Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes (original) (raw)
Abstract
Imprinting of the maternally-expressed Igf2r gene is controlled by an intronic imprint control element (ICE) known as Region2 that contains the promoter of the noncoding Air RNA, whose transcript overlaps the silenced paternal Igf2r promoter in an antisense orientation. Two novel imprinted genes, Slc22a2 and Slc22a3 are described here that lie 110 and 155 kb 3′ to Igf2r and that are not overlapped by the Air transcript but are regulated by the _Igf2r_-ICE, as previously shown for Igf2r. These results identify a new cluster of imprinted genes whose repression by the bidirectional action of the Region2-ICE is independent of transcript overlap by the Air RNA.
Keywords: Slc22a3–Orct3–EMT, Slc22a2–Orct2, Igf2r, genomic imprinting
_Cis_-acting epigenetic mechanisms that regulate gene expression are of critical importance in understanding how gene regulation is controlled in development and disease, but are currently understood only at a basic level. Of the many mammalian systems currently used to investigate epigenetic mechanisms, parental-specific gene expression induced by genomic imprinting has the best potential to identify relevant DNA sequences and modifications, since both the active and silent parental alleles are retained in the same nuclear environment and can be directly compared (for reviews, see Reik and Walter 2001; Sleutels and Barlow 2001). At present, 50 imprinted genes have been identified with approximately equal numbers of maternally and paternally expressed examples (Beechey et al. 2000). This may represent a near complete collection, since the total number of imprinted genes, predicted to be in the range of 200, should now be reduced by 60%, following the reappraisal of the total number of genes in the mammalian genome (Claverie 2001).
A large body of evidence indicates that imprinted genes are regulated by _cis_-linked DNA imprint control elements (ICEs) that are epigenetically modified by an imprint in either the maternal germ line (for maternally imprinted genes) or paternal germ line (for paternally imprinted genes). Experimental data also indicate that DNA methylation is the imprint or at least is involved in maintaining the imprint for most but not all genes (Caspary et al. 1998; Jackson-Grusby et al. 2001). In contrast to early expectations, the data clearly show that imprints can act either as epigenetic activators to activate one allele of an imprinted gene, or as epigenetic repressors to silence one allele (Obata et al. 1998; Kato et al. 1999; Jackson-Grusby et al. 2001). The observation that imprinted genes are clustered within a chromosomal region has led to the suggestion that the ICE may also serve as an epigenetic domain regulator that controls the imprinted expression of more than one imprinted gene. To date, seven clusters have been identified in the mouse genome that are also conserved in the human genome (Beechey et al. 2000). The clusters show generally similar behavior whereby a majority of the clustered genes are expressed from one parental chromosome and only a minority from the reciprocal parental chromosome. Two well-known examples are the Prader-Willi syndrome (PWS) region (human 15q11–15 syntenic to mouse 7D) that contains 11 paternally expressed transcripts and one maternally expressed transcript, and the Beckwith-Weidemann syndrome (BWS) region (human 11p15 syntenic to mouse 7C) that contains 13 maternally expressed transcripts and four paternally expressed transcripts (Beechey et al. 2000).
Genomic deletions in mice and humans have confirmed that imprinting of more than one gene can be regulated by an ICE, but these data also show that not all genes present in one cluster are regulated by the same element. For example, in the mouse 7C cluster homologous to the human BWS region, deletion of the H19 ICE that lies close to the H19 promoter causes reexpression of the silent allele of the downstream H19 gene and two upstream genes, Igf2 and Ins2, but does not affect imprinted expression of the more distal upstream Mash2, Cdkn1, and Kcnq1 genes (Caspary et al. 1998; Thorvaldsen et al. 1998). Deletion of the mouse PWS ICE that maps to the Snrpn gene promoter shows a similar effect on multiple genes but, in contrast, leads to silencing of the active allele of flanking upstream and downstream imprinted genes (Yang et al. 1998). Similar regulation has been observed following spontaneous ICE deletions in the human PWS region that resulted in a complete loss of expression of at least five imprinted paternally expressed transcripts located across a 1.5 Mb domain (Ohta et al. 1999). In addition, an in vitro model system has shown that targeted deletion of the LIT1 (KCNQ1OT1) promoter from a paternal copy of human chromosome 11 and subsequent analysis in a hamster monochromosomal hybrid cell results in reexpression of the silent allele of the upstream CDKN1 and SMS4 genes and of the downstream KCNQ1 promoter, but did not affect the more distal IGF2/H19 genes (Horike et al. 2000).
