well as the other three cold dNTPs. The samples were subsequently digested with _Hae_III, and separated on 8% nondenaturing polyacrylamide gels.
Acknowledgments
We thank all members of the Cellular Physiology Laboratory at RIKEN for helpful discussions and encouragement. This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan, and from the Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Corporation. This work was also supported by a grant from the Bioarchitect Research Project of RIKEN.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sugasawa@postman.riken.go.jp; FAX 81-48-462-4673.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.866301.
References
- Asahina H, Kuraoka I, Shirakawa M, Morita EH, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat Res. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Bootsma D, Kraemer KH, Cleaver J, Hoeijmakers JHJ. Nucleotide excision repair syndromes: Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic basis of inherited disease. New York, NY: McGraw-Hill Book Co.; 1997. [Google Scholar]
- Burns J, Guzder S, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- Buschta-Hedayat N, Buterin T, Hess MT, Missura M, Naegeli H. Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA. Proc Natl Acad Sci. 1999;96:6090–6095. doi: 10.1073/pnas.96.11.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clungston C, McLaughlin K, Kenny M, Brown R. Binding of human single-stranded DNA binding protein to DNA damaged by the anticancer drug cis-diaminedichloroplatinum (II) Cancer Res. 1992;52:6375–6379. [PubMed] [Google Scholar]
- Cooper PK, Nouspikel T, Clarkson SG, Leadon SA. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- Coverley D, Kenny MK, Lane DP, Wood RD. A role for the human single-stranded DNA binding protein HSSB/RPA in an early stage of nucleotide excision repair. Nucleic Acids Res. 1992;20:3873–3880. doi: 10.1093/nar/20.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue BA, Yin S, Taylor J-S, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997a;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Moggs JG, Hwang JR, Egly J-M, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997b;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- Hanawalt P, Mellon I. Stranded in an active gene. Curr Biol. 1993;3:67–69. doi: 10.1016/0960-9822(93)90156-i. [DOI] [PubMed] [Google Scholar]
- He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision step of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- Hess MT, Naegeli H, Capobianco M. Stereoselectivity of human nucleotide excision repair promoted by defective hybridization. J Biol Chem. 1998;273:27867–27872. doi: 10.1074/jbc.273.43.27867. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53 dependent and is involved in global genome repair. Proc Natl Acad Sci. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S, Shimizu M, Kamiya H, Ohtsuka E. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6-4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J Am Chem Soc. 1996;118:7642–7643. [Google Scholar]
- Jones CJ, Wood RD. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- Kim J-K, Patel D, Choi B-S. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: Implication in mutagenesis and repair activity. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- Kusumoto, R., Masutani, C., Sugasawa, K., Iwai, S., Araki, M., Uchida, A., Mizukoshi, T., and Hanaoka, F. 2001. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat. Res. (in press). [DOI] [PubMed]
- Le Page F, Kwoh EE, Avrutskaya A, Gentil A, Leadon SA, Sarasin A, Cooper PK. Transcription-coupled repair of 8-oxoguanine: Requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- Li R-Y, Calsou P, Jones CJ, Salles B. Interactions of the transcription/DNA repair factor TFIIH and XP repair proteins with DNA lesions in a cell-free repair assay. J Mol Biol. 1998;281:211–218. doi: 10.1006/jmbi.1998.1949. [DOI] [PubMed] [Google Scholar]
- Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homolog of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Araki M, Sugasawa K, van der Spek PJ, Yamada A, Uchida A, Maekawa T, Bootsma D, Hoeijmakers JHJ, Hanaoka F. Identification and characterization of XPC-binding domain of hHR23B. Mol Cell Biol. 1997;17:6915–6923. doi: 10.1128/mcb.17.12.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Mu D, Park CH, Reardon JT, Sancar A. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J Biol Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- McAteer K, Jing Y, Kao J, Taylor J-S, Kennedy MA. Solution-state structure of a DNA dodecamer duplex containing a cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA. J Mol Biol. 1998;282:1013–1032. doi: 10.1006/jmbi.1998.2062. [DOI] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mitchell DL, Nairn RS. The biology of the (6-4) photoproduct. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Yarema KJ, Essigmann JM, Wood RD. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Szymkowski DE, Yamada M, Karran P, Wood RD. Differential human nucleotide excision repair of paired and mispaired cisplatin-DNA adducts. Nucleic Acids Res. 1997;25:480–490. doi: 10.1093/nar/25.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Sancar A. Model for XPC-independent transcription-coupled repair of pyrimidine dimers in humans. J Biol Chem. 1997;272:7570–7573. doi: 10.1074/jbc.272.12.7570. [DOI] [PubMed] [Google Scholar]
- Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997a;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Wakasugi M, Hsu DS, Sancar A. Characterization of reaction intermediates of human excision repair nuclease. J Biol Chem. 1997b;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- Murata T, Iwai S, Ohtsuka E. Synthesis and characterization of a substrate for T4 endonuclease V containing a phosphorodithioate linkage at the thymine dimer site. Nucleic Acids Res. 1990;18:7279–7286. doi: 10.1093/nar/18.24.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli H. Enzymology of human nucleotide excision repair. In: Smith P, Jones C, editors. DNA recombination and repair. Oxford, UK: Oxford University Press; 1999. pp. 99–137. [Google Scholar]
- Nocentini S, Coin F, Saijo M, Tanaka K, Egly J-M. DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor IIH. J Biol Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Mu D, Sancar A. Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JHJ, Chambon P, Egly J-M. DNA repair helicase: A component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers JHJ, Egly J-M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2393. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji MKK, Eker APM, Wood RD. DNA repair defect in xeroderma pigmentosum group C and complementing factor from HeLa cells. J Biol Chem. 1994;269:22749–22757. [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei Y-F, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Masutani C, Uchida A, Maekawa T, van der Spek PJ, Bootsma D, Hoeijmakers JHJ, Hanaoka F. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol Cell Biol. 1996;16:4852–4861. doi: 10.1128/mcb.16.9.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JMY, Masutani C, Maekawa T, Uchida A, van der Spek PJ, Eker APM, Rademakers S, Visser C, Aboussekhra A, et al. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol Cell Biol. 1997;17:6924–6931. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JMY, Masutani C, Iwai S, van der Spek PJ, Eker APM, Hanaoka F, Bootsma D, Hoeijmakers JHJ. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornaletti S, Reines D, Hanawalt PC. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J Biol Chem. 1999;274:24124–24130. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A, Venema J, Meschini R, van Zeeland AA, Mullenders LHF. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblast. EMBO J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, van Hoffen A, Natarajan AT, van Zeeland AA, Mullenders LHF. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Nucleic Acids Res. 1990;18:443–448. doi: 10.1093/nar/18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, van Hoffen A, Karcagi V, Natarajan AT, van Zeeland AA, Mullenders LHF. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci. 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Order of assembly of human DNA repair excision nuclease. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Reardon J, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272:16030–16034. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- Wobbe CR, Dean FB, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC–HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]