In contrast to the finding of clusters of imprinted genes, the mouse Igf2r gene that encodes an intracellular transport receptor has been proposed to be a solo maternally expressed imprinted gene (Schweifer et al. 1997; Verhaagh et al. 1999). Paternal repression of the Igf2r promoter is regulated by a 3.7 kb ICE called Region2 (Wutz et al. 1997, 2001). Region2 is located within intron 2 of the Igf2r gene and contains a 1.5 kb maternally methylated CpG-island that is the promoter for an antisense RNA named Air that overlaps the Igf2r promoter. The Air RNA is specifically expressed from the paternal unmethylated ICE, but not from the maternal methylated ICE. The finding that the mature Air RNA is a 108 kb-long, nonspliced, repeat-rich, noncoding RNA has led to the proposal that this may not be a legitimate gene and instead may represent genomic RNA (Lyle et al. 2000). An additional transcript in this region corresponding to the Mas locus and previously described as imprinted has now been shown to be part of the Air transcript where it overlaps the 3′ Mas exons (Lyle et al. 2000). No other imprinted genes have been identified on mouse chromosome 17, and analysis of mice with maternal or paternal disomy for proximal parts of this chromosome does not predict their existence (Beechey et al. 2000).
We recently identified a new gene, Slc22a3 (also known as Orct3/EMT), whose promoter is located 155 kb from the 3′ end of the Igf2r gene, which lies in close association with two related genes, Slc22a2/Orct2 and Slc22a1/Orct1 (Fig. 1; Verhaagh et al. 1999). The Slc22a1, Slc22a2, and Slc22a3 genes all encode membrane-spanning transporter proteins that show a highly tissue-specific expression pattern in the embryo and adult, in contrast to the ubiquitous expression of the Igf2r gene (Verhaagh et al. 1999; Wutz et al. 2001). We show here by expression analysis that the genes Slc22a2 and Slc22a3 are imprinted and maternally expressed in the placenta. Imprinted expression of Slc22a2 and Slc22a3 shows tissue and temporal specificity, in contrast to the widespread imprinted expression of the Igf2r gene. Despite their contrasting gene expression and imprinted expression profiles, and despite the fact that they are not overlapped by the antisense Air transcript, targeted deletion of the Region2 ICE results in derepression of the paternal alleles of Slc22a2 and Slc22a3, as shown previously for the Igf2r gene (Wutz et al. 2001). These results identify the presence of a new cluster of imprinted genes spanning more than 400 kb on proximal mouse chromosome 17, the majority of which show maternal-specific expression. Furthermore, the data show that the _Igf2r_-ICE generates a long-range effect that acts in a bidirectional manner to repress upstream and downstream genes.
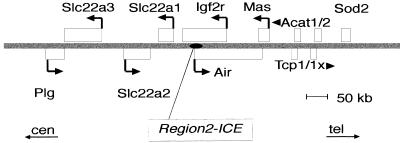
Figure 1.
Map of all known genes within 600 kb of the mouse Igf2r locus relative to centromere (cen) and telomere (tel). Region2 is shown as a black oval; genes are shown as white boxes which represent the locus size. Arrows and arrowheads indicate the direction of transcription when known.
Results and Discussion
The position of genes analyzed in this study is shown in Figure 1. The Igf2r, Air, Slc22a2, and Slc22a3 gene loci span ∼400 kb. The Igf2r promoter lies 30 kb downstream of Region2 and is overlapped by the Air RNA. The Slc22a2 and Slc22a3 gene promoters lie 170 kb and 210 kb upstream of Region2 and are not overlapped by Air. The Thp (hairpin) mouse mutant used for the analyses below carries a 3 cM deletion that includes the entire region shown in Figure 1. Paternal inheritance of the Thp allele (+/Thp) is viable, whereas maternal inheritance (the maternal allele is written on the left-hand side; Thp/+) is lethal during 16.5–18.5 days post-conception (dpc) due to the absence of Igf2r expression. Parental allele-specific expression can be analyzed before the lethal period (Wutz et al. 2001).
Slc22a3 is a new imprinted gene linked to the Igf2r locus
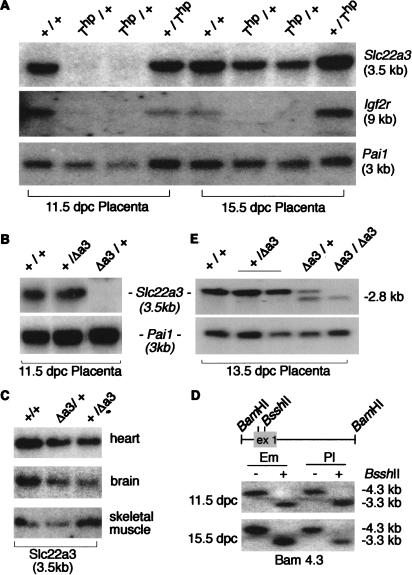
Imprinted expression of Slc22a3 was analyzed in placentas of embryos heterozygous for a paternally or maternally inherited Thp allele. Slc22a3 expression is restricted to placenta in development but shows widespread expression in adult organs (Verhaagh et al. 1999). Figure 2A shows that at 11.5 dpc, Slc22a3 was absent in Thp/+ placentas, whereas +/Thp placentas showed expression equal to wild-type placentas (+/+). Thus Slc22a3 shows maternal-specific expression at 11.5 dpc. In contrast, at 15.5 dpc, expression is detected in both paternal and maternal Thp heterozygotes, and thus Slc22a3 is expressed from both parental alleles at this stage, as reported previously (Verhaagh et al. 1999). These results show that Slc22a3 imprinted expression is restricted to an early time window in development, in contrast to the imprinted expression of Igf2r (Fig. 2A).
Figure 2.
The Slc22a3 gene is imprinted in placenta. (A) Blot of placental RNA from Thp/+, +/Thp, and +/+ offspring isolated at 11.5 and 15.5 dpc. The two (Thp/+) lanes are independent samples. The same blot was hybridized with Igf2r and Pai1. (B) RNA blot of 11.5-dpc placentas isolated from wild-type mice (+/+) and mice with a maternally (Δa3/+) or paternally (+/Δa3) inherited Slc22a3 targeted allele. (C) RNA blot of adult heart, skeletal muscle, and brain RNA isolated from wild-type mice (+/+) and mice with a maternally (Δa3/+) or paternally (+/Δa3) inherited Slc22a3 targeted allele. (D) Methylation analysis of the Slc22a3 CpG-island. A 4.3-kb genomic _Bam_HI fragment that contains Slc22a3 exon 1 (gray box) embedded in a CpG-island is shown (top) with the positions of two methyl-sensitive _Bss_HII restriction enzyme sites. The complete 4.3 kb _Bam_HI fragment was used as a probe on a DNA blot of genomic DNA of 11.5-dpc and 15.5-dpc wild-type embryos and placentas digested with _Bam_HI (−) or _Bam_HI and _Bss_HII (+). Only a 3.3-kb genomic fragment was detected upon digestion with _Bss_HII, showing that the Slc22a3 CpG-island is unmethylated. (E) RNA blot of 13.5-dpc placental RNAs isolated from wild-type mice (+/+) with a maternally (Δa3/+) or paternally (+/Δa3) inherited targeted allele, and homozygous targeted mice (Δa3/Δa3). The 2.8-kb Slc22a3 RNA is specific for the targeted allele.
Thp/+ placentas show increased growth from 13.5 dpc (Ludwig et al. 1996). To exclude the possibility that absence of Igf2r leads directly to loss of Slc22a3 expression, we also analyzed imprinted expression in placentas of embryos heterozygous for a targeted deletion of the Slc22a3 gene. Mice with a null mutation of Slc22a3 (Δa3) show impaired transport of the neurotoxin MPP+ in placenta and adult heart, but lack any obvious biological or cytological phenotype and are viable and fertile (Zwart et al. 2001). Figure 2B shows that Slc22a3 expression is imprinted at 11.5 dpc (cf. Δa3/+ with +/Δa3) in placentas with a wild-type Igf2r allele. The Δa3 mutant mice were further used to test imprinted expression in adult organs. In Figure 2C, adult brain, heart, and skeletal muscle from maternal (Δa3/+) and paternal (+/Δa3) heterozygous knockout mice show equal levels of expression, equivalent to 50% of wild-type expression (quantitated by PhosphorImager; data not shown), showing an absence of imprinting in adult tissues. These results show that Slc22a3 is imprinted in a tissue-specific fashion in placenta, and in a manner temporally restricted to early postimplantation stages. Several examples of placental- or extraembryonic-specific imprinted expression have been described: for example, the human WT1 gene (Jinno et al. 1994) and the mouse genes Ins2, Tssc4, Obph1, and Kcnq1 (Deltour et al. 1995; Caspary et al. 1998; Engemann et al. 2000; Paulsen et al. 2000). Temporal changes of imprinted expression in placenta have also been reported for Mash2 and Ins2 (Deltour et al. 1995; Caspary et al. 1998).
The Slc22a3 promoter is embedded in a CpG-island (Verhaagh et al. 1999; GenBank accession no. AF078748). To investigate whether the Slc22a3 CpG-island is methylated in a manner that correlates with imprinted expression in placenta, we performed a methylation-sensitive DNA blot analysis. Complete digestion of two _Bss_HII sites (Fig. 2D) and _Sac_II and _Hpa_II sites (present at several sites in the CpG-island promoter; data not shown) was observed in both 11.5 dpc and 15.5 dpc embryonic, placental, and adult tail genomic DNA. Thus, the Slc22a3 CpG-island is nonmethylated on both alleles in expressing and nonexpressing tissues, irrespective of the imprinted expression of Slc22a3 in early placenta.
Targeted inactivation of Slc22a3 removed a 4.3-kb fragment containing the complete CpG-island and exon 1. This deletion abolished Slc22a3 gene expression in Δa3/Δa3 adult tissues and in 11.5-dpc placentas (Fig. 2B; Zwart et al. 2001). However, in placentas from 12.5 dpc onward, a 2.8-kb aberrant, nontranslated Slc22a3 transcript that uses exons 3–11 was identified in mice carrying the targeted allele (Zwart et al. 2001). Because the original Slc22a3 gene promoter (P1) was deleted, an alternative promoter sequence (P2) must be employed to drive expression of the 2.8-kb transcript. Figure 2E shows that expression of the 2.8-kb transcript is specific to the targeted allele and is expressed only from the maternal allele in 13.5-dpc placenta (cf. lanes Δa3/+ and +/Δa3). Note that the Δa3/+ lane shows maternal expression of the 2.8-kb transcript and paternal expression of the 3.5-kb transcript, indicating the partial depression of the P1 promoter at this stage. The aberrant 2.8-kb transcript is expressed one day later than the 3.5-kb wild-type mRNA, and imprinted expression persists until 15.5 dpc in most but not all offspring, indicating a temporal difference between the P1 and P2 Slc22a3 promoters (Fig. 2; data not shown).
The flanking Slc22a2 gene is imprinted in placenta
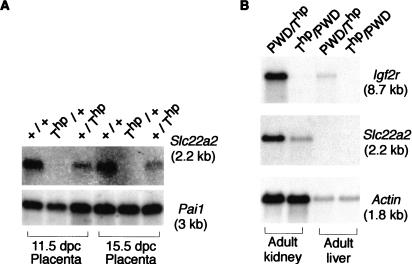
The finding that both the Slc22a3 wild-type P1 and the aberrant P2 promoter were imprinted in early placenta prompted us to reexamine imprinted expression of other genes flanking Igf2r. We had shown previously in 14.5-dpc embryos that the Plg, Tcp-1, and Sod2 genes (Fig. 1) were not imprinted; however, placental tissues were not tested (Barlow et al. 1991). We retested these genes and found no imprinted expression in 11.5-dpc embryos, nor in 11.5-dpc or 15.5-dpc placentas (data not shown). Other genes in this region include the Mas1 gene, initially described as imprinted in embryonic tissue, but later shown to be expressed solely in adult brain and testis and to lack any imprinted expression (Schweifer et al. 1997; Lyle et al. 2000), and the Slc22a1 and Slc22a2 genes that lie between Slc22a3 and Igf2r. Slc22a1 is not expressed during embryonic development, and expression in adults has been shown to be biallelic (Schweifer et al. 1997). Slc22a2 expression had been reported previously only in adult kidney and brain (Mooslehner and Allen 1999). However, hybridization of wild-type placental RNA with an Slc22a2 cDNA probe detects abundant expression (Fig. 3A). Analysis of embryos heterozygous for the Thp allele shows that in both 11.5-dpc and 15.5-dpc placentas, Slc22a2 was repressed on the paternal allele (Thp/+), but showed normal expression from the maternal allele (+/Thp). Imprinted maternal-specific Slc22a2 expression was also confirmed at these two developmental ages in placenta of mice carrying a targeted Slc22a2 null mutation that are wild-type for the Igf2r allele (H. Jonkers and A. Schinkel, unpubl.). Methylation analysis of the Slc22a2 promoter region in 13.5-dpc placental DNA from Thp/+ reciprocal crosses demonstrated an absence of parental-specific methylation at _Hpa_II sites (data not shown). Paternal expression of Slc22a2 in later placental stages could not be examined in Thp/+ offspring due to lethality at 16.5 dpc; however, adult mice can be obtained in crosses with the PWD Mus m. musculus subspecies (Forejt and Gregorova 1992). Figure 3B shows that while imprinting of Igf2r is maintained in adult kidney, the Slc22a2 gene is expressed from the paternal allele. However, comparison with Actin indicates that expression of the paternal Slc22a2 allele is reduced compared to the maternal allele.
Figure 3.
Slc22a2 is imprinted in placenta but not in adult tissue. (A) RNA blot of 11.5- and 15.5-dpc wild-type (+/+) and maternal T-hairpin (Thp/+) and paternal T-hairpin (+/Thp) placentas. The blot was hybridized successively with Slc22a2 and Pai1. (B) RNA blot of adult liver and kidney from maternal (Thp/PWD) and paternal (PWD/Thp) placentas. The blot was hybridized successively with Igf2r, Slc22a2, and Actin.
In development, imprinting of Slc22a2 and Slc22a3 is restricted to placenta. Neither gene is expressed in embryonic tissues assessed by nonamplification techniques, although low levels of Slc22a2 can be detected by RT–PCR in early embryos (R. Zwart, unpubl.; Mooslehner and Allen 1999; Verhaagh et al. 1999). Slc22a3 shows a loss of imprinted expression in late placenta, contrasting with Slc22a2, which maintains imprinted expression at this stage. The Igf2r gene differs from Slc22a2 and Slc22a3 in that it shows imprinted expression in embryos that is maintained in adult tissues (Fig. 3B; Forejt and Gregorova 1992). Adult brain has been reported as showing a loss of Igf2r imprinting (Hu et al. 1999). However, whereas the paternal Igf2r allele is reexpressed to some extent in brain, it remains repressed compared to the maternal allele when analyzed using nonamplification techniques (Schweifer et al. 1997). Therefore, these three genes show differences in both the regulation of their imprinted expression and in their general expression patterns. The biological significance of Slc22a2 and Slc22a3 imprinting is not clear at this time, as neither gene has an essential developmental function in embryos or adults (Zwart et al. 2001; H. Jonker and A. Schinkel, pers. comm.). Taken together, these results identify a new cluster of imprinted genes in the proximal part of mouse chromosome 17.
Control of imprinted expression by Region2
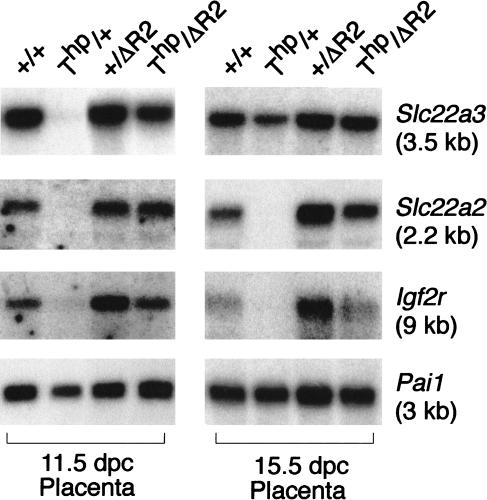
The imprinted expression of the two Slc22a3 P1 and P2 promoters and that of the Slc22a2 gene resemble that of the Igf2r gene in that all transcripts show maternal-specific expression. The imprinted expression of Igf2r has been shown, using targeted deletion in mice, to be controlled by the Region2 ICE that lies in intron 2 (Wutz et al. 2001). To investigate whether this ICE also regulates imprinted expression of the Slc22a3 and Slc22a2 genes, we performed gene expression analysis in placentas from double heterozygote crosses of T-hairpin (Thp) and Region2 deletion (ΔR2) mice (Fig. 4). Thp/+ placentas showed no expression of the Slc22a3, Slc22a2, or Igf2r genes at 11.5 dpc, confirming the absence of expression from the paternal copy of these genes at this timepoint in this genetic background. When a Region2 deletion was introduced on the paternal allele (Thp/ΔR2), paternal expression of the three genes Slc22a3 (P1 promoter), Slc22a2, and Igf2r was apparent to a level which approximates the maternal expression observed in the wild-type placentas (+/+). The Slc22a3 P2 promoter was not tested, because the tight linkage (210 kb) would have required an excessive breeding program to place the Slc22a3 P2 promoter in cis to the Region2 deletion. At 15.5 dpc, Slc22a3 is expressed from the paternal allele in (Thp/+) placenta, indicating that it has also lost its imprinted expression in placenta in this genetic background. Imprinted expression of the Slc22a2 and Igf2r genes is maintained at this later timepoint in this genetic background, and is still dependent on the presence of Region2 (15.5 dpc, Thp/ΔR2).
Figure 4.
The Region2_-_ICE regulates imprinting of Slc22a2 and Slc22a3. RNA blot of 11.5 and 15.5 dpc wild-type (+/+), maternal T-hairpin (Thp/+), paternal Region2 deletion (+/ΔR2), and maternal T-hairpin/paternal Region2 deletion double heterozygote (Thp/ΔR2) placentas. The blot was hybridized successively with Igf2r, Slc22a2, Slc22a3, and Pai1.
Region2 is a bidirectional silencer
The data presented here show that the Region2 ICE is a bidirectional silencer on the paternal allele, able to simultaneously repress a cluster of genes in a 400-kb domain. The silencing activity of Region2 on the paternal chromosome correlates with absence of methylation and the presence of Air RNA. In contrast, expression of the Igf2r/Slc22a2/Slc22a3 cluster on the maternal chromosome correlates with Region2 methylation and Air repression (Wutz et al. 1997). The presence of a methylation imprint on the active maternal allele indicates that interestingly, this allele is epigenetically activated by DNA methylation. This interpretation contrasts with results obtained in vitro based on demethylating agents (Hu et al. 1999), but is consistent with results obtained in vivo based on mouse mutants lacking the maintenance methyltransferase gene (Jackson-Grusby et al. 2001). A large number of imprinted genes have now been shown to be epigenetically activated by imprints (for review, see Sleutels and Barlow 2001).
The earlier demonstration that deletion of the Region2 ICE containing the Air promoter leads to derepression of the Igf2r promoter suggested several models to explain its action (Wutz et al. 1997). The expression–competition model proposed that competition for limiting transcription factors, or for the use of shared _cis_-acting elements, could prevent coexpression of the Igf2r and Air promoter in cis. However, the identification of four additional examples of imprinted protein-coding genes overlapped by antisense noncoding RNAs on their silent allele [_UBE3A-as_ overlapping _UBE3A_ (Rougeulle et al. 1998), KCNQ1OT1 overlapping KCNQ1 (Smilinich et al. 1999), Nesp-as overlapping Nesp (Wroe et al. 2000), and Cop2-as overlapping Cop2 (Lee et al. 2000)] indicated a function for sense/antisense transcription, for example, by promoter occlusion or by the formation of double-stranded interfering RNA on one chromosome. The demonstration here of the bidirectional action of Region2 is a surprise, and although it does not directly exclude a function for the Air RNA, it does indicate that neither the antisense orientation nor the physical overlapping of coding/noncoding transcripts is necessary for repression, at least of Slc22a2 and Slc22a3 (see Fig. 1). While all three genes are regulated by the Region2 ICE, it remains possible that a dual mechanism operates for transcriptional silencing, and a role for the Air RNA in silencing Igf2r is not yet excluded. In vitro deletion of the antisense KCNQ1OT1 promoter in a hamster/human hybrid cell line leads to derepression of upstream and downstream genes, indicating that it may also be a bidirectional silencer (Horike et al. 2000).
The recent demonstration that imprinting of the Igf2/H19 genes is caused by the insulator CTCF protein binding to the unmethylated maternal allele at the H19 ICE, thus preventing interaction between the Igf2 promoter and enhancer, showed that repressed genes lie on one side of the ICE and indicates that repression is unidirectional (for review, see Thorvaldsen and Bartolomei 2000). It has also recently been proposed that that ‘epigenetic regulation of access to shared enhancers’ may be a common feature of imprinting mechanisms (Bell et al. 2001). Although enhancers for none of the genes studied here have been identified, the location of the Slc22a2 and Slc22a3 genes on the opposite side of the Region2 ICE from the Igf2r promoter shows the bidirectional action of Region2 and argues against the generality of the shared enhancer model. The data presented here are compatible with a model that the Region2 ICE has a long-range effect on the paternal allele leading to silencing of upstream and downstream susceptible gene promoters. The details of the silencing action are not yet known but may involve chromatin changes such as indicated by the observation that a 1 Mb domain around the Igf2r locus shows a parental-specific replication asynchrony whereby the paternal allele replicates before the maternal (Simon et al. 1999). Since the early replicating allele is predicted to indicate a less compact chromatin structure, this pattern reflects the paternal-specific expression of Air and may provide a possible explanation as to why Region2 ICE fails to silence the paternal Air promoter.
The data presented here show the existence of a cluster of three imprinted and maternally expressed protein-coding genes on proximal mouse chromosome 17 that are silenced on the paternal chromosome by the unmethylated Region2 ICE. Although these results cannot directly exclude a role for Air expression, because the 3.7 kb deleted in the ΔR2 allele included the Air promoter, they do show clearly that silencing mechanisms in genomic imprinting can act bidirectionally on several genes in a subchromosomal domain.
Materials and methods
Mice
All mouse strains were maintained on an FVB/N background. Genotyping as described: Thp, ΔR2 (Wutz et al. 1997), Δa3 (Zwart et al. 2001). Timed matings (vaginal plug at 0.5 dpc) were used to collect samples at different developmental stages.
Expression analysis
RNA was isolated using LiCl/urea and analyzed using standard formaldehyde gels and hybridization techniques. Probes: Igf2r, exon 3 to exon 6 RT–PCR fragment; Slc22a2, 626 bp RT–PCR fragment (bp 989–1605; AJ006036); Slc22a3, 2.7 kb _Eco_RI cDNA fragment detects the wild-type and aberrant transcript (bp 1–2766; AF078750); Pai1, 500 bp cDNA fragment (Zwart et al. 2001); Plg, 526 bp RT–PCR fragment (bp 2129–2655; NM008877); Sod2, 1-kb cDNA fragment, and Tcp-1, 0.8-kb cDNA fragment (Barlow et al. 1991).
Acknowledgments
We thank N. Bosnie and members of the animal facility for mouse management, J. Forejt for Thp/PWD tissues, M. Buitelaar for technical assistance, E. Wagner and A. Berns for reading the manuscript, K. Mooslehner for providing the Slc22a2 promoter map and hybridization probes, and A. Berns for constant support and encouragement. This research was supported by the Dutch Cancer Society (KWF).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dbarlow@imb.oeaw.ac.at; FAX 43-662-639-6129.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.206201.
References
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Beechey, C.V., Cattanach, B.M., and Selley, R.L. 2000. MRC Mammalian Genetics Unit, Harwell, Oxfordshire. World Wide Web Site—Mouse Imprinting Data and References (URL: http://www.mgu.har.mrc.ac.uk/).
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM. Gene number. What if there are only 30,000 human genes? Science. 2001;291:1255–1257. doi: 10.1126/science.1058969. [DOI] [PubMed] [Google Scholar]
- Deltour L, Montagutelli X, Guenet JL, Jami J, Paldi A. Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev Biol. 1995;168:686–688. doi: 10.1006/dbio.1995.1114. [DOI] [PubMed] [Google Scholar]
- Engemann S, Strodicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J. Sequence and functional comparison in the Beckwith-Wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum Mol Genet. 2000;9:2691–2706. doi: 10.1093/hmg/9.18.2691. [DOI] [PubMed] [Google Scholar]
- Forejt J, Gregorova S. Genetic analysis of genomic imprinting: An imprintor-1 gene controls inactivation of the paternal copy of the mouse Tme locus. Cell. 1992;70:443–450. doi: 10.1016/0092-8674(92)90168-c. [DOI] [PubMed] [Google Scholar]
- Horike S, Mitsuya K, Meguro M, Kotobuki N, Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y, Oshimura M. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum Mol Genet. 2000;9:2075–2083. doi: 10.1093/hmg/9.14.2075. [DOI] [PubMed] [Google Scholar]
- Hu JF, Balaguru KA, Ivaturi RD, Oruganti H, Li T, Nguyen BT, Vu TH, Hoffman AR. Lack of reciprocal genomic imprinting of sense and antisense RNA of mouse insulin-like growth factor II receptor in the central nervous system. Biochem Biophys Res Commun. 1999;257:604–608. doi: 10.1006/bbrc.1999.0380. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jinno Y, Yun KK, Nishiwaki K, Kubota T, Ogawa O, Reeve AE, Niikawa N. Mosaic and polymorphic imprinting of the WT1 gene in humans. Nat Genet. 1994;6:305–309. doi: 10.1038/ng0394-305. [DOI] [PubMed] [Google Scholar]
- Kato Y, Rideout WM, 3rd, Hilton K, Barton SC, Tsunoda Y, Surani MA. Developmental potential of mouse primordial germ cells. Development. 1999;126:1823–1832. doi: 10.1242/dev.126.9.1823. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Park CW, Hahn Y, Park J, Lee J, Yun JH, Hyun B, Chung JH. Mit1/Lb9 and Copg2, new members of mouse imprinted genes closely linked to Peg1/Mest(1) FEBS Lett. 2000;472:230–234. doi: 10.1016/s0014-5793(00)01461-7. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Eggenschwiler J, Fisher P, D'Ercole AJ, Davenport ML, Efstratiadis A. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- Mooslehner KA, Allen ND. Cloning of the mouse organic cation transporter 2 gene, Slc22a2, from an enhancer-trap transgene integration locus. Mamm Genome. 1999;10:218–224. doi: 10.1007/s003359900976. [DOI] [PubMed] [Google Scholar]
- Obata Y, Kaneko-Ishino T, Koide T, Takai Y, Ueda T, Domeki I, Shiroishi T, Ishino F, Kono T. Disruption of primary imprinting during oocyte growth leads to the modified expression of imprinted genes during embryogenesis. Development. 1998;125:1553–1560. doi: 10.1242/dev.125.8.1553. [DOI] [PubMed] [Google Scholar]
- Ohta T, Gray TA, Rogan PK, Buiting K, Gabriel JM, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll DJ, et al. Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet. 1999;64:397–413. doi: 10.1086/302233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M, El-Maarri O, Engemann S, Strodicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet. 2000;9:1829–1841. doi: 10.1093/hmg/9.12.1829. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Cardoso C, Fontés M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- Schweifer N, Valk PJ, Delwel R, Cox R, Francis F, Meier-Ewert S, Lehrach H, Barlow DP. Characterization of the C3 YAC contig from proximal mouse chromosome 17 and analysis of allelic expression of genes flanking the imprinted Igf2r gene. Genomics. 1997;43:285–297. doi: 10.1006/geno.1997.4816. [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Barlow DP. The origins of genomic imprinting in mammals. In: Wu C-t, Dunlap JC., editors. Advances in genetics, Volume 46, Homology effects. San Diego, CA: Academic Press; 2001. (in press). [DOI] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Bartolomei MS. Molecular biology. Mothers setting boundaries. Science. 2000;288:2145–2146. doi: 10.1126/science.288.5474.2145. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes & Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaagh S, Schweifer N, Barlow DP, Zwart R. Cloning of the mouse and human solute carrier 22a3 (Slc22a3/SLC22A3) identifies a conserved cluster of three organic cation transporters on mouse chromosome 17 and human 6q26-q27. Genomics. 1999;55:209–218. doi: 10.1006/geno.1998.5639. [DOI] [PubMed] [Google Scholar]
- Wroe SF, Kelsey G, Skinner JA, Bodle D, Ball ST, Beechey CV, Peters J, Williamson CM. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc Natl Acad Sci. 2000;97:3342–3346. doi: 10.1073/pnas.050015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- Wutz A, Theussl HC, Dausman J, Jaenisch R, Barlow DP, Wagner EF. Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development. 2001;128:1881–1887. doi: 10.1242/dev.128.10.1881. [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- Zwart R, Verhaagh S, Buitelaar M, Popp-Snijders C, Barlow DP. Impaired activity of the extraneuronal monoamine transporter system known as Uptake-2 in Orct3/Slc22a3-deficient mice. Mol Cell Biol. 2001;21:4188–4196. doi: 10.1128/MCB.21.13.4188-4196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